Allosteric activation of the co-receptor BAK1 by the EFR receptor kinase initiates immune signaling
Figures
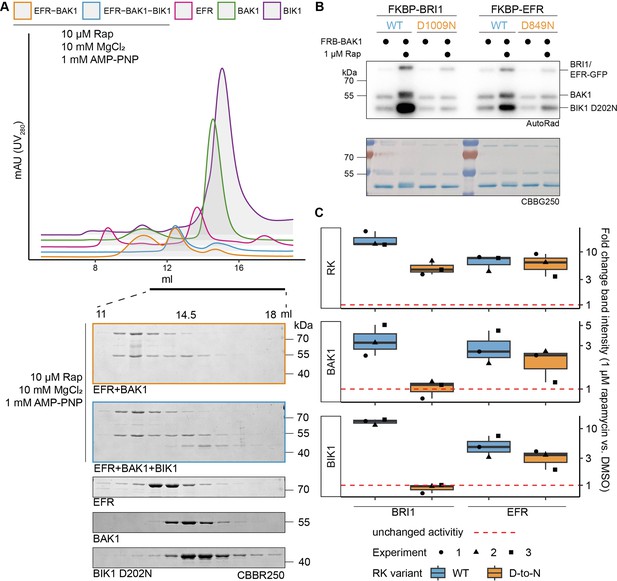
EFR facilitates BIK1 trans-phosphorylation by BAK1 non-catalytically.
The kinase domains of coRK BAK1 and the RKs BRI1 and EFR were tagged with RiD domains and purified from E. coli λPP cells. (A) Recombinantly expressed RiD-tagged kinase domains were mixed together at equimolar ratios (2 µM), with or without addition of 10 µM Rap as well as 10 mM MgCl2 and 1 mM AMP-PNP. (B) RK and coRK were mixed at an equimolar ratio at 50 nM, kinase-dead BIK1D202N substrate was added at 500 nM. Reactions were carried out at RT for 10 min with 0.5 μCi [γ-³²P]ATP, 100 μM ATP and 2.5 mM each of MgCl2 and MnCl2. Addition of 1 μM Rap enhanced transphosphorylation of BIK1 by EFR and BRI1. Kinase-dead BRI1 failed to enhance BIK1 transphosphorylation, but kinase-dead EFR retained some ability to do so. A similar trend was observed for (auto)phosphorylation of BAK1 itself. (C) Quantification of band intensities over three independent experiments of which a representative is shown in B.
-
Figure 1—source data 1
Raw data for gel images in Figure 1A.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig1-data1-v1.zip
-
Figure 1—source data 2
Raw data for autoradiography in Figure 1B.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig1-data2-v1.zip
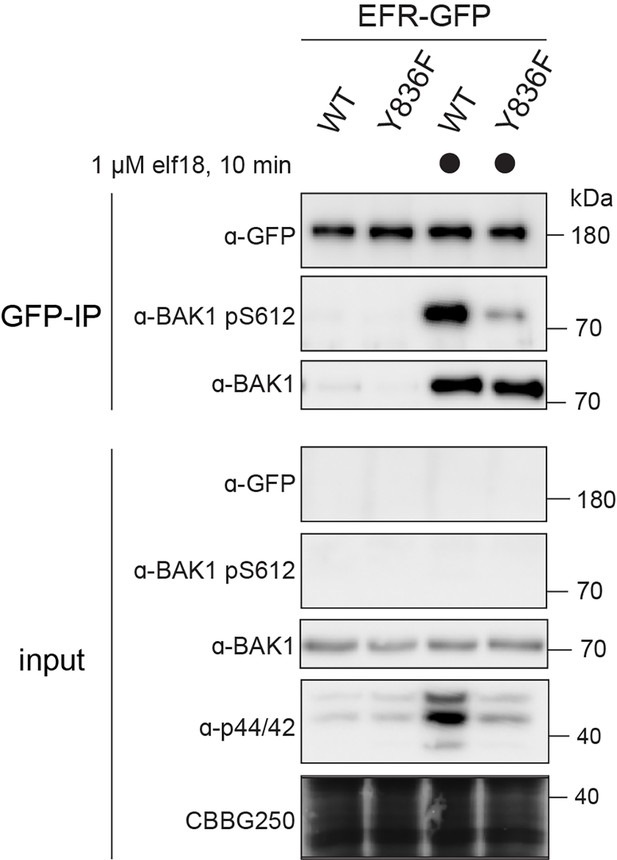
EFRY836F compromises ligand-induced receptor complex activation.
Two-week old seedlings were mock treated or treated with 1 µM elf18 for 10 min. Immunoprecipitation was then performed with anti-GFP, and the resulting immunoprecipitates probed for BAK1 S612 phosphorylation. Phosphorylated BAK1 was found to co-immunoprecipitate with WT EFR-GFP but hardly with EFRY836F-GFP, indicating ligand-induced receptor complex activation.
-
Figure 1—figure supplement 1—source data 1
Raw data for immunoblots in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig1-figsupp1-data1-v1.zip
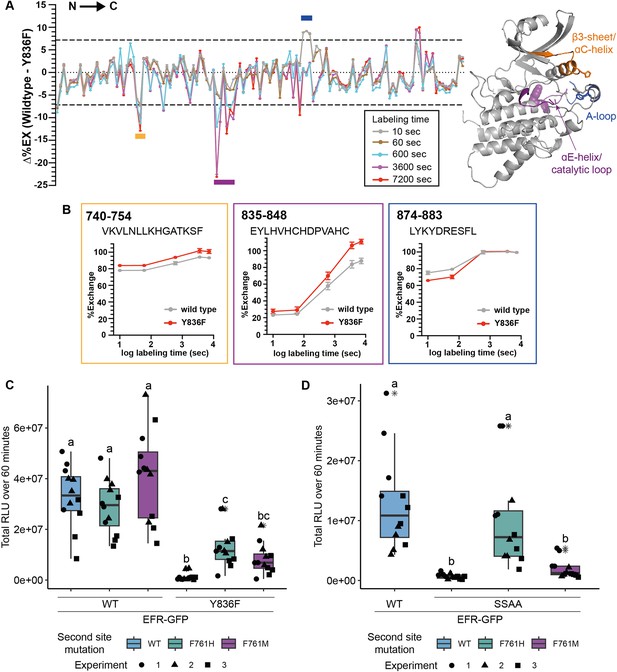
EFRY836F and EFRSSAA impair the active kinase conformation, which is required for signaling function.
