A chemically induced attenuated strain of Candida albicans generates robust protective immune responses and prevents systemic candidiasis development
Figures
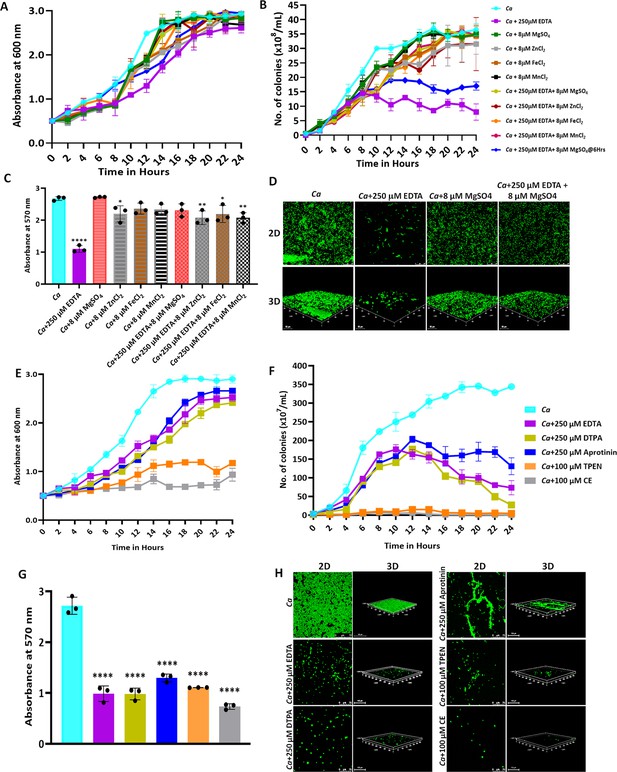
Effect of metal chelators on the growth and biofilm of C. albicans.
(A) C. albicans cells were cultured in YPD media at 30 °C for 24 hr without (cyan blue) and with the indicated concentration of EDTA (purple), MgSO4 (Green), ZnCl2 (Grey), FeCl2 (Brown), and MnCl2 (Black), EDTA+MgSO4 (Lime green), EDTA+MgSO4@6 hr (dark blue), EDTA+ZnCl2 (Maroon), EDTA+FeCl2 (Orange), and EDTA+MnCl2 (Pink). Optimal absorbance was measured at 600 nm in different intervals of incubation. (B) The cultures from the above-mentioned experiment was diluted and plated on YPD plate. Colonies were counted and plotted to determine the CFU efficiency. (C) Pre-formed C. albicans biofilm was treated with EDTA and divalent metals as mentioned. Their effect on biofilm was observed post-24 hr treatment by crystal violet staining and estimating at 570 nm. (D) Effect of EDTA and divalent metals on C. albicans biofilm was again observed by acridine orange staining and visualization under a ×40 magnification using a CLSM. Similarly, the effect of other metal chelators like DTPA (Lime green), Aprotinin (blue), TPEN (orange) and CE (grey) on the growth (E), CFU (F), and biofilm formation of C. albicans analyzed by crystal violet staining (G) and CLSM (H). Mean values from three independent experiments considered and error bar represents SEM. p values *<0.05, **<0.01 and ****<0.0001 were significant as determine by one-way ANOVA.
-
Figure 1—source data 1
Effect of metal chelators on the growth, CFU, and biofilm of C. albicans.
- https://cdn.elifesciences.org/articles/93760/elife-93760-fig1-data1-v1.zip
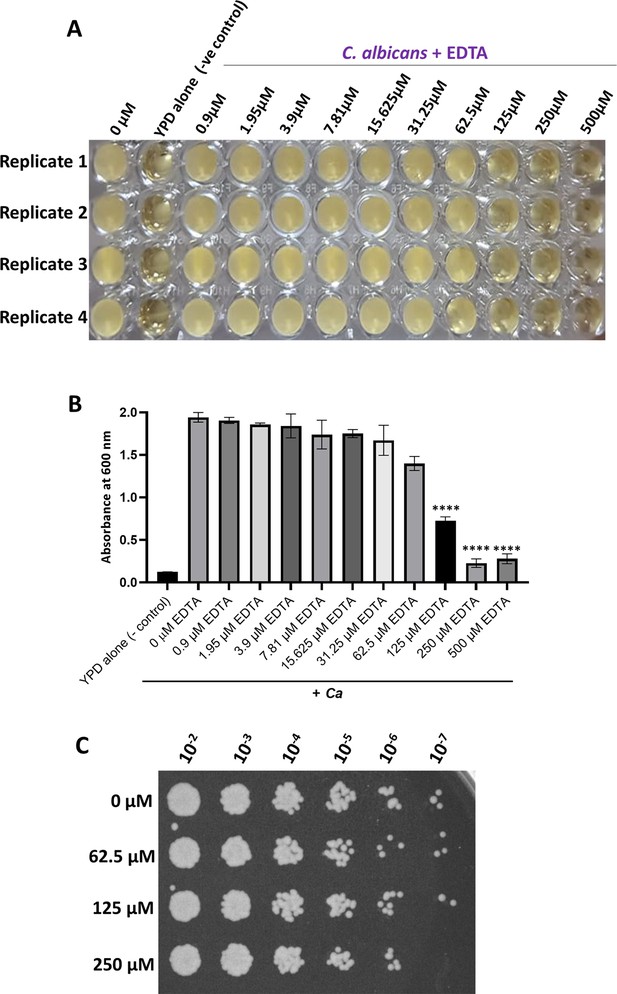
Identification of a fungistatic concentration of EDTA.
(A) Various concentrations of EDTA ranging from 0.9 µM to 500 µM was added to the C. albicans culture with a total volume of 150 µL in a 96-well plate and allowed to grow overnight at 30 °C under static condition. YPD media alone and without EDTA treatment culture as controls were also taken. After 24 hr, the image of the 96-well polystyrene plate was taken. (B) After 24 hr, the absorbance was measured at OD600 nm using a Perkin-Elmer plate reader and plotted using GraphPad Prism software version 8.0. p value as indicated ****<0.0001 was determined by one-way ANOVA. (C) C. albicans cells treated without and with 62.5 µM, 125 µM, and 250 µM concentration of EDTA for 24 hr and indicated dilutions were spotted on YPD-agar plate. The plate was incubated at 30 °C for 24 hr and imaged using a Chemidoc imaging system.
