Cell-autonomous timing drives the vertebrate segmentation clock’s wave pattern
Figures
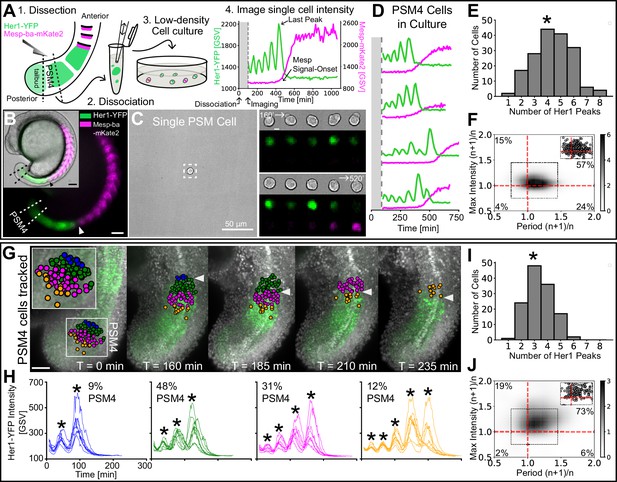
Cell-autonomous oscillatory dynamics reproduce those underlying wave pattern in the embryo.
(A) Experimental design: (1) Dissection of posterior-PSM quarter (PSM4) (dashed lines) from a Tg(her1-YFP;mesp-ba-mKate2) 15 somite-stage embryo, (2) Dissociation into single cells, (3) Culture at low-density, and (4) Her1-YFP and Mesp-ba-mKate2 imaging over time. (B) Her1-YFP and Mesp-ba-mKate2 in a 15 somite-stage Tg(her1-YFP;mesp-ba-mKate2) embryo (bright-field inset). Arrowheads mark the recently formed somite (S1). Dissection lines surround PSM4. Scale bar 100 µm. (C) One cell per field of view imaged. Boxed region over time (scale bar 5 µm). Intensity trace shown in A. (D) Representative intensity traces of PSM4 cells in culture. (E) Number of Her1-YFP peaks (*mean ± SD, 4.4±1.6 peaks) per PSM4 cell in culture (N=11 embryos, n=174 cells). (F) Density plot of ratios of successive Her1-YFP periods and peak intensity (cyclen+1/cyclen) for PSM4 cells in culture (n=421 successive cycle ratios, circles in inset). Percent of total ratios per quadrant is indicated. (G,H) PSM4 cells in a Tg(her1-YFP;h2b- mCherry) embryo tracked until somite formation (arrowhead) (N=2 embryos, n=128 PSM4 cells). Scale bar 100 µm. Cells contributing to the same somite are identically coloured in the embryo and representative Her1-YFP intensity traces (* peaks). Percentage of tracked PSM4 cells contributing to a given somite is shown. (I) Number of Her1-YFP peaks (*mean ± SD, 3.4±1.0 peaks) per PSM4 cell in the embryo. (J) As described in F with PSM4 cells in the embryo (n=179 total successive cycles).
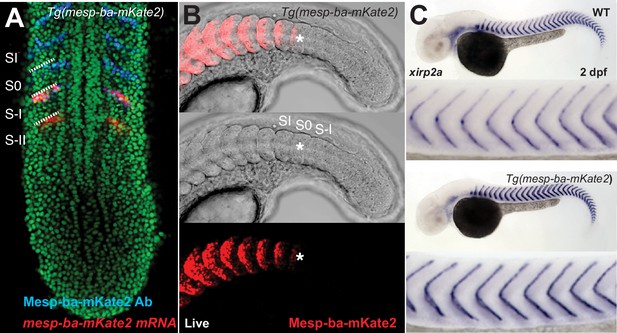
Mesp-ba-mKate2 transgenic expression and detection.
(A) Detection of mesp-ba-mKate2 mRNA (red, in situ hybridization) and Mesp-ba-mKate2 (blue, anti-mKate2 antibody) in a fixed Tg(mesp-ba-mKate2) 10 somite-staged embryo (nuclei labelled green). mesp-ba-mKate2 was detected only in the rostral half of pre-segments (S–II and S–I) in the anterior PSM as expected from endogenous mesp-ba expression (Cutty et al., 2012). Mesp-ba-mKate2 was first detected by antibodies to mKate2 in the rostral half of S-I, where it persisted in the newly forming somite (S0) and formed somites (SI). (B) Mesp-ba-mKate2 signal was first detected in live Tg(mesp-ba-mKate2) embryos within the rostral half of S0 (*), after which it remained in the rostral half of the formed somites (21 somite-stage embryo). (C) Boundary formation was normal as detected by in situ hybridization for the boundary marker xirp2a in Tg(mesp-ba-mKate2) embryos compared to wildtype (WT) at 2 days post-fertilization (dpf).