(A) (left) HDX-MS results for unphosphorylated EFR and EFRY836F protein. The difference in percent H/D exchange in wild type EFR and EFRY836F is expressed as the Δ%EX (wild type EFR – EFRY836F), with the positive and negative Δ%EX indicating more stabilized and destabilized regions in EFRY836F, respectively, compared to wild-type EFR. The Δ%EX values at different labeling time points are shown as colored lines, as indicated in the figure. The horizontal dotted black lines indicate the 98% confidence interval for the Δ%EX data (±7.18%, corresponding to ±0.4 Da difference between wild type and Y836F percent exchange) calculated as described previously (Houde et al., 2011). Regions with Δ%EX values that exceed this confidence limit are indicated as colored bars in the figure, including the β3-αC loop (orange), the catalytic loop plus part of αE (purple), and the A-loop (blue). These regions are colored in the AlphaFold2-derived model of the EFR kinase domain shown at right, in which Y836 is shown as a purple sphere. All data are the average of three independent biological repeats (n=3) with three technical repeat experiments each. A summary of the HDX-MS analysis is presented in Table 3. (B) HDX-MS analysis of representative peptides from regions with significantly different HD exchange. Frames are color-coded according to regions in A. Amino acid range of the peptides in full length EFR are indicated in the top left corner and the sequence below. (C, D) Secondary site mutation EFR F761[H/M] partially restores function of EFRY836F (C) and EFRSSAA (D). Full length EFR and its variants were expressed transiently in N. benthamiana and their function was tested in an oxidative burst assay. EFR F761H partially restored oxidative bursts of EFRY836F and EFRSSAA. Outliers are in indicated by asterisk in addition to the outlier itself and are included in statistical analysis; Statistical test: Kruskal-Wallis test (p<2.2*10–16 in C, p=1.163*10–7 in D), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like lowercase letter designations are not statistically different.
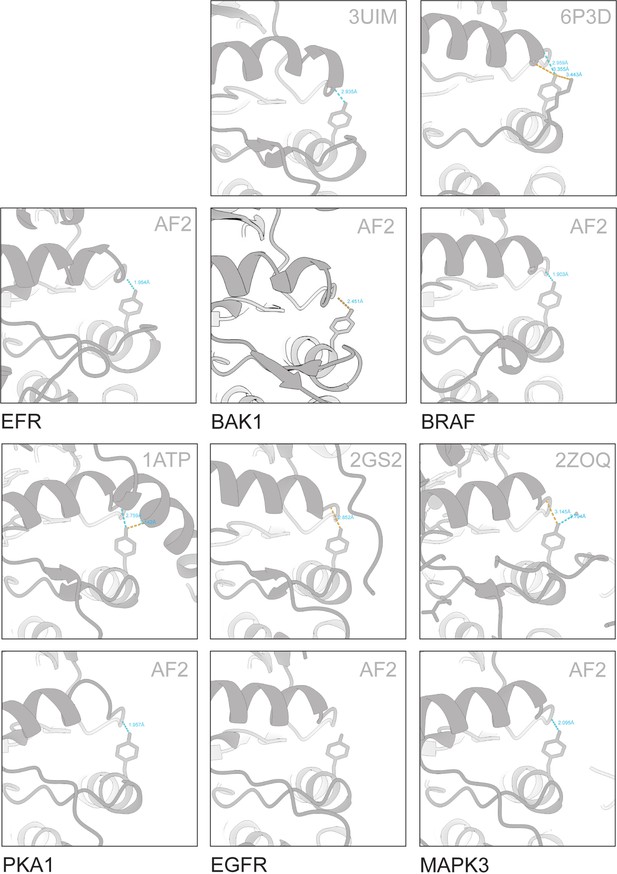
VIa-Tyr forms H-bonds with the αC-β4 loop in various predicted and solved structures.
Solved structures were retrieved from PDB. AlphaFold2 models for kinases in their active conformation were retrieved from Faezov and Dunbrack, 2023. BAK1 and EFR models were predicted by AlphaFold2, using the complete intracellular domain. H-bonds were predicted in ChimeraX and distances are indicated.
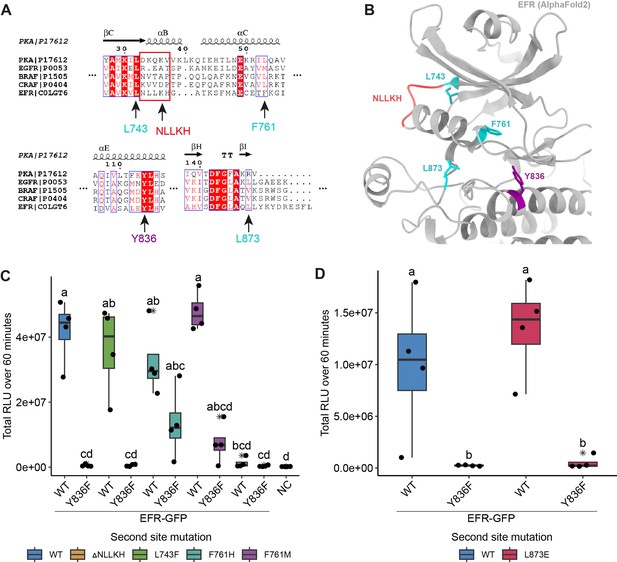
Rational design of activating mutations in EFR and screen for functional recovery of EFRY836F.
(A) Alignments of EFR with human kinases containing oncogenic, kinase activating mutations that stabilize the αC-helix-in active-like conformation (described in Foster et al., 2016; Hu et al., 2015). Homologous sites in EFR are indicated by arrows with the residue number. (B) Structural model of the EFR kinase domain from AlphaFold2 with homologous sites identified in the sequence alignment from A highlighted in teal (missense mutation) or red (deletion). EFR Y836 at the C-terminal end of the αE-helix is colored purple. (C) Screening of the homology-based putatively activating EFR mutations for restoration of EFRY836F function in N. benthamiana. All putative activating mutations were functional at WT-like level except EFRΔNLLKH. Only EFRF761[H/M] could functionally recover EFRY836F as the oxidative burst was partially restored. (D) EFRL873E showed a WT-like oxidative burst but EFRL873E/Y836F did not restore the oxidative burst. Outliers are in indicated by asterisk in addition to the outlier itself and are included in statistical analysis; Statistical test: Kruskal-Wallis test (p=9.319*10–6 in C, p=0.01242 in D), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like lowercase letter designations are not statistically different.
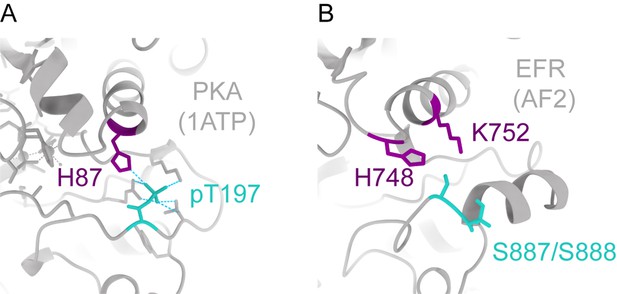
EFR A-loop phosphorylation sites may coordinate with basic residues from the β3-αC loop and αC-helix.
(A) In PKA (1ATP), the A-loop phosphorylation on T197 coordinates with H87 from the αC-helix. (B) In EFR (AlphaFold2 (AF2) model), there are two basic residues extending downwards from β3-αC loop (H748) and αC-helix (K752) that may coordinate with A-loop phosphorylation on S887 or S888.