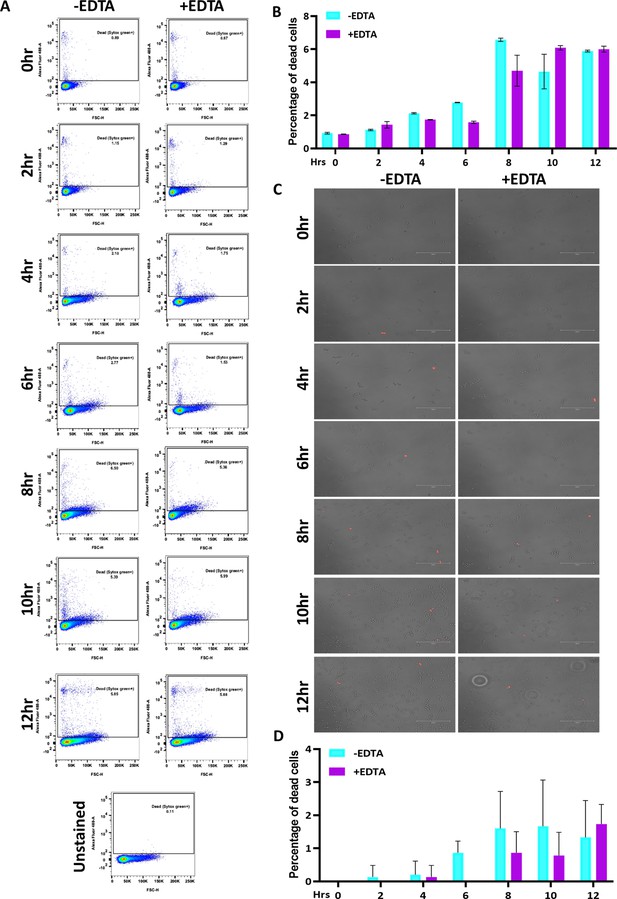
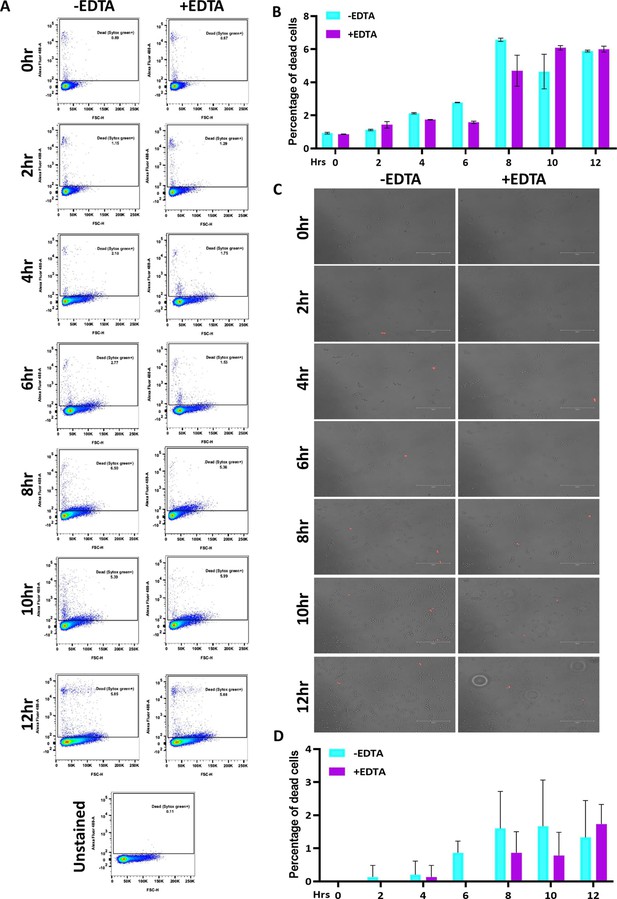
Effect of 250 μM EDTA on cell viability.
C. albicans pre-culture was diluted to an OD600nm=0.5 and allowed to grow in the absence and presence of 250 μM EDTA at 30 °C and 200 rpm up to 12 hr. (A) At mentioned time points (0–12 hr), cells were harvested, stained with SYTOX Green and analyzed by flow cytometry (blue laser; excitation at 488 nm). The data acquisition was performed using the BD LSR Fortessa Flow cytometer and analysis was carried out by FlowJo version 8.1 software. Analysed data exported in JPEG format. Acquisition profile of unstained cells was also shown. (B) The percentage of dead cells population from two technical replicates was plotted using GraphPad Prism software version 8.0. No statistical significance difference was found between treated and untreated data. (C) Similarly, the harvested cells were stained with PI and images were captured using a fluorescence microscope (EVOS imaging system; Thermo Fisher Scientific) at ×40 magnification and the red dots indicated the dead cells. (D) The population of dead cells was determined from 15 microscopy frames for each time interval and plotted using GraphPad Prism. p value that was non-significant as determined by two-way ANOVA.
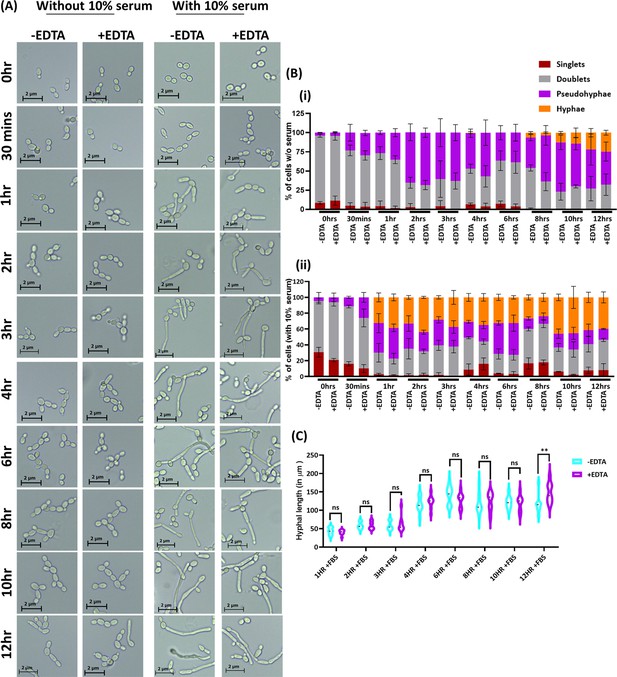
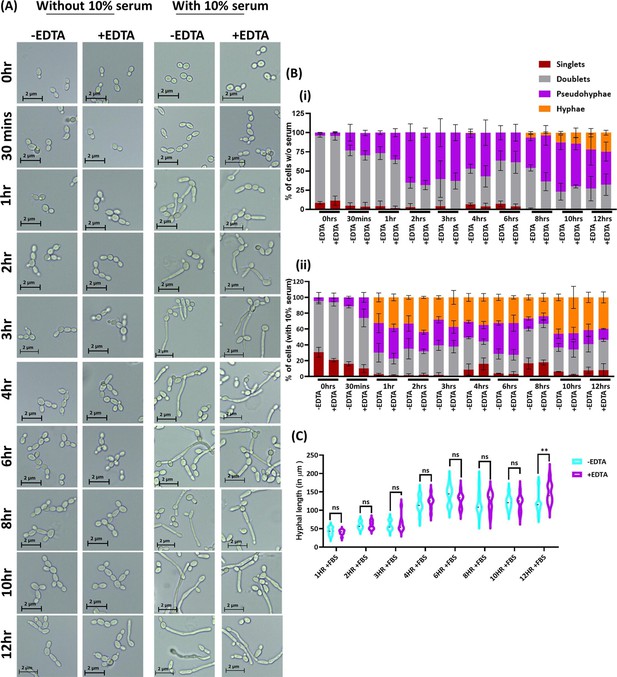
Effect of EDTA on C.albicans morphology.
C. albicans pre-culture was diluted to an OD600nm=0.5 and allowed to grow in the absence and presence of 250 μM EDTA at 30 °C and 200 rpm up to 12 hr. Similar experiment was also carried with 10% serum and morphological transition was induced at 37 °C. (A) At various time intervals, cell morphology was observed using a ×40 Leica DM500 microscope (scale bar of 2 µm). (B) The percentages of singlet, doublet, pseudohyphae, and hyphae cells were quantified without (i) and with serum (ii). (C) The length of serum-induced germ tubes (n=60) in µm was measured using ImageJ software. p value as indicated **<0.01 or non-significance as determined by two-way ANOVA.
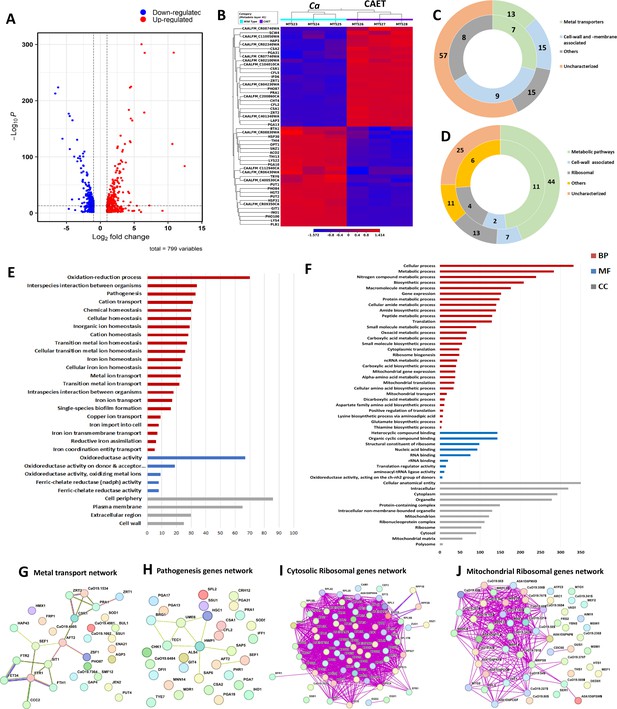
Transcriptomics analyses C.albicans cells upon EDTA treatment.