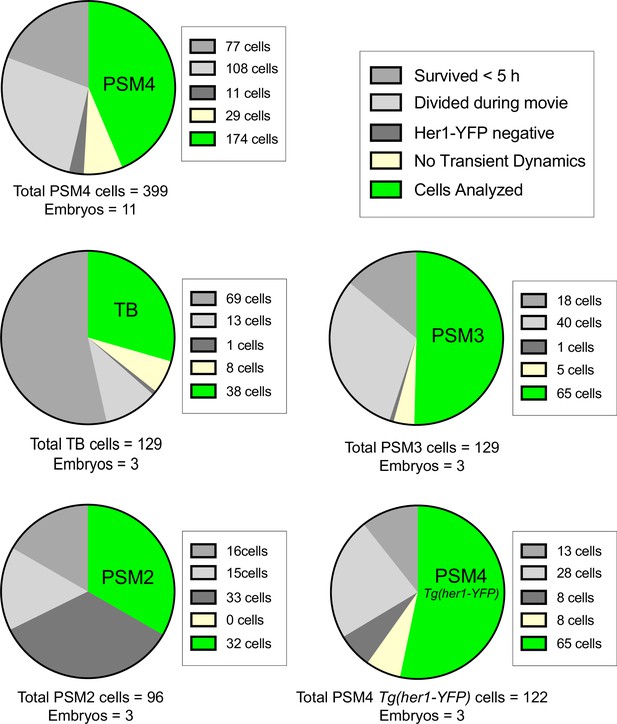
Cell culture analysis criteria.
Dissociated cells originating from different anteroposterior positions (PSM2, PSM3, PSM4 and TB) in Tg(her1-YFP;mesp-ba-mKate2) embryos, or PSM4 from Tg(her1-YFP) control embryos. Analysis criteria was as follows: (1) Single cells alone in the field of view were selected at the start of imaging; (2) Cells dying before 5 hr post-dissociation were excluded from analysis; (3) Cells that survived >5 hr but divided during the movie were excluded; (4) From the remaining cells, those not expressing Her1-YFP or failing to oscillate and arrest before cell death were excluded (No Transient Dynamics); (5) Transient Dynamics were then analyzed in the remaining cells. Note that PSM2 cells listed as Her1-YFP negative may have already arrested before imaging started.

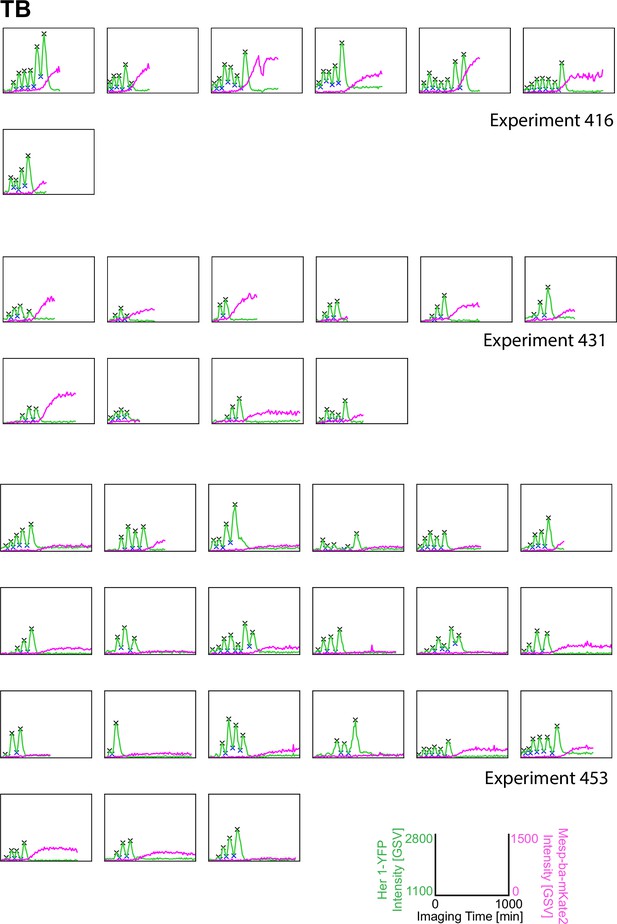
Her1-YFP and Mesp-ba-mKate2 intensity traces from PSM4 cells in culture.