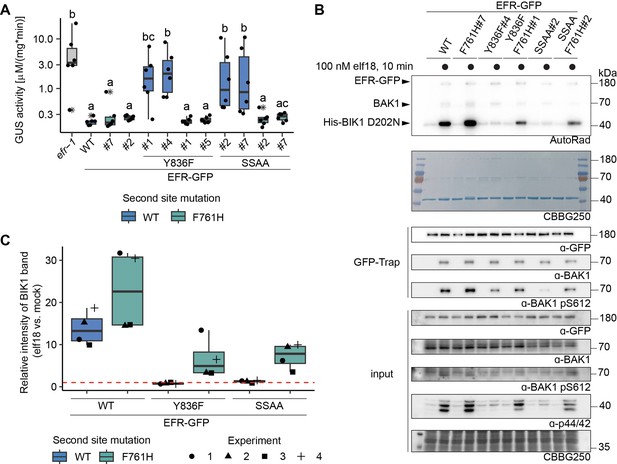
EFRF761H/Y836F and EFRF761H/SSAA recover receptor complex activation.
(A) In infection assays, GUS activity was high in the positive control efr-1 line. GUS activity level was reduced in the EFRWT and EFRF761H complementation lines, but much less so in the EFRY836F and EFRSSAA complementation lines. By contrast, EFRF761H/Y836F and EFRF761H/SSAA complementation lines displayed substantially repressed GUS activity. Each experiment was repeated three times with similar results. Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (P=5.704*10–7), Dunn’s post-hoc test with Benjamin-Hochberg correction (P ≤ 0.05) Groups with like lowercase letter designations are not statistically different. (B) In IP kinase assays, ligand-induced interaction of EFRWT and EFRF761H with BAK1 increased transphosphorylation of BIK1D202N, but this was abolished for EFRY836F and EFRSSAA. Both EFRF761H/Y836F and EFRF761H/SSAA showed partially restored BIK1D202N trans-phosphorylation as well as BAK1 S612 phosphorylation (across four replicates for EFRF761H/SSAA and in two out of four replicates for EFRF761H/Y836F). Samples were also probed for MAPK phosphorylation for effective ligand treatment. Treatment: 100 nM elf18 for 10 min. (C) Quantification of BIK1D202N band intensity observed in autoradiographs from the four independent replicates performed. Dotted red line indicates unchanged band intensity in mock vs. elf18 treatment.
-
Figure 3—source data 1
Raw data for autoradiography and immunoblotting in Figure 3B.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig3-data1-v1.zip
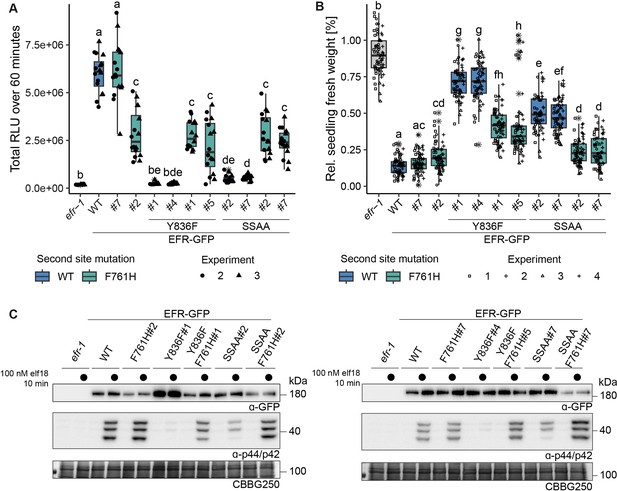
Multiple immune signaling branches are partially restored in EFRF761H/Y836F and EFRF761H/SSAA.
Stable transgenic complementation lines in the Arabidopsis efr-1 background were generated and physiological experiments conducted in the T3 generation (except for EFRF761H /Y836F#5, which is a double insertion line in T2 generation). (A) In the oxidative burst assay, EFR F761H restored oxidative burst in EFRF761H/Y836F and EFRF761H/SSAA complementation lines. Two independent experiments were merged into one graph as WT controls showed comparable total oxidative burst. A third independent experiment was performed with similar results. Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (p<2.2*10–16), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like letter designations are not statistically different. Like oxidative burst assays, EFR F761H restored SGI (B) and MAPK activation (C) in EFRF761H/Y836F and EFRF761H/SSAA complementation lines. For SGI assays, four independent experiments wtih 5 nM elf18 treatment are shown. Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (p<2.2 *10–16), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like letter designations are not statistically different. For MAPK activation assays, a representative experiment is shown. Similar results were obtained in three more experiments.
-
Figure 3—figure supplement 1—source data 1
Raw data for immunoblots shown in Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig3-figsupp1-data1-v1.zip
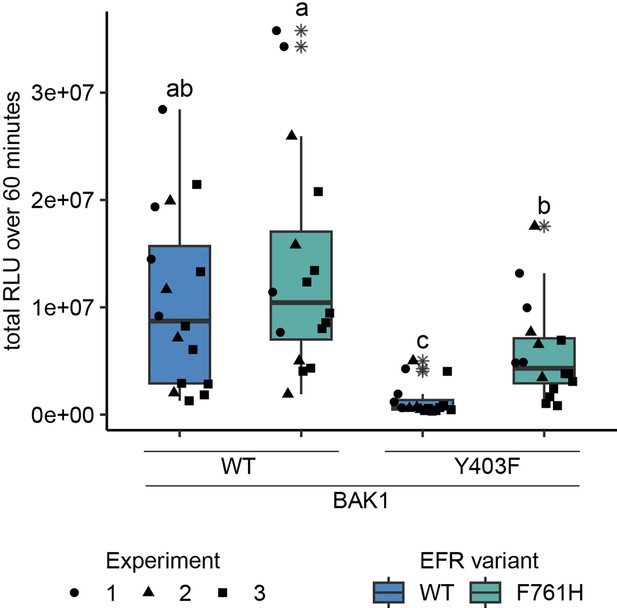
EFRF761H recovers BAK1Y403F function.
The cytoplasmic domains of BAK1 and EFR variants with fused RiD-tags were transiently expressed in N. benthamiana and leaf discs were treated with Rap to induce dimerization. EFR and EFRF761H induced a similar total oxidative burst when BAK1 was co-expressed. The co-expression of BAK1Y403F and EFR diminished the oxidative burst, which was restored partially when EFRF761H was co-expressed. Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (p<8.516 *10–7), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like letter designations are not statistically different.
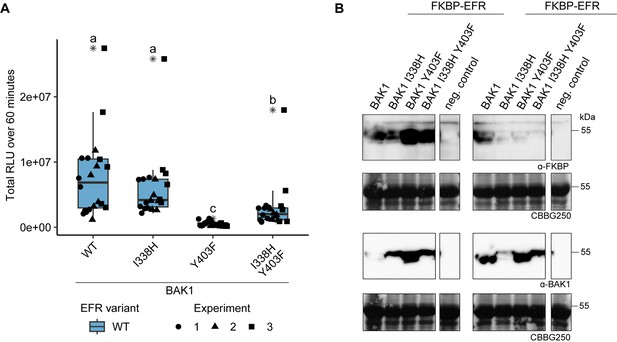
Function of BAK1 Y403F is partially recovered by the secondary mutation I338H.