(A) A Volcano plot depicting differentially expressed genes (DEGs) in CAET cells. The red dots indicate upregulated genes (411) and blue dots indicates the downregulated genes (388). (B) Heat map showing relative abundance of top 25 significantly upregulated and downregulated genes in Ca and CAET. RNA sequencing was carried out in triplicates. Red colour indicates upregulation and blue indicates downregulation as denoted by the Z score. (C) Double donut chart indicates top 100 significantly upregulated genes categorized into four main groups that is, metal transporters (light green), cell wall- and membrane-associated genes other than metal transporters (blue), others (includes drugs resistance-associated, morphology-associated, biosynthetic and catalytic processes; represented in grey), and uncharacterized genes (cream). Inner donut depicts the number of DEGs from the above categories associated with virulence and pathogenesis in C. albicans as reported by published literatures. (D) Double donut chart depicting top 100 significant downregulated gene categorized as metabolic pathways (green), cell-wall associated (blue), ribosomal (grey), others (includes morphology-associated, resistance-associated; represented in yellow), and uncharacterized (cream). Outer circle represents the total number of genes in each category and inner circle indicates the virulence-related genes out of the total number of genes in that specific category. (E) Gene ontology enrichment analysis for 411 upregulated DEGs. (F) Gene ontology enrichment analysis for 388 downregulated DEGs were plotted. Both in (E) and (F), x-axis represents the number of DEGs and y-axis represents various processes like BP (biological processes), MF (molecular functions), and CC (cellular components) indicated in maroon, blue, and grey colors, respectively. (G) STRING analyses showing relationship between DEGs. Interaction amongst 15 connected out of 31 upregulated DEGs involved in metal transport and homeostasis is shown. (H) STRING analysis shows 9 connected out of 33 upregulated DEGs involved in pathogenesis. (I) Among the down-regulated DEGs, 70 connected out of 74 DEGs belonging to cytosolic ribosomal genes as determined by STRING. (J) 38 connected out of 56 DEGs belonging to mitochondrial ribosomal components were connected by as determined STRING.
-
Figure 2—source data 1
Transcriptomic analyses of CAET.
- https://cdn.elifesciences.org/articles/93760/elife-93760-fig2-data1-v1.zip
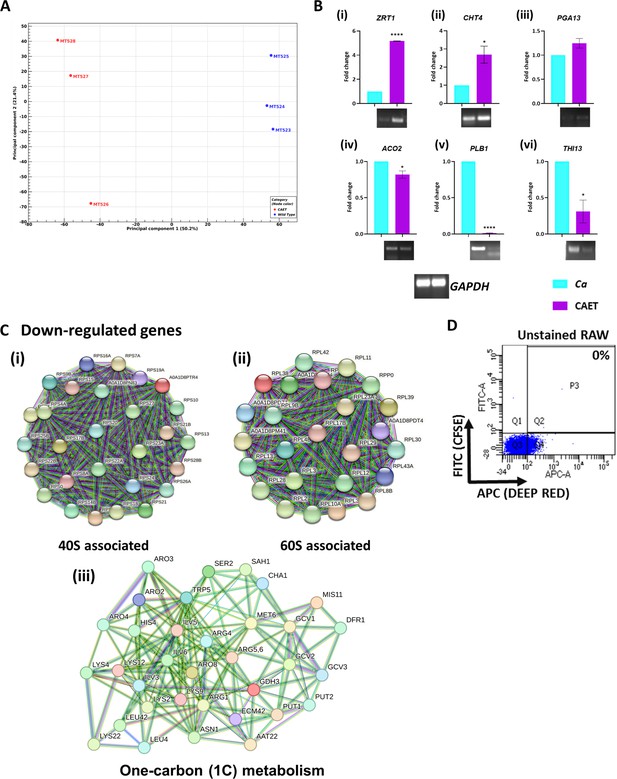
Transcriptomic analyses of CAET and validation.
(A) Principal Component Analyses (PCA) was performed for proper grouping of three biological replicates represented as MT523, MT524, and MT525 as one cluster for Ca and MT526, MT527, and MT528 for CAET for another. The plot scores indicate the principal component 1 vs. principal component 2 signifying the largest variable vs. second largest variable. (B) Gene expression was performed using qRT-PCR and semi-quantitative assay for selected upregulated genes obtained from the heat map that is, (i) metal ion associated gene ZRT1, (ii) cell wall associated gene CHT4 (chitin-specific gene), and (iii) host-pathogen interaction-related gene PGA13. Similarly, the downregulated genes selected are (iv) aconitase ACO2, (v) virulence-associated gene PLB1, and (vi) thiamine synthesis-associated gene THI13 expression were analysed. GAPDH considered as control. p values *<0.05 and ****<0.0001 were as determined by unpaired t test. (C) STRING analysis showing only the clusters related to 40 S (i), 60 S subunits (ii) and one-carbon metabolism (iii). An interaction score of 0.7 was considered for all the networks. (D) Gating strategy of unstained RAW cells alone was shown.
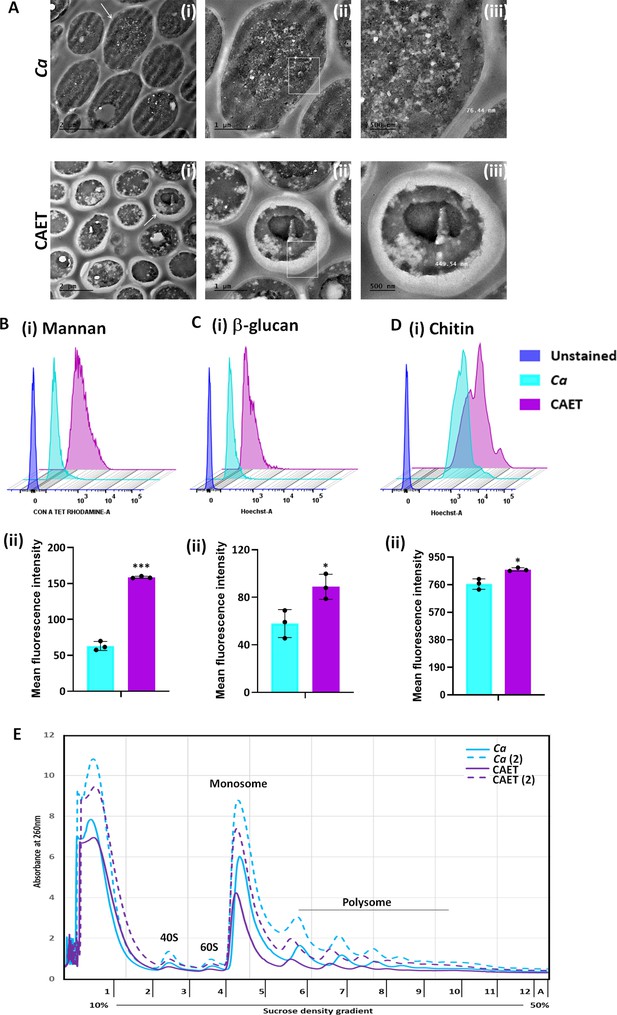
ffect of EDTA on the cell wall and polysome of C. albicans.
(A) Showing TEM images of ultrathin sections of untreated (Ca) and EDTA-treated (CAET) C. albicans at different resolution. Samples visualized at ×14,000 magnification, arrow indicates the marked cell, and box indicates the focus area where thickness was measured. Scale bar = 2 µm for intact cells (i); scale bar = 1 µm for a single cell (ii); scale bar = 500 nm for zoomed in image of an individual cell wall (iii). (B) C. albicans cells were stained with Con A and analyzed by flow cytometry (i) and mean fluorescence intensity was measured to estimate the mannan level (ii). (C) C. albicans cells were stained with aniline blue and analyzed by flow cytometry (i) and mean fluorescence intensity was measured to estimate the β-glucan level (ii). (D) C. albicans cells were stained with calcofluor white and analyzed by flow cytometry (i) and mean fluorescence intensity was measured to estimate the chitin level (ii). Mean average of three independent biological replicates with error bars is shown. p values *<0.05 and ***<0.001 as determined by unpaired t-test. (E) Cell-free total ribosomes were isolated by taking two different concentrations (pellet size of 200 µL and 400 µL) of C. albicans cells (Ca and CAET) and fractionated using sucrose gradient centrifugation. Fractions were analysed and plotted. The position and transition of ribosome subunits, monosome and polysome peaks were as shown.