(A) The RiD system was utilized to test the recovery of BAK1 Y403F by transient expression in N. benthamiana. The Y403F mutation in BAK1 diminished the oxidative burst, whereas BAK1 I338H displayed near WT-like responses. Combining the I338H and Y403F mutations, however, led to a partial recovery of oxidative burst. Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (p<2.247 *10–11), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like letter designations are not statistically different. (B) SDS-PAGE analysis of protein levels in experiments 2 and 3 of A is shown. Protein accumulation data for experiment 1 was not collected.
-
Figure 4—figure supplement 1—source data 1
Raw data for immunoblots shown in Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig4-figsupp1-data1-v1.zip
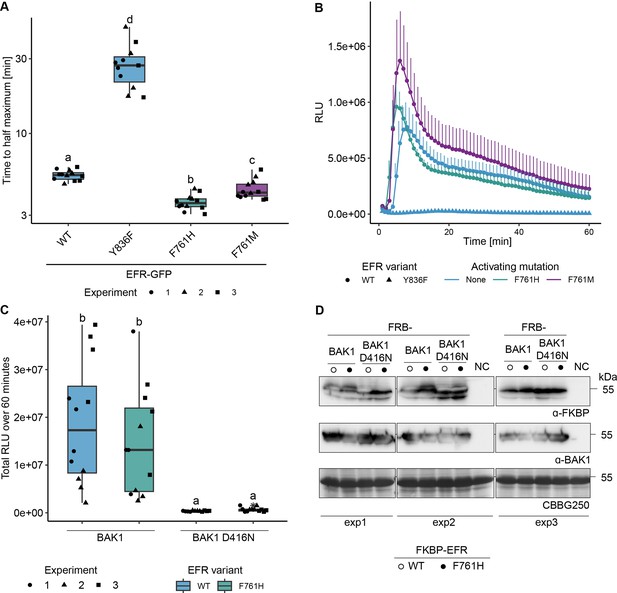
EFR F761H accelerates the onset of the oxidative burst but requires the catalytic activity of BAK1.
(A) Quantification of the time until the oxidative burst reaches its half maximum from experiments presented in Figure 2B. Both putative activating mutations, F761H and F761M accelerate the onset of the oxidative burst. (B) Time resolved oxidative burst assay. Presented curves are from replicate number three as a representative example. Graphs in A and B are based on data presented in Figure 2B. Error bars represent standard error of the mean (n=4). (C) EFR F761H requires the catalytic activity of BAK1 to induce the oxidative burst. Data from three independent experiments is merged in one graph. (D) Protein accumulation of the RiD-tagged protein related to panel C. Statistical analysis in A and C: Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (p=1.686*10–8 in A, p=5.89910–8 in C), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like letter designations are not statistically different.
-
Figure 4—figure supplement 2—source data 1
Raw data for immunoblots shown in Figure 4—figure supplement 2D.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig4-figsupp2-data1-v1.zip
Protein accumulation for the oxidative burst assay in Figure 4.
Leaf discs were collected after the oxidative burst assay and protein was extracted by boiling in SDS-loading buffer followed by immunoblotting. Non-infiltrated leaf discs served as negative control.
-
Figure 4—figure supplement 3—source data 1
Raw data for immunoblots shown in Figure 4—figure supplement 3.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig4-figsupp3-data1-v1.zip
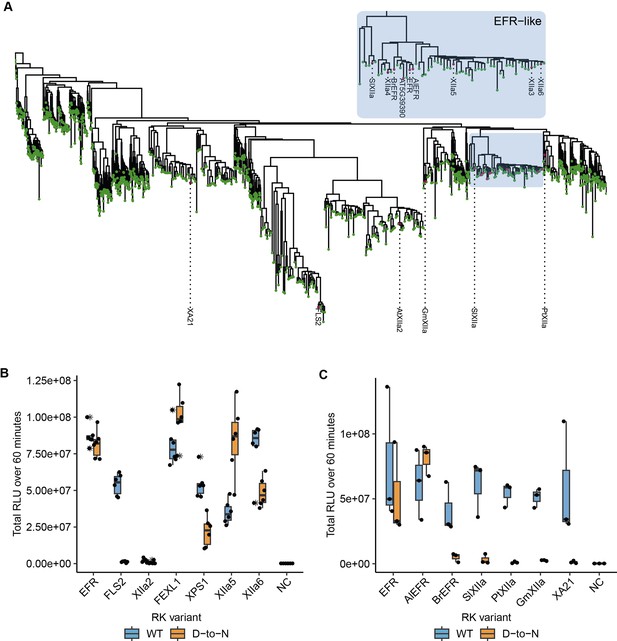
Related EFR kinases from LRR-RK XIIa in the Arabidopsis genus can function independent of their calatytic activity.
(A) Phylogenetic analysis of LRR-RK subfamily XIIa. Selected LRR-RK XIIa kinase domains are labeled and highlighted with purple points. The EFR-like clade contains all Arabidopsis XIIa kinases except FLS2 and XIIa2 and also selected XIIa kinases from Arabidopsis lyrata and Brassica rapa. (B, C) The ectodomain of EFR was fused to the transmembrane and intracellular domain of selected LRR-RK XIIa members to create elf18-responsive chimeras for testing the immune signaling function and catalytic dependency of the related kinase domains. The chimeras were transiently expressed in N. benthamiana and tested in oxidative burst assays. All Arabidopsis LRR-RK XIIa members induced an oxidative burst except XIIa2, the closest FLS2 related kinase in the subfamily. Catalytic dependency of the kinase domains appears to vary from kinase to kinase, with catalytically dead versions of EFR, FEXL1 and XIIa5 inducing a WT-like oxidative burst and XPS1 and XIIa6 displaying a reduced oxidative burst. FLS2 kinase dead exhibited a diminished oxidative burst. Experiments were repeated three times with similar results.
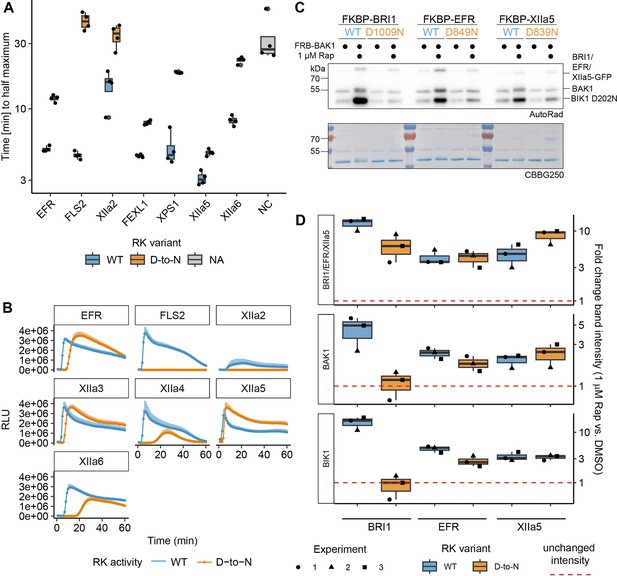
XIIa5D839N exhibits largely XIIa5WT-like characteristics.