-
Figure 3—source data 1
Ultrastructure of C. albicans cells upon EDTA treatment and estimation of cell wall thickness, various components of cell wall, and ribosome levels.
- https://cdn.elifesciences.org/articles/93760/elife-93760-fig3-data1-v1.zip
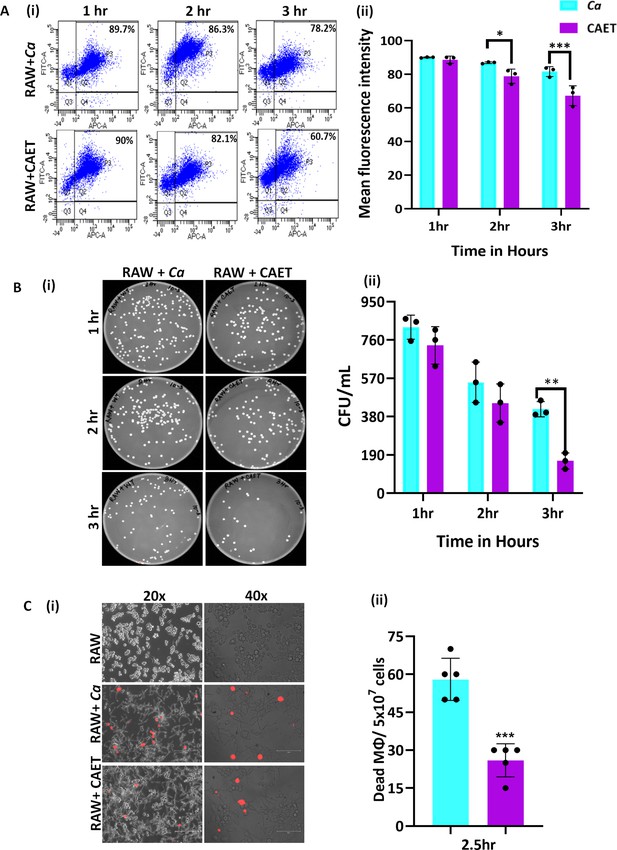
C.albicans-Macrophage interaction.
(A) Deep red-stained RAW 264.7 murine macrophage cells were co-cultured with C. albicans (Ca and CAET) in 1:1 ratio. After each time point (1 hr, 2 hrs and 3 hrs) cells were pooled down and double positive cells (CFSE-FITC channel and deep red-APC channel; Q2, P3) were analysed by flow cytometry (i). Mean florescence intensity of three independent biological replicates were plotted using the GraphPad Prism software (ii). (B) From a similar co-culture experiment, C. albicans cells (Ca and CAET) were retrieved from macrophage cells and plated on YPD/Chloramphenicol plate by taking appropriate dilutions. The plates were incubated at 30 °C for 48 hr. The plates were imaged (i) and colony-forming unit (CFU) was determined (ii). (C) A similar co-culture of macrophage and C. albicans (Ca and CAET) were carried out for 2.5 hr and the dying macrophages were observed by propidium iodide staining (Red dots). Images were captured with fluorescence microscope (EVOS imaging system; Thermo Fisher Scientific) at ×20 and ×40 magnifications (i) and the population of dead macrophages were determined (ii). p values *<0.05, **<0.01, and ***<0.01 as determined by two-way ANOVA.
-
Figure 4—source data 1
Estimation of phagocytosis, fungal cells clearance and macrophage killing.
- https://cdn.elifesciences.org/articles/93760/elife-93760-fig4-data1-v1.zip
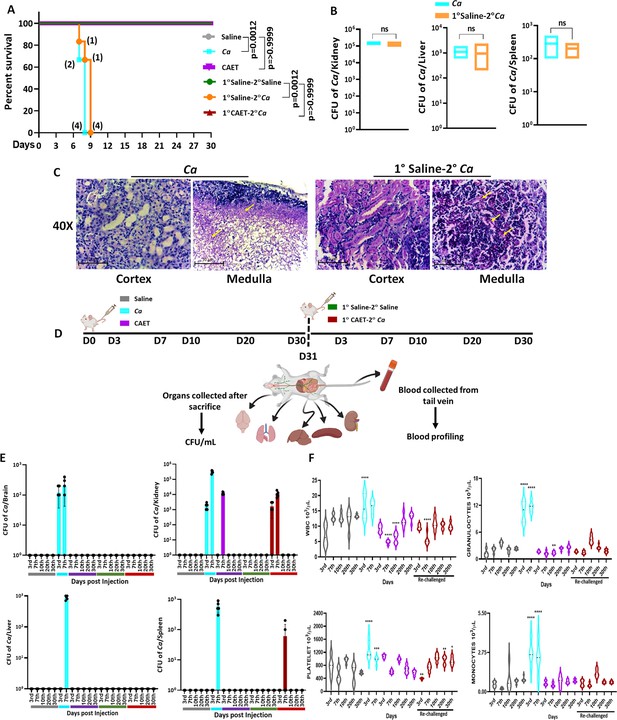
Systemic candidiasis development and progression in C. albicans challenged mice.
(A) Mice (n=6/category) were injected intravenously with 5x105 C. albicans cells (Ca: cyan blue and CAET: purple) and saline as control (grey) and their survivability was monitored for 30 days. In a similar set of experiment, mice (n=6/category) were first immunized with CAET (represented in brown) or sham vaccinated with saline (represented in orange), and after 30 days, they were further re-challenged with C. albicans. Their survivability was monitored for another 30 days and a survival curve was plotted. The number of sacrificed mice were mentioned on the particular days in the survival plot. 1° and 2° suggest primary and secondary challenges, respectively. Statistical significance of the survival curve was determined using Mantel-Cox test. p values were as mentioned, otherwise they were non-significant. Y-axis was represented in Log rank scale. (B) Fungal load in the kidney, liver, and spleen of sacrificed mice in Log10 scale was determined by CFU analyses. No statistical significant difference was observed between these groups. (C) PAS-staining for one of the kidneys carried out to visualize the fungal burden in the cortex and medulla regions under ×40 magnification. (D) To check the disease progression, a kinetic experiment was planned as indicated in the schematic diagram. A group of mice were injected intravenously with 5x105 C. albicans cells (Ca: cyan blue, CAET: purple, and 1° CAET 2°Ca: brown) and saline controls (grey and green) at 0 day, and five mice from each group were sacrificed on each day mentioned (3d, 7th, 10th, 20thh, and 30th day). Approximately 20 µl blood was drawn from the lateral tail vein of the mice prior to sacrifice. (E) Fungal load in the brain, liver, kidney, and spleen in Log scale was determined by CFU analyses. (F) Blood parameters such as WBC, Granulocytes, Platelets, and Monocytes levels were analyzed on the above-mentioned days and graphs were plotted using the GraphPad Prism software. The statistical analyses between saline and fungal infected mice groups of same day sacrificed were carried out. p values **<0.01, **<0.01, ***<0.00, and ****<0.0001 as determined by two-way ANOVA.
-
Figure 5—source data 1
Virulence and vaccine potentials of Ca Vs CAET.
- https://cdn.elifesciences.org/articles/93760/elife-93760-fig5-data1-v1.zip
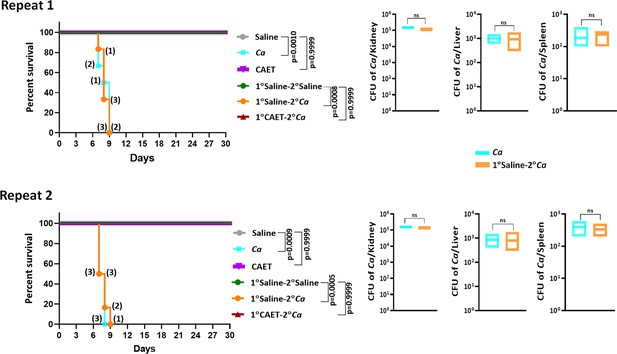
Systemic candidiasis development in C.albicans challenged mice.
Mice (n=6/category) were injected intravenously with 5x105 C. albicans cells (untreated Ca: cyan blue and CAET: purple) and saline as control (grey) and their survivability was monitored for 30 days. In a similar set of experiment, mice (n=6/category) were first immunized with CAET (represented in brown) or sham vaccinated with saline (represented in orange), and after 30 days, they were further re-challenged with C. albicans. Their survivability was monitored for another 30 days and a survival curve was plotted. 1° and 2° suggest primary and secondary challenges, respectively. Mice suffered due to severe infections were euthanized and fungal load in vital organs were determined by CFU analyses. Statistical significance of the survival curve was determined using Mantel-Cox test. p values were as mentioned and others were non-significant. Y-axis was represented in Log rank scale. Two repeat studies were shown.
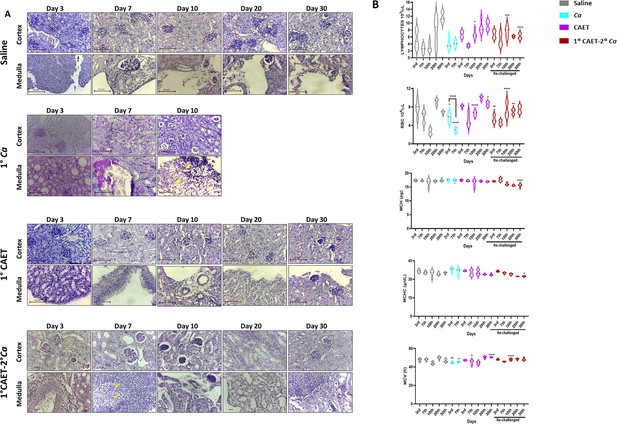
Kinetic analyses of fungal cells load and blood profiles of mice challenged with Ca and CAET.
(A) PAS-stained kidney sections of different mice groups (saline, Ca, CAET and 1°CAET 2°Ca) from the experiment in Fig.Figure 5D showing fungal burden at different times of sacrifice post inoculation. (B) Lymphocytes, RBC, MCHC, MCH, and MCV (fl) present in total blood of various mice groups sacrificed on various days from the experiment in Figure 5D was estimated. p values *<0.05, **<0.01, ***<0.001, and ****<0.0001 as determined by two-way ANOVA.
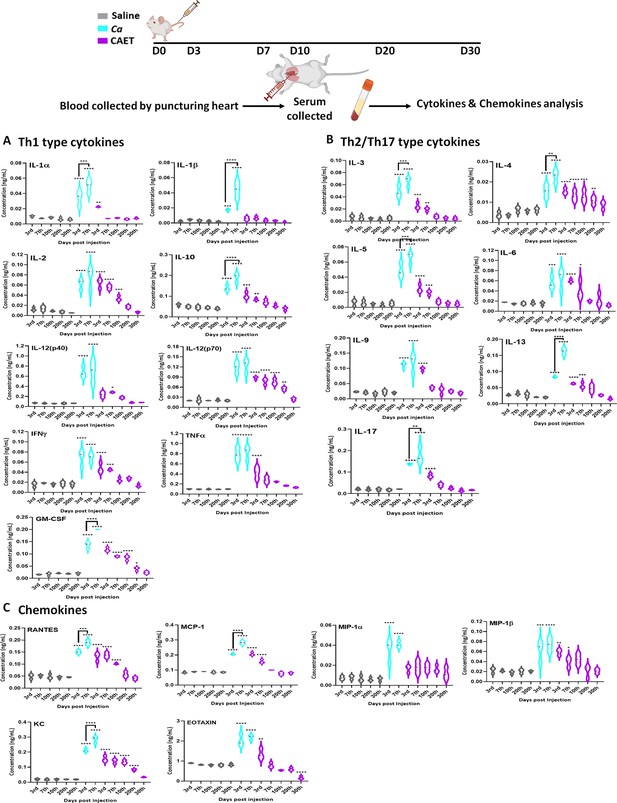
Cytokine and chemokine estimation in infected mice.
The mice (n=3/category) were inoculated with Ca or CAET or saline and sacrificed on various days as indicated and cytokine and chemokine levels in the blood serum were quantified by using Bio-plex Pro cytokine multiplex kit. (A) A panel of Th-1 cytokines, that is IL-1α, IL-1β, IL2, IL-10, IL-12(p40), IL-12(p70), IFN-γ, TNF-α, and GM-CSF, (B) Th-2/Th-17 cytokines, that is IL-3, IL-4, IL-5, IL-6, IL-9, IL-13, and IL-17, and (C) Chemokines that is RANTES, MCP-1, MIP-1α, MIP-1β, KC, and EOTAXIN. The statistical analyses between saline and fungal infected mice groups of same day sacrificed were carried out. p values **<0.01, **<0.01, ***<0.00, and ****<0.0001 as determined by two-way ANOVA.
-
Figure 6—source data 1
Cytokines and chemokines estimation in fungal infected mice.
- https://cdn.elifesciences.org/articles/93760/elife-93760-fig6-data1-v1.zip
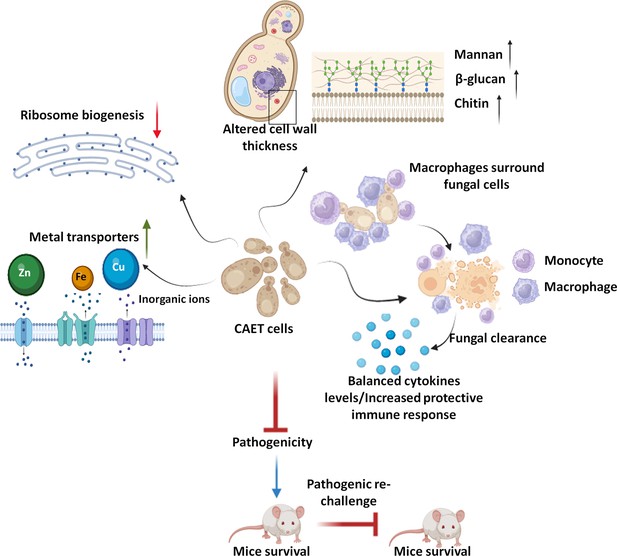
Model depicting the attributes of CAET cells.
EDTA alters the cell wall thickness by altering its composition. Metal transporters and several cell wall and membrane associated genes get upregulated. To mitigate the essential metal scarcity, genes involved in ribosome biogenesis and one-carbon metabolism were down-regulated. CAET cells get phagocytosed efficiently and eliminated faster by macrophages. CAET-infected mice survived and induced robust host immune responses to protect the lethal rechallenge. Thus, CAET is a potential live whole-cell vaccine candidate.
Tables
A brief summary of RNA sequencing reads obtained from Illumina Novaseq.
| Sample Name (Sequencing ID) | Read Length (bp) | Total raw reads | Reads after rRNA removal | Average Length after trim | Read after trimming | % known mRNA | % Unknown mRNA |
|---|---|---|---|---|---|---|---|
| Wild type (MT523) | 151×2 | 21,937,416 | 19,222,356 (87.62%) | 143.22 | 19,221,728 | 98.06 | 1.94 |
| Wild type (MT524) | 151×2 | 20,794,992 | 18,475,070 (88.84%) | 141.71 | 18,474,428 | 98.36 | 1.68 |
| Wild type (MT525) | 151×2 | 20,564,218 | 17,699,794 (86.07%) | 140.89 | 17,699,180 | 98.3 | 1.7 |
| CAET (MT526) | 151×2 | 22,376,466 | 20,284,442 (90.65%) | 140.79 | 20,283,776 | 98.15 | 1.85 |
| CAET (MT527) | 151×2 | 21,412,804 | 20.073,926 (97.49%) | 142.14 | 20,073,230 | 98.72 | 1.28 |
| CAET (MT528) | 151×2 | 20,435,938 | 19,391,726 (94.89%) | 142.25 | 19,391,142 | 98.85 | 1.5 |
List of top 100 upregulated genes in EDTA treated C. albicans cell (CAET).