(A, B) Catalytic site mutation of XIIa5 exhibited the least delayed onset of oxidative burst. In A, quantification of the time to reach the half maximum of the oxidative burst is shown. The underlying data are the same as used for total oxidative burst in the main figure. In B, the actual oxidative burst curves are presented as average of six individual plants transiently expressing the indicated chimeric protein. Error bars represent standard error of the mean (n=6). (C) A catalytic site mutation of XIIa5 did not negatively affect BIK1 trans-phosphorylation or BAK1 autophosphorylation. Experiments were performed as described in Figure 1. (D) Quantification of band intensities on autoradiographs from three independent experiments are shown. BRI1D1009N and EFRD849N displayed results similar to Figure 1B and C. In contrast to EFRD849N, for which BIK1 and BAK1 relative band intensities slightly decreased compared to wild type EFR, BIK1D202N and BAK1 relative band intensities were wild-type-like for XIIa5D839N.
-
Figure 5—figure supplement 1—source data 1
Raw data for autoradiography shown in Figure 5—figure supplement 1C.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig5-figsupp1-data1-v1.zip
Protein accumulation of EFR-XIIa chimeras in N. benthamiana.
All constructs exhibited detectable protein accumulation in transiently transformed N. benthamiana leaves. Similar protein accumulation was observed in 3 replicates for (A). Protein accumulation for constructs in (B) was tested once.
-
Figure 5—figure supplement 2—source data 1
Raw data for autoradiography shown in Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/92110/elife-92110-fig5-figsupp2-data1-v1.zip
Tables
Homology-based design of putative intragenic suppressor mutations for EFR.
The list contains the residue number of EFR and the analogous oncogenic mutation in BRAF, as well as a short description of the mode of action of the oncogenic mutation. See Figure 2—figure supplement 1B for structural locations.
| EFR mutation | Analogous oncogenic mutation | Mode of action | Source |
|---|---|---|---|
| L743F | BRAF L485F | Extended hydrophobic interaction network along the αC-helix | Hu et al., 2015 |
| F761[H/M] | BRAF L505[F/H/M] | Enforcement of hydrophobic interactions in the regulatory spine | Hu et al., 2015 |
| ΔNLLKH | BRAF ΔNVTAP | Shortening of β3-αC-helix loop, pulling ‘in’ the αC-helix | Foster et al., 2016 |
| L873E | BRAF V600E | Upward bending of C-terminal αC-helix end | Hu et al., 2015 |
Protein expression conditions.
| Protein | T after induction [°C] | t of expression [h] | pH of extraction buffer |
|---|---|---|---|
| For in vitro kinase assay | |||
| EFR/EFR D849N | 30 | 4 | 8.0 |
| BAK1 | 30 | 4 | 8.0 |
| BRI1/BRI1 D1009N | 18 | Over night | 8.0 |
| FLS2/FLS2 D997N | 18 | Over night | 8.0 |
| BIK1 D202N | 18 | Over night | 7.5 |
| For HDX-MS | |||
| EFR/EFR Y836F | 30 | 3 | 8.0 |
Summary table of HDX-MS analysis.
| Data Set | Wild-type EFR | EFRY836F |
|---|---|---|
| HDX reaction details | 20 mM HEPES, 100 mM NaCl, pD 7.4 | 20 mM HEPES, 100 mM NaCl, pD 7.4 |
| HDX time course | 0, 10, 60, 600, 3600, and 7200 s | 0, 10, 60, 600, 3600, and 7200 s |
| HDX control samples | 8 M urea-D4 for fully deuterated standard | 8 M urea-D4 for fully deuterated standard |
| Back-exchange | 46% | 49% |
| # of Peptides | 143 | 153 |
| Sequence coverage | 100% | 100% |
| Ave peptide length / Redundancy | 12.7/5.44 | 13.3/6.06 |
| Replicates | n=3 biological repeats (3 technical/n) | n=3 biological repeats (3 technical/n) |
| Repeatability (Ave SD) | 0.129 Da (2.28 %) | 0.111 Da (2.09 %) |
| Significant differences in HDX (98% CL) | 0.395 Da (7.18 %) | |
Extinction coefficients and molecular weights retrieved from ProtParam and used for determination of protein concentration.
| Protein | Extinction coefficient (reduced state)[Abs 0.1% (=1 g/l)] | Molecular weight[kDa] |
|---|---|---|
| FKBP-EFR-mEGFP | 7.40 | 80 |
| FKBP-BRI1-mEGFP | 8.26 | 83 |
| FKBP-FLS2-mEGFP | 6.56 | 79 |
| FKBP-XIIa5-mEGFP | 7.59 | 79.5 |
| FRB-BAK1 | 15.07 | 55 |
| 6xHis-TEV-BIK1D202N | 9.45 | 47 |
| 6xHis-TEV-EFR(684–1031) | 6.97 | 41.5 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Arabidopsis thaliana) | Col-0 | ther | wild-type ecotype | |
| Strain, strain background (Nicotiana benthamiana) | WT | Lab stock | Wild-type strain | |
| strain, strain background (Escherichia coli) | BL21(DE3) pLPP | Amid Biosciences | Cat#: BLPP-201 | Co-expression of lambda phosphatase |
| Strain, strain background (Escherichia coli) | BL21(DE3)-V2R-pACYC LamP | PMID: 20436473 | Co-expression of lambda phosphatase | |
| Strain, strain background (Agrobacterium tumefaciens) | GV3101 | Lab stock | ||
| Strain, strain background (Arabidopsis thaliana) | efr-1 | PMID: 16713565 | ||
| Genetic reagent (Arabidopsis thaliana) | EFR-GFP | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | F761H#2 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | F761H#7 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | Y836F#1 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | Y836F#4 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | F761H/Y836F#1 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | F761H/Y836F#5 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | SSAA#2 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | SSAA#7 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | F761H/SSAA#2 | This paper | See Materials and methods | |
| Genetic reagent (Arabidopsis thaliana) | F761H/SSAA#7 | This paper | See Materials and methods | |
| Sequence-based reagent | EFR-f | This paper | PCR primer | atcgGAAGACaaAATGA AGCTGTCCTTTTCACTTG |
| Sequence-based reagent | EFR Y836F-f | This paper | PCR primer | ggagtTtctgcacgtt cattgtcatgaccctgta |
| Sequence-based reagent | EFR SSAA-f | This paper | PCR primer | Gtttgctgctgctggt gtcagaggcaccat |
| Sequence-based reagent | EFR F761H-f | This paper | PCR primer | acgatgtcgtatacccttgt gggtttcacattccgccata |
| Sequence-based reagent | EFR F761M-f | This paper | PCR primer | tacgatgtcgtatacccttc atggtttcacattccgccata |