| Metal Transporters | ||||
|---|---|---|---|---|
| No. of DEGs | Name of DEGs (13) | Fold Change | FDR p-value | Function |
| 1 | ZRT1 | 5644.9942 | 5.20999E-82 | Zinc ion transporter; Hyphal inducer |
| 2 | PRA1 | 1829.3467 | 1.3833E-282 | Zinc sequester; pH regulated antigen |
| 3 | CFL5 | 151.1291 | 4.16014E-13 | Ferric reductase |
| 4 | CFL2 | 94.2481 | 1.5889E-282 | Ferric reductase; Virulence |
| 5 | PHO87 | 90.3067 | 1.0754E-176 | Phosphate transporter; Virulence |
| 6 | ZRT2 | 68.1720 | 2.5856E-297 | Zinc uptake; Biofilm inducer |
| 7 | CSA2 | 26.6647 | 0.000886698 | Hyphal inducer; Heme utilization protein; Biofilm inducer |
| 8 | CSR1 | 23.2254 | 2.0725E-222 | zinc homeostasis, Filamentation inducer; |
| 9 | CSA1 | 22.5038 | 3.4486E-166 | Hyphal inducer; Iron homeostasis; Biofilm inducer; |
| 10 | HAP3 | 18.4511 | 5.412E-13 | Iron homeostasis; |
| 11 | CFL4 | 8.4663 | 3.22997E-11 | Uptake of iron metal ion |
| 12 | FRE9 | 7.5052 | 1.41623E-45 | Ion transport, Ferric reductase |
| 13 | ARH2 | 5.7589 | 1.18159E-60 | Involved in heme biosynthesis |
| Cell-wall and -membrane associated | ||||
| Name of the DEGs (15) | Fold Change | FDR p-value | Function | |
| 1 | SCW4 | 28.9947 | 0.000794714 | Glucanase activity |
| 2 | PGA13 | 26.3347 | 8.3797E-163 | Cell wall integrity; Morphogenesis, Virulence |
| 3 | PGA31 | 22.0969 | 1.25365E-18 | Cell wall integrity |
| 4 | CHT4 | 20.7121 | 4.5927E-126 | Chitin synthesis and degradation |
| 5 | RIM9 | 12.4158 | 4.4708E-121 | Promote growth under alkaline condition |
| 6 | ALS4 | 10.0218 | 1.4683E-110 | Enhance cell surface adhesion |
| 7 | CSH1 | 9.5387 | 1.2788E-107 | Maintain cell integrity; Virulence |
| 8 | MRV8 | 8.7526 | 0.007934383 | Biofilm inducer; Caspofungin tolerance |
| 9 | HGT5 | 7.5433 | 2.42539E-86 | Glucose transporter |
| 10 | CDA2 | 7.2664 | 1.39323E-07 | Chitin deacetylase |
| 11 | PRN4 | 6.2037 | 5.00644E-35 | Protein with similarity to pirins |
| 12 | GLX3 | 5.9890 | 3.73497E-61 | Binds human IgE; Induce biofilm |
| 13 | PGA7 | 5.6247 | 2.14119E-46 | GPI-linked hyphal surface antigen; Induce spider biofilm |
| 14 | RBT5 | 5.5414 | 7.36268E-45 | GPI-linked cell wall protein; utilize haemin and haemoglobin; biofilm induced |
| 15 | JEN1 | 5.1930 | 4.63735E-06 | Localized on plasma membrane and induced by lactic acid |
| Others | ||||
| Name of the DEGs (15) | Fold Change | FDR p-value | Function | |
| 1 | LAP3 | 20.7648 | 6.7063E-221 | Aminopeptidase; Bleomycin resistance |
| 2 | IFD6 | 18.4736 | 2.2359E-181 | Azole resistance; Biofilm-inducer |
| 3 | ATO1 | 13.3199 | 6.23932E-08 | pH neutralizer in macrophage phagolysosome |
| 4 | NOP6 | 10.5223 | 3.90346E-07 | Ribosome biosynthesis |
| 5 | RAS2 | 10.2667 | 8.69395E-36 | Filamentous growth enhancer; cAMP pathway regulator |
| 6 | HST6 | 8.6960 | 6.13855E-09 | Transport lipid; antifungal drugs resistance; Encode ABC transporters |
| 7 | STE23 | 8.3030 | 3.41151E-92 | Involved in processing of mating pheromones |
| 8 | FGR46 | 7.4482 | 1.72544E-07 | Filamentous Growth Regulator |
| 9 | POL93 | 7.0291 | 1.77132E-69 | Encode reverse transcriptase, protease and integrase; Induce biofilm |
| 10 | HPD1 | 5.9926 | 6.53346E-22 | Involved in degradation of toxic propionyl-CoA; Induce spider biofilm |
| 11 | AOX2 | 5.6936 | 7.2209E-114 | Induce biofilm formation |
| 12 | SOU2 | 5.5746 | 0.006421918 | L-sorbose utilization; biofilm induced |
| 13 | GPD1 | 5.4805 | 2.01281E-74 | Involved in glycerol biosynthesis |
| 14 | CAG1 | 5.3023 | 2.20049E-05 | Role in mating pheromone response |
| 15 | FDH1 | 5.2775 | 2.01281E-74 | xidization of formate to CO2 |
| Uncharacterized | ||||
| Name (57 DEGs) | Fold Change | FDR p-value | Function | |
| 1 | CAALFM_C200860CA | 1600.2301 | 4.4708E-121 | Uncharacterized |
| 2 | CAALFM_C401340WA | 583.6099 | 0.000537249 | Uncharacterized |
| 3 | CAALFM_CR08740WA | 64.8421 | 0.035751787 | Uncharacterized |
| 4 | CAALFM_CR02340WA | 53.5609 | 3.38443E-05 | Uncharacterized |
| 5 | CAALFM_CR07740WA | 42.2094 | 1.62356E-12 | Uncharacterized |
| 6 | CAALFM_C602100WA | 32.1762 | 0.000435707 | Uncharacterized |
| 7 | CAALFM_C104010CA | 21.9880 | 4.8225E-124 | Uncharacterized |
| 8 | CAALFM_C604230WA | 19.1498 | 1.67518E-34 | Uncharacterized |
| 9 | CAALFM_C110050WA | 18.1320 | 0.000124176 | Uncharacterized |
| 10 | CAALFM_CR06510WA | 17.4047 | 6.21185E-37 | Uncharacterized |
| 11 | CAALFM_CR06500CA | 13.6055 | 2.25687E-88 | Uncharacterized |
| 12 | CAALFM_CR08310CA | 10.6593 | 4.21036E-20 | Uncharacterized |
| 13 | CAALFM_CR06570CA | 10.6284 | 6.6067E-77 | Uncharacterized |
| 14 | CAALFM_C204430WA | 9.9253 | 0.003875235 | Uncharacterized |
| 15 | CAALFM_C303570CA | 9.6111 | 5.27224E-07 | Uncharacterized |
| 16 | CAALFM_C306040WA | 9.4663 | 1.09545E-56 | Uncharacterized |
| 17 | CAALFM_C106860WA | 9.4073 | 0.000100814 | Uncharacterized |
| 18 | CAALFM_C602480WA | 9.3877 | 1.58803E-89 | Uncharacterized |
| 19 | CAALFM_C302790WA | 9.1272 | 2.43382E-06 | Uncharacterized |
| 20 | CAALFM_C503430WA | 8.6904 | 0.000101716 | Uncharacterized |
| 21 | CAALFM_C402930WA | 8.6463 | 2.18312E-11 | Uncharacterized |
| 22 | CAALFM_C403340CA | 8.6294 | 0.000183681 | Uncharacterized |
| 23 | CAALFM_C701010WA | 8.4791 | 3.30179E-61 | Uncharacterized |
| 24 | CAALFM_C202580WA | 8.4163 | 0.001492629 | Uncharacterized |
| 25 | CAALFM_C405830WA | 8.3518 | 3.67121E-05 | Uncharacterized |
| 26 | CAALFM_C304210WA | 8.1065 | 3.03712E-07 | Uncharacterized |
| 27 | CAALFM_C406150CA | 7.9704 | 0.014291503 | Uncharacterized |
| 28 | CAALFM_C303370CA | 7.9704 | 1.1929E-135 | Uncharacterized |
| 29 | CAALFM_C204450WA | 7.8462 | 6.43154E-11 | Uncharacterized |
| 30 | CAALFM_C202180WA (ZRT3) | 7.7272 | 3.31888E-71 | Uncharacterized |
| 31 | CAALFM_C100270WA | 7.1398 | 4.2578E-73 | Uncharacterized |
| 32 | CAALFM_C401860CA | 7.0743 | 1.14226E-09 | Uncharacterized |
| 32 | CAALFM_C302360CA | 6.7969 | 5.99145E-37 | Uncharacterized |
| 34 | CAALFM_C600980CA | 6.6896 | 7.44423E-52 | Uncharacterized |
| 35 | CAALFM_CR06310WA | 6.6535 | 0.00233344 | Uncharacterized |
| 36 | CAALFM_C202750CA | 6.5793 | 0.000307212 | Uncharacterized |
| 37 | CAALFM_C200760CA | 6.4769 | 1.48506E-85 | Uncharacterized |
| 38 | CAALFM_C104690CA | 6.2900 | 1.03644E-05 | Uncharacterized |
| 39 | CAALFM_C304350CA | 6.2289 | 1.2123E-25 | Uncharacterized |
| 40 | CAALFM_C207620WA | 6.1661 | 6.18529E-06 | Uncharacterized |
| 41 | CAALFM_CR01910CA | 6.0167 | 0.000328533 | Uncharacterized |
| 42 | CAALFM_C702280WA | 6.0140 | 0.037653707 | Uncharacterized |
| 43 | CAALFM_C307070CA | 5.9226 | 2.70125E-54 | Uncharacterized |
| 44 | CAALFM_C405900CA | 5.9168 | 1.1249E-116 | Uncharacterized |
| 45 | CAALFM_CR07260CA | 5.8779 | 0.013967914 | Uncharacterized |
| 46 | CAALFM_C106620CA | 5.8775 | 0.016118237 | Uncharacterized |
| 47 | CAALFM_C109300CA | 5.8320 | 2.75478E-11 | Uncharacterized |
| 48 | CAALFM_CR09530CA | 5.7885 | 2.74225E-78 | Uncharacterized |
| 49 | CAALFM_C209850CA | 5.7740 | 3.08818E-07 | Uncharacterized |
| 50 | CAALFM_C601940WA | 5.5926 | 0.006011195 | Uncharacterized |
| 51 | CAALFM_C100310WA | 5.5729 | 6.50774E-57 | Uncharacterized |
| 52 | CAALFM_C601810WA | 5.5637 | 1.97617E-12 | Uncharacterized |
| 52 | CAALFM_C110060CA | 5.4134 | 4.01975E-08 | Uncharacterized |
| 54 | CAALFM_CR07300WA | 5.2272 | 7.10193E-05 | Uncharacterized |
| 55 | CAALFM_C402330CA | 5.2137 | 6.40082E-05 | Uncharacterized |
| 56 | CAALFM_C603240WA | 5.2105 | 9.75776E-08 | Uncharacterized |
| 57 | CAALFM_C305840WA (FMO1) | 5.1777 | 0.000145963 | Uncharacterized |
List of top 100 downregulated genes in EDTA treated C. albicans cell (CAET).
| Metabolic pathways | ||||
|---|---|---|---|---|
| No. of DEGs | Name (44 DEGs) | Fold Change | FDR p-value | Function |
| 1 | PHO100 | 100.3586958 | 1.7869E-210 | encode an enzyme phosphomonoesterase |
| 2 | PLB1 | 75.16562361 | 3.7897E-221 | Virulence; Phospholipase activity; Lipid metabolism |
| 3 | THI4 | 48.25803524 | 2.1564E-130 | Involved in thiamine biosynthesis |
| 4 | THI13 | 25.45919657 | 8.2924E-175 | Regulate IL-10 and IL-12 production |
| 5 | GIT1 | 22.7136005 | 3.2311E-112 | functions as proton symporter |
| 6 | HSP31 | 18.09639 | 2.39778E-23 | Involved in diauxic shift reprogramming |
| 7 | PHO84 | 17.26103731 | 4.3071E-163 | High-affinity inorganic phosphate/H+symporter |
| 8 | PUT1 | 14.48749801 | 7.5039E-99 | Involved in amino acid metabolism |
| 9 | INO1 | 10.74384531 | 1.5532E-128 | Encodes inositol-1-phosphate synthase |
| 10 | OPT1 | 10.46218378 | 1.83956E-90 | Involved in peptide transportation |
| 11 | PUT2 | 8.132948215 | 1.989E-99 | Amino acid metabolism regulator |
| 12 | LYS22 | 7.895086763 | 2.77628E-95 | Homocitrate synthase activity and lysine auxotrophy |
| 13 | ACO2 | 7.333020332 | 2.21018E-91 | Putative aconitate hydratase 2 |
| 14 | SNZ1 | 7.225024726 | 2.98178E-78 | Involved in pyridoxine (vitamin B6) synthesis |
| 15 | BTA1 | 7.201443893 | 2.2825E-11 | Encodes the betaine lipid synthase |
| 16 | LYS4 | 6.846020558 | 1.77724E-70 | Involve amino acid metabolism |
| 17 | CAN2 | 6.021560973 | 3.95846E-54 | Import arginine |
| 18 | FCY24 | 5.986276905 | 4.80688E-58 | Recruit vitamin B6 transport |
| 19 | THI6 | 5.891998871 | 3.5842E-104 | Thiamine biosynthesis |
| 20 | LEU1 | 5.775957021 | 5.4161E-99 | Catalyzes leucine biosynthesis pathway |
| 21 | GIS2 | 5.726657785 | 2.59522E-83 | Encodes the homologue of mammalian CNBP |
| 22 | LYS12 | 5.682928227 | 1.2519E-72 | Involved in dehydrogenation of homoisocitrate |
| 23 | CHA1 | 5.516083404 | 4.36052E-27 | Involved in nutrient acquisition/metabolism |
| 24 | FET99 | 5.171473494 | 4.85608E-18 | Involved in p-phenylenediamine oxidase; Iron transporter |
| 25 | GCV2 | 5.124244872 | 8.8508E-108 | Catabolises glycine; Induction in elevated CO2 |
| 26 | GIT3 | 4.705321982 | 1.43209E-52 | Transport glycerophosphodiester metabolites into cells |
| 27 | CYC1 | 4.533120319 | 7.41108E-12 | Encode cytochrome c |
| 28 | HXT5 | 4.370444946 | 3.87863E-30 | Uptake fructose, glucose and mannose |
| 29 | ILV3 | 4.326855778 | 2.72309E-49 | Amino acid metabolism regulator |
| 30 | OPT3 | 4.085475749 | 1.8761E-38 | Transport of sulfur-containing compounds |
| 31 | GAL1 | 4.015748921 | 6.75558E-71 | Executes metabolic functions in carbon source uptake |
| 32 | GCY1 | 3.986027968 | 4.12996E-66 | Confers hypersensitivity to toxic ergosterol analog |
| 33 | GAL10 | 3.818893745 | 1.55917E-65 | Induce biofilm; Utilize the source of galactose |
| 34 | GDH2 | 3.769049401 | 8.49692E-47 | Catalyzes deamination of glutamate to alpha-ketoglutarate |
| 35 | GCV1 | 3.737254451 | 2.09928E-39 | Catabolises glycine; Induction in elevated CO2 |
| 36 | DFR1 | 3.498716699 | 3.23053E-05 | Catalyses of 7,8-dihydrofolate to 5,4,7,8-tetrahydrofolate |
| 37 | GDH3 | 3.475852592 | 9.07648E-30 | Encode NADP+-dependent glutamate dehydrogenases |
| 38 | CTP1 | 3.456185992 | 2.97824E-16 | Involved in transportion of citrate |
| 39 | TPO4 | 3.351032169 | 4.97048E-29 | Bcr1- associated repression in RPMI a/alpha biofilms |
| 40 | OPI3 | 3.327563871 | 1.89288E-15 | Biosynthesis of phosphatidylcholine |
| 41 | EGD2 | 3.290389697 | 2.54237E-30 | GlcNAc-induced protein |
| 42 | LYS2 | 3.228201144 | 3.15144E-40 | lysine biosynthesis; biofilm induced |
| 43 | NUP | 3.137825901 | 7.10193E-05 | Involved in transportion of purine nucleosides and thymidine |
| 44 | ARO3 | 3.04435509 | 1.66463E-45 | aromatic amino acid synthesis; |
| Cell-wall associated | ||||
| Name (7 DEGs) | Fold Change | FDR p-value | Function | |
| 1 | HGT2 | 22.87250932 | 1.9154E-170 | Encode cell- wall associated proteins |