| Sequence-based reagent | EFR V845F-f | This paper | PCR primer | gcttaatatcacagtgagc gaaagggtcatgacaatgaacgtg |
| Sequence-based reagent | EFR L743F-f | This paper | PCR primer | gctttaggaggttgaaaac tttaaccgcgacgagttta |
| Sequence-based reagent | EFR L873E-f | This paper | PCR primer | ctcagGAGctctataaata cgatcgagaatcctttcta |
| Sequence-based reagent | EFR dNLLKH-f | This paper | PCR primer | taaagttttgggagcgacga aaagctttatggcggaatgtg |
| Sequence-based reagent | EFR D849N-f | This paper | PCR primer | cactgtAatattaagcca agcaacattcttctagacgat |
| Sequence-based reagent | BAK1-f | This paper | PCR primer | atcgGAAGACaaAATGGA ACGAAGATTAATGATCC |
| Sequence-based reagent | BAK1 Y403F-f | This paper | PCR primer | cttgcgtttttacatgat cattgcgacccaaaga |
| Sequence-based reagent | BAK1 I338H-f | This paper | PCR primer | gagatgCAtagtatggcg gttcacagaaacttgct |
| Sequence-based reagent | BAK1 D416N-f | This paper | PCR primer | catcgaAatgtgaaagctg caaatattttgttggatgaag |
| Sequence-based reagent | EFRprom-f | This paper | PCR primer | atcgGgtctctTACGCGTCTC aGTAGatctagacgat taagtaattgagca |
| Sequence-based reagent | BAK1prom-f | This paper | PCR primer | atcgGgtctctTACGCGTCTCa CCTAtgtcgtgaaaagggcac |
| Sequence-based reagent | XIIa2_Tmicd-f | This paper | PCR primer | atcgGCTCTTCgAAGGTT CTTCTACCGGTTCTGTTAT |
| Sequence-based reagent | XIIa3_Tmicd-f | This paper | PCR primer | atcgGCTCTTCgAAGG TTGCGATTGGGGTCA |
| Sequence-based reagent | XIIa4_Tmicd-f | This paper | PCR primer | atcgGCTCTTCgAAGATAA TCACCATTTGTGTCAGTG |
| Sequence-based reagent | XIIa5_Tmicd-f | This paper | PCR primer | atcgGCTCTTCgAAGGTT GTGATTGGAGTTAGCGT |
| Sequence-based reagent | XIIa6_Tmicd-f | This paper | PCR primer | atcgGCTCTTCgAAGGTTG CAATTTTAGTAAGCATAGG |
| Sequence-based reagent | FLS2_Tmicd-f | This paper | PCR primer | atcgGCTCTTCgAAGGTCA TCCTGATTATTCTTGGATCA GCCGCGGCaCTTCTTCTTGTCCTG |
| Sequence-based reagent | AlXIIa_D850N-f | This paper | PCR primer | atcgGCTCTTCAGTa ATATTAAGCCAAGCAACG |
| Sequence-based reagent | BrXIIa_D846N-f | This paper | PCR primer | atcgGCTCTTCAGTaA TCTTAAGCCAAGCAAC |
| Sequence-based reagent | GmXIIa_D829N-f | This paper | PCR primer | atcgGCTCTTCAGTaATA TTAAGCCAAGCAACATT |
| Sequence-based reagent | PtXIIa_D848N-f | This paper | PCR primer | atcgGCTCTTCAGT aatctgaagccaagcaa |
| Sequence-based reagent | SlXIIa_D883N-f | This paper | PCR primer | atcgGCTCTTCAGTaATAT AAAACCACAGAACATTCT |
| Sequence-based reagent | XIIa2_D803N-f | This paper | PCR primer | atcgGCTCTTCAGTaATC TCAAACCGAGCAATATCC |
| Sequence-based reagent | XIIa3_D838N-f | This paper | PCR primer | atcgGCTCTTCACaATCT TAAGCCAAGCAACATACT |
| Sequence-based reagent | XIIa4_D856N-f | This paper | PCR primer | atcgGCTCTTCAGTaATATT AAGCCAAGCAATATTCTACTA |
| Sequence-based reagent | XIIa5_D839N-f | This paper | PCR primer | atcgGCTCTTCAGCaA TCTTAAGCCAAGCAACGT |
| Sequence-based reagent | XIIa6_D840N-f | This paper | PCR primer | atcgGCTCTTCAGCaATC TCAAGCCAAGCAACG |
| Sequence-based reagent | FLS2icd_D997N-f | This paper | PCR primer | atcgGCTCTTCAGTaATC TGAAGCCAGCTAATATACT |
| Sequence-based reagent | FLS2_D997N-f | This paper | PCR primer | GTTCATTGTaATCTGAAGC CAGCTAATATACTCCTTGACA |
| Sequence-based reagent | EFR_ecto-f | This paper | PCR primer | atcgGGTCTCaAATGAAG CTGTCCTTTTCACTTG |
| Sequence-based reagent | EFR_SapIdom-f | This paper | PCR primer | GTTATGAgGAGCTTCAT AGTGCAACAAGTCGCTTC |
| Sequence-based reagent | EFR_Esp3Idom-f | This paper | PCR primer | TTCCCGTgTCTTTCGGG AAGCTTTTGAACTTGC |
| Sequence-based reagent | ccdB cassette-f | This paper | PCR primer | atgcGGTCTCAGAAGgGAAG AGCAAAGCTGAACGA GAAACGTAAAAT |
| Sequence-based reagent | pET28_lin-f | This paper | PCR primer | tgagatccggctgctaa |
| Sequence-based reagent | pETGG cloning cassette-f | This paper | PCR primer | AGAAGGAGATATACCAATGAGAGACCAAAGCTGAA |
| Sequence-based reagent | P641-BsaI_mut1-f | This paper | PCR primer | agagacCaaagctgaacga gaaacgtaaaatgatataaata |
| Sequence-based reagent | P641-BsaI_mut2-f | This paper | PCR primer | gccagtGgtctcttc tggtcgtgactggg |
| Sequence-based reagent | 6xHis-FKBP-f | This paper | PCR primer | atcgCGTCTCtTACGGGTCT CaAATGggcagcagccatcatcatc atcatcacagcagcggcGGA GTGCAGGTGGAAAC |
| Sequence-based reagent | 6xHis-FRB-f | This paper | PCR primer | atcgCGTCTCtTACGGGTCTCaA ATGggcagcagccatcatcatcatcatc acagcagcggcATCCT CTGGCATGAGATGT |
| Sequence-based reagent | EFR_icd (stop)-f | This paper | PCR primer | atcgCGTCTCtTACGGGTCT CaAGGTaagaggaaaaag aaaaacaatgcc |
| Sequence-based reagent | EFR_icd (no stop)-f | This paper | PCR primer | atcgCGTCTCtTACGGGTCTCa AGGTaagaggaaa aagaaaaacaatgcc |
| Sequence-based reagent | BAK1_icd (stop)-f | This paper | PCR primer | atcgCGTCTCtTACGGGTCTC aAGGTgcttggtggcgaagg |
| Sequence-based reagent | FLS2_icd (no stop)-f | This paper | PCR primer | atcgCGTCTCtTACGGGTCTCaAG GTACCTGTTGCAAGAAA AAAGAAAAAAAG |
| Sequence-based reagent | BRI1_icd (no stop)-f | This paper | PCR primer | atcgGGTCTCaAGGT GGTAGAGAGATGA GGAAGAGA |
| Sequence-based reagent | BRI1icd_D1009N-f | This paper | PCR primer | atcgGGTCTCaAaACATG AAATCCAGTAATGTGTTGC |
| Sequence-based reagent | EFR-r | This paper | PCR primer | cgatGAAGACttCGAAcc CATAGTATGCATGTCCG |
| Sequence-based reagent | EFR Y836F-r | This paper | PCR primer | gtgcagaAactccaaagc tgaagccacatctat |
| Sequence-based reagent | EFR SSAA-r | This paper | PCR primer | gcagcagcaaactggttt agaaaggattctcgatcg |
| Sequence-based reagent | EFR F761H-r | This paper | PCR primer | tatggcggaatgtgaaacc cacaagggtatacgacatcgt |
| Sequence-based reagent | EFR F761M-r | This paper | PCR primer | tatggcggaatgtgaaaccat gaagggtatacgacatcgta |
| Sequence-based reagent | EFR V845F-r | This paper | PCR primer | cacgttcattgtcatgaccctt tcgctcactgtgatattaagc |
| Sequence-based reagent | EFR L743F-r | This paper | PCR primer | taaactcgtcgcggttaaa gttttcaacctcctaaagc |
| Sequence-based reagent | EFR L873E-r | This paper | PCR primer | tatagagCTCctgagcca aaccaaagtcactaacatg |
| Sequence-based reagent | EFR dNLLKH-r | This paper | PCR primer | tcgtcgctcccaaaactttaa ccgcgacgagtttattct |
| Sequence-based reagent | EFR D849N-r | This paper | PCR primer | ggcttaatatTacagtgag ctacagggtcatgaca |
| Sequence-based reagent | BAK1-r | This paper | PCR primer | cgatGAAGACttAAGCc cTTATCTTGGACCCGAGG |
| Sequence-based reagent | BAK1 Y403F-r | This paper | PCR primer | catgtaaaaacgca agccctcttgcagatccca |
| Sequence-based reagent | BAK1 I338H-r | This paper | PCR primer | ccatactaTGcatctca acctctgtctggaactgc |
| Sequence-based reagent | BAK1 D416N-r | This paper | PCR primer | ctttcacatTtcgatgaata atctttgggtcgcaatg |
| Sequence-based reagent | EFRprom-r | This paper | PCR primer | atcgGgtctctCAGACGTCTC aCATTgtcgattataaaaa gataaaagaaaggtt |
| Sequence-based reagent | BAK1prom-r | This paper | PCR primer | atcgGgtctctCAGACGTCT CaCATTtttatcctcaagag attaaaaacaaac |
| Sequence-based reagent | XIIa2_Tmicd-r | This paper | PCR primer | cgatGCTCTTCtCGAAccT GAACTAGCTTCTCCTTGTG |
| Sequence-based reagent | XIIa3_Tmicd-r | This paper | PCR primer | cgatGCTCTTCtCGAAccA CGTCTGGCTGTTCTCC |
| Sequence-based reagent | XIIa4_Tmicd-r | This paper | PCR primer | cgatGCTCTTCtCGAAccAG TCTCCTCGTCTCTGAAA |
| Sequence-based reagent | XIIa5_Tmicd-r | This paper | PCR primer | cgatGCTCTTCtCGAAccACGCC AAGTCGTTCTACTGGCTT TAAAGAACCTCTCTCTGAT TGAGATCAACTCCTTG |
| Sequence-based reagent | XIIa6_Tmicd-r | This paper | PCR primer | cgatGCTCTTCtCGAAccAC GTCTAGGTGTTCTTCTG |
| Sequence-based reagent | FLS2_Tmicd-r | This paper | PCR primer | cgatGCTCTTCtCGAAccA ACTTCTCGATCCTCGTTAC |
| Sequence-based reagent | AlXIIa_D850N-r | This paper | PCR primer | atcgGCTCTTCAtACA GTGAGCTACAGGGT |
| Sequence-based reagent | BrXIIa_D846N-r | This paper | PCR primer | atcgGCTCTTCAtACAG TGAGCTATTTGGTCAT |
| Sequence-based reagent | GmXIIa_D829N-r | This paper | PCR primer | atcgGCTCTTCAtACA GTGAACTACGGCCT |
| Sequence-based reagent | PtXIIa_D848N-r | This paper | PCR primer | atcgGCTCTTCAtACa atgaatgatgggcatg |
| Sequence-based reagent | SlXIIa_D883N-r | This paper | PCR primer | atcgGCTCTTCAtACA GTGAATCATGGGTGTT |
| Sequence-based reagent | XIIa2_D803N-r | This paper | PCR primer | atcgGCTCTTCAtACAGT GAACAACTTTTACAGGTGA |
| Sequence-based reagent | XIIa3_D838N-r | This paper | PCR primer | atcgGCTCTTCATtGCAA TGAGCTATAGGCTCATGA |
| Sequence-based reagent | XIIa4_D856N-r | This paper | PCR primer | atcgGCTCTTCAtACA GTGGGCTATAGGGTTGT |
| Sequence-based reagent | XIIa5_D839N-r | This paper | PCR primer | atcgGCTCTTCAtGCAA TGAGCTATAGGTTCATGACA |
| Sequence-based reagent | XIIa6_D840N-r | This paper | PCR primer | atcgGCTCTTCAtGCAAT GAGCTATAGGCTCATGAC |
| Sequence-based reagent | FLS2icd_D997N-r | This paper | PCR primer | atcgGCTCTTCAtACAA TGAACGATGGGAAAACC |
| Sequence-based reagent | FLS2_D997N-r | This paper | PCR primer | CTTCAGATtACAATGAAC GATGGGAAAACCATATCCAGA |
| Sequence-based reagent | EFR_ecto-r | This paper | PCR primer | atcgGGTCTCacTTCT TTCTAACTGACAGAGGC |
| Sequence-based reagent | EFR_SapIdom-r | This paper | PCR primer | TGAAGCTCcTCATAACT TACCTTCTCATGGAACATCC |
| Sequence-based reagent | EFR_Esp3Idom-r | This paper | PCR primer | GAAAGAcACGGGAAGT TCTCCACTCAACATATTTGTTT |
| Sequence-based reagent | ccdB cassette-r | This paper | PCR primer | tacgGGTCTCACGAAGA AGAGCactggctgtgtataaggga |
| Sequence-based reagent | pET28_lin-r | This paper | PCR primer | ggtatatctccttct taaagttaaaca |
| Sequence-based reagent | pETGG cloning cassette-r | This paper | PCR primer | AGCAGCCGGATCTCAAAGCAGAGACCACTGGC |
| Sequence-based reagent | P641-BsaI_mut1-r | This paper | PCR primer | cagctttGgtctctcg tacggcctcctgt |
| Sequence-based reagent | P641-BsaI_mut2-r | This paper | PCR primer | agagacCactggctgtg tataagggagcctga |
| Sequence-based reagent | 6xHis-FKBP-r | This paper | PCR primer | atcgCGTCTCtCAGAGGTC TCaACCTGAGCCGCTTTCC |
| Sequence-based reagent | 6xHis-FRB-r | This paper | PCR primer | atcgCGTCTCtCAGAGGTCT CaACCTGAGCCGCTCTTT |
| Sequence-based reagent | EFR_icd (stop)-r | This paper | PCR primer | atcgCGTCTCtCAGAGGTCTC aAAGCttacatagtatgcatgtccgtatt |
| Sequence-based reagent | EFR_icd (no stop)-r | This paper | PCR primer | atcgCGTCTCtCAGAGGTCTC aAAGCttacatagtatgcatgtccgtatt |
| Sequence-based reagent | BAK1_icd (stop)-r | This paper | PCR primer | atcgCGTCTCtCAGAGGTC TCaAAGCttatcttggacccgaggg |
| Sequence-based reagent | FLS2_icd (no stop)-r | This paper | PCR primer | atcgCGTCTCtCAGAGGTCTCaC GAAccAACTTCTCGATCCTCGTTACG |
| Sequence-based reagent | BRI1_icd (no stop)-r | This paper | PCR primer | atcgGGTCTCaCGAAccTAA TTTTCCTTCAGGAACTTCTTTTATA |
| Sequence-based reagent | BRI1icd_D1009N-r | This paper | PCR primer | atcgGGTCTCaGTtTCTGT GGATGATATGCGGAC |
| Antibody | GFP-HRP; mouse monoclonal | Santa Cruz Biotechnology | Cat#: sc-9996 | Dilution: 1:1000 |
| Antibody | BAK1; rabbit polyclonal | PMID: 21693696 | Dilution: 1:10000 | |
| Antibody | BAK1 pS612; rabbit polyclonal | PMID: 30177827 | Dilution: 1:2000 | |
| Antibody | FKBP12; mouse monoclonal | Santa Cruz Biotechnology | Cat#: sc-133067 | Dilution: 1:500 |
| Antibody | p44/42; rabbit polyclonal | Cell Signaling Technology | Cat#: 9101 | Dilution: 1:1000 |
| Antibody | Anti-rabbit-HRP; goat polyclonal | Sigma-Aldrich | Cat#: A0545 | Dilution: 1:10000 |
| Antibody | Anti-mouse-HRP; goat polyclonal | Sigma-Aldrich | Cat#: A0168 | Dilution: 1:5000 |
| Recombinant DNA reagent | p641-EFRprom(BsaI) | This paper | See Materials and methods | |
| Recombinant DNA reagent | 35 S_omegaEnhancer | PMID: 21364738 | ||
| Recombinant DNA reagent | pICH41258-nMyr-FKBP | PMID: 33964457 | ||
| Recombinant DNA reagent | pICH41258-nMyr-FRB | PMID: 33964457 | ||
| Recombinant DNA reagent | pICSL01003-GFP | PMID: 21364738 | ||
| Recombinant DNA reagent | pICSL01003-mEGFP | Mark Youles, TSL | ||
| Recombinant DNA reagent | HSP18t | Mark Youles, TSL | ||