| 2 | PGA10 | 9.817718799 | 2.80343E-96 | Involved in Iron acquisition |
| 3 | TRY6 | 8.936709002 | 5.07311E-24 | Transcriptional regulator in biofilm |
| 4 | HSP30 | 6.401066175 | 8.86109E-17 | Stress-protective function on plasma membrane |
| 5 | ECM331 | 3.900822094 | 5.76083E-24 | Involved in cell wall biogenesis |
| 6 | ACS2 | 3.097921844 | 3.19601E-24 | antigenic during human and murine infection |
| 7 | STB3 | 3.038270294 | 5.50968E-41 | caspofungin induced |
| Ribosomal | ||||
| Name (13 DEGs) | Fold Change | FDR p-value | Function | |
| 1 | RPS28B | 5.090570767 | 1.40419E-10 | Autoregulates the decapping of its own mRNA machinery |
| 2 | RPS42 | 4.347730989 | 1.02472E-32 | Enhance tolerance to fluconazole |
| 3 | RDN18 | 4.047975479 | 1.9867E-13 | component of the small (40 S) ribosomal subunit; |
| 4 | RPS12 | 3.944266266 | 1.51774E-29 | pre-rRNA processing and polysome content |
| 5 | RPP1B | 3.606949912 | 2.33237E-25 | Involved in regulation of translation elongation |
| 6 | RPS21B | 3.588439098 | 1.10333E-29 | Regulated by Nrg1, Tup1 |
| 7 | RPS13 | 3.497233295 | 3.88662E-35 | _ |
| 8 | ASC1 | 3.385291153 | 3.46635E-25 | Required for virulence in mice |
| 9 | RPL18 | 3.234852275 | 1.52941E-28 | repressed upon phagocytosis by murine macrophage |
| 10 | RPL9B | 3.193159155 | 3.27531E-29 | repressed upon phagocytosis by murine macrophages |
| 11 | RPL5 | 3.102732401 | 2.25382E-30 | repressed upon phagocytosis by murine macrophages |
| 12 | RPP2B | 3.083956604 | 5.93521E-16 | possibly involved in regulation of translation elongation |
| 13 | RPS27 | 3.03019245 | 2.15647E-21 | repressed upon phagocytosis by murine macrophage |
| Others | ||||
| Name (11 DEGs) | Fold Change | FDR p-value | Function | |
| 1 | PGA45 | 5.590917256 | 6.40747E-43 | Putative GPI-anchored protein of unknown function |
| 2 | RBT7 | 4.226949465 | 1.5772E-09 | Encode secreted RNase T2 |
| 3 | PEX4 | 3.972508718 | 1.24715E-12 | Spider biofilm induction |
| 4 | RME1 | 3.697681605 | 7.1604E-68 | Development of fluconazole resistance |
| 5 | THI20 | 3.271494397 | 5.58989E-53 | Spider biofilm induced |
| 6 | ASM3 | 3.235903328 | 1.59858E-31 | Possible Kex2 substrate |
| 7 | BFR1 | 3.226066337 | 1.87417E-16 | Protein involved in the maintenance of normal ploidy |
| 8 | TUF1 | 3.224254282 | 7.29461E-36 | Encodes GTPase mitochondrial elongation factor Tu |
| 9 | GAL7 | 3.207467422 | 7.9819E-49 | downregulated by hypoxia |
| 10 | SSP96 | 3.203024284 | 1.12259E-06 | F-12/CO2 early biofilm induced |
| 11 | CYB5 | 3.179133812 | 1.7903E-06 | induced in high iron |
| Uncharacterized | ||||
| Name (25 DEGs) | Fold Change | FDR p-value | Function | |
| 1 | CAALFM_C400530CA | 25.251715 | 3.77714E-07 | Uncharacterized |
| 2 | CAALFM_CR08830WA | 13.160107 | 3.7788E-05 | Uncharacterized |
| 3 | CAALFM_CR06430WA | 6.746508 | 0.002469228 | Uncharacterized |
| 4 | CAALFM_CR09350CA | 6.189599 | 1.40099E-55 | Uncharacterized |
| 5 | CAALFM_C112940CA | 6.061342 | 0.037846274 | Uncharacterized |
| 6 | CAALFM_C404230WA | 5.563465 | 2.28386E-82 | Uncharacterized |
| 7 | CAALFM_C106870CA | 5.0202 | 0.017180492 | Uncharacterized |
| 8 | CAALFM_C306240CA (MRPL39) | 4.970367 | 0.002579226 | Uncharacterized |
| 9 | CAALFM_C500130CA (YML34) | 4.786879 | 5.51407E-07 | Uncharacterized |
| 10 | CAALFM_C204770WA | 4.55791 | 0.001066926 | Uncharacterized |
| 11 | CAALFM_C103620CA | 4.161708 | 4.50987E-09 | Uncharacterized |
| 12 | CAALFM_C501540WA (RPS11A) | 3.909799 | 3.0519E-17 | Uncharacterized |
| 13 | CAALFM_C503290CA (MRP10) | 3.907568 | 1.74642E-10 | Uncharacterized |
| 14 | CAALFM_C503480CA | 3.871642 | 1.41077E-32 | Uncharacterized |
| 15 | CAALFM_C208180CA | 3.782614 | 0.001381795 | Uncharacterized |
| 16 | CAALFM_C400990WA | 3.6619335 | 0.000850431 | Uncharacterized |
| 17 | CAALFM_C403500CA | 3.65632 | 0.012820924 | Uncharacterized |
| 18 | CAALFM_C204110WA | 3.438488 | 1.6049E-10 | Uncharacterized |
| 19 | CAALFM_C703560WA | 3.37104 | 3.35062E-18 | Uncharacterized |
| 20 | CAALFM_C108770WA | 3.243523 | 1.51784E-29 | Uncharacterized |
| 21 | CAALFM_CR03110WA (MSE1) | 3.172182 | 6.38954E-14 | Uncharacterized |
| 22 | CAALFM_CR03470WA | 3.162178 | 2.39698E-48 | Uncharacterized |
| 23 | CAALFM_CR08480CA (RPS29A) | 3.142855 | 2.18259E-06 | Uncharacterized |
| 24 | CAALFM_CR02380CA | 3.093999 | 0.000634778 | Uncharacterized |
| 25 | CAALFM_C400230WA | 3.0457946 | 8.36614E-08 | Uncharacterized |
Additional files
-
Supplementary file 1
List of top 100 upregulated genes in EDTA treated C. albicans cell (CAET) with detailed information.
- https://cdn.elifesciences.org/articles/93760/elife-93760-supp1-v1.xlsx
-
Supplementary file 2
GO annotation analyses of upregulated genes and downregulated genes.
(A) GO annotation analysis for 411 upregulated genes and (B) 388 downregulated genes.
- https://cdn.elifesciences.org/articles/93760/elife-93760-supp2-v1.xlsx
-
Supplementary file 3
STRING cluster analyses of DEGs.
(A) STRING cluster analysis for 411 upregulated genes, (B) 388 downregulated genes, and (C) 74 downregulated genes out of 388 DEGs specific to 40 S and 60 S ribosomal subunits.
- https://cdn.elifesciences.org/articles/93760/elife-93760-supp3-v1.xlsx
-
Supplementary file 4
STRING cluster analysis for 33 DEGs specific to pathogenesis.
- https://cdn.elifesciences.org/articles/93760/elife-93760-supp4-v1.xlsx
-
Supplementary file 5
List of top 100 downregulated genes in EDTA treated C. albicans cell (CAET) with detailed information.
- https://cdn.elifesciences.org/articles/93760/elife-93760-supp5-v1.xlsx
-
Supplementary file 6
Oligonucleotides and PCR conditions.
- https://cdn.elifesciences.org/articles/93760/elife-93760-supp6-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93760/elife-93760-mdarchecklist1-v1.docx