| Recombinant DNA reagent | p641-6xHis-FKBP | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641-6xHis-FRB | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR Y836F | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR SSAA | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR F761H | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR F761M | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR dNLLKH | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR L743F | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR L873E | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR F761H Y836F | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR F761M Y836F | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR dNLLKH Y836F | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR L743F Y836F | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR L873E Y836F | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR D849N | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR F761H D849N | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR F761H SSAA | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL01005-EFR F761M SSAA | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICH41308-BAK1 | This paper | See Materials and MethodsSee Materials and methods | |
| Recombinant DNA reagent | pICH41308-BAK1 Y403F | This paper | See Materials and MethodsSee Materials and methods | |
| Recombinant DNA reagent | pICSL86955 | PMID: 21364738 | ||
| Recombinant DNA reagent | pICSL86955-pEFR::EFR-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR Y836F-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR SSAA-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR F761H-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR F761M-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR dNLLKH-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR L743F-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR L873E-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR F761H Y836F-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR F761M Y836F-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR dNLLKH Y836F-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR L743F Y836F -GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR Y836F L873E-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR F761H SSAA-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-pEFR::EFR F761M SSAA-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641 | PMID: 31300661 | ||
| Recombinant DNA reagent | p641-EFR_icd | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641-EFR F761H_icd | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641-EFR D849N_icd | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641-EFR F761H D849N_icd | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641-BAK1_icd | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641-BAK1 Y403F_icd | This paper | See Materials and methods | |
| Recombinant DNA reagent | p641-BAK1 D416N_icd | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-ccdB-GFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa2icd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa2 D803N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa3icd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa3 D838N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa4icd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa4 D856N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa5icd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa5 D839N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa6icd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XIIa6 D840N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-FLS2icd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-FLS2 D997N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-EFRicd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-EFR D849N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XA21icd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-XA21 D803N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Aralyicd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Araly D850N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Brapaicd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Brapa D846N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Solycicd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Solyc D883N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Poptricd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Poptr D848N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-Glymaicd-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL86955-35S::EFRecto-GlymaD829N-mEGFP::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-EFRicd-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-EFRicd D849N-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-BRI1-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-BRI1 D1009N-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-FLS2-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-FLS D997N-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-XIIa5-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FKBP-XIIa5 D839N-mEGFP | This paper | See Materials and methods | |
| Recombinant DNA reagent | pETGG-6xHis-FRB-BAK1 | This paper | See Materials and methods | |
| Recombinant DNA reagent | pET28a(+)–6xHis-BIK1 D202N | This paper | See Materials and methods | |
| Recombinant DNA reagent | pC1a1-35S::FRB-BAK1_icd::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pC1a1-35S::FRB-BAK1 Y403F_icd::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pC1a1-35S::FRB-BAK1 D416N_icd::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pC1a2-35S::FKBP-EFR_icd::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pC1a2-35S::FKBP-EFR F761H_icd::HSP18t | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL4723 | PMID: 21364738 | See Materials and methods | |
| Recombinant DNA reagent | pICSL4723-FRB-BAK1-FKBP-EFR | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL4723-FRB-BAK1 D416N-FKBP-EFR | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL4723-FRB-BAK1-FKBP-EFR F761H | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL4723-FRB-BAK1 D416N-FKBP-EFR F761H | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL4723-FRB-BAK1 Y403F-FKBP-EFR | This paper | See Materials and methods | |
| Recombinant DNA reagent | pICSL4723-FRB-BAK1 Y403F-FKBP-EFR F761H | This paper | See Materials and methods |