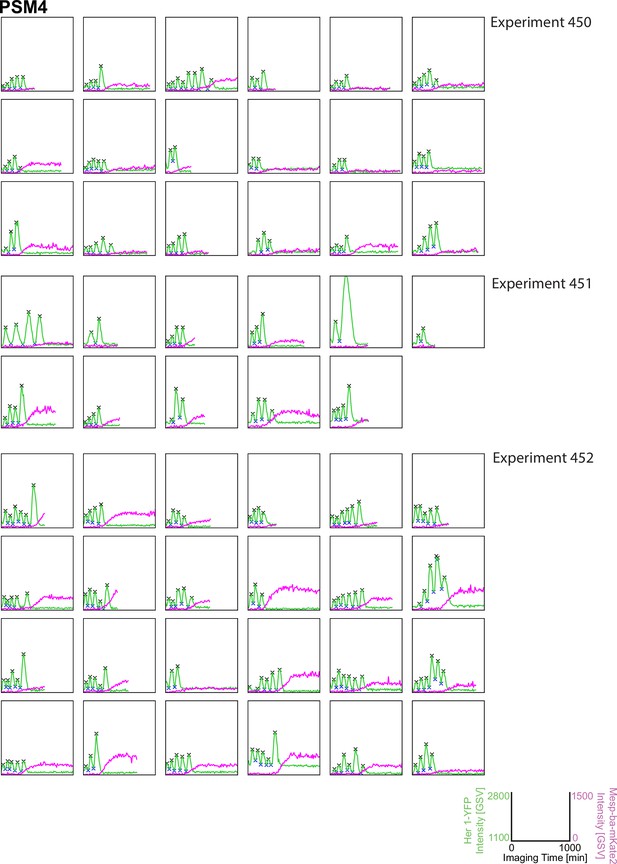
Her1-YFP and Mesp-ba-mKate2 intensity traces from single cells with peaks and troughs marked (X) and grouped by experiment number. N=11 experiments (n=174 cells) combined from Figure 1—figure supplement 3–6.

Her1-YFP and Mesp-ba-mKate2 intensity traces from PSM4 cells in culture.
Her1-YFP and Mesp-ba-mKate2 intensity traces from single cells with peaks and troughs marked (X) and grouped by experiment number. N=11 experiments (n=174 cells) combined from Figure 1—figure supplements 3–6.
Her1-YFP and Mesp-ba-mKate2 intensity traces from PSM4 cells in culture.
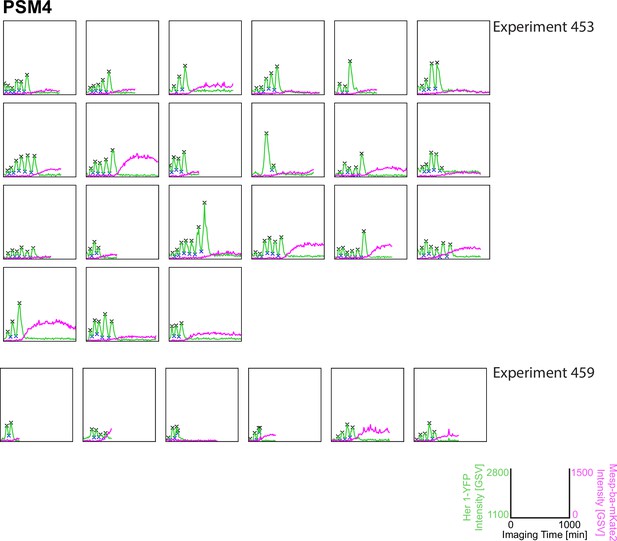
Her1-YFP and Mesp-ba-mKate2 intensity traces from single cells with peaks and troughs marked (X) and grouped by experiment number. N=11 experiments (n=174 cells) combined from Figure 1—figure supplements 3–6.
Her1-YFP and Mesp-ba-mKate2 intensity traces from PSM4 cells in culture.
Her1-YFP and Mesp-ba-mKate2 intensity traces from single cells with peaks and troughs marked (X) and grouped by experiment number. N=11 experiments (n=174 cells) combined from Figure 1—figure supplements 3–6.
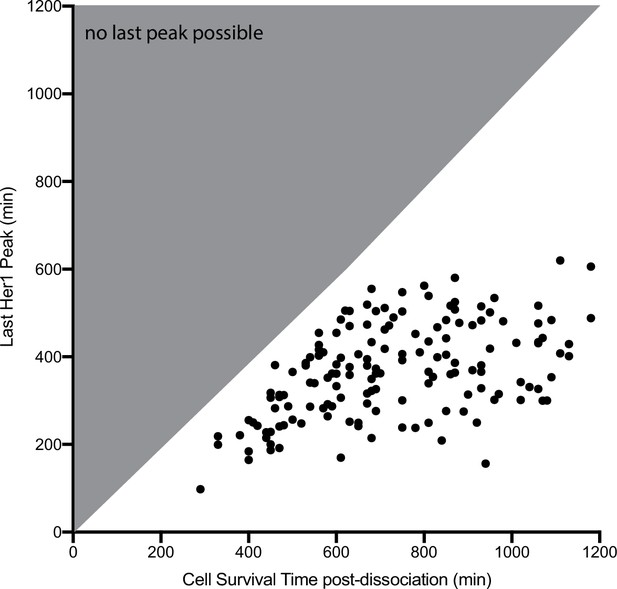
Time of cell death and oscillatory arrest in single cells.
Time of PSM4 cell death in culture and Her1-YFP last peak time in single cells. The grey triangle marks the region in which a last peak is not possible because the cells are already dead. Cells dying before 5 hr post-dissociation were not analyzed. Cells alive at the end of imaging (>1200 min, n=20 cells) are not shown. Mean cell survival time was 787 min, SD ±262 min.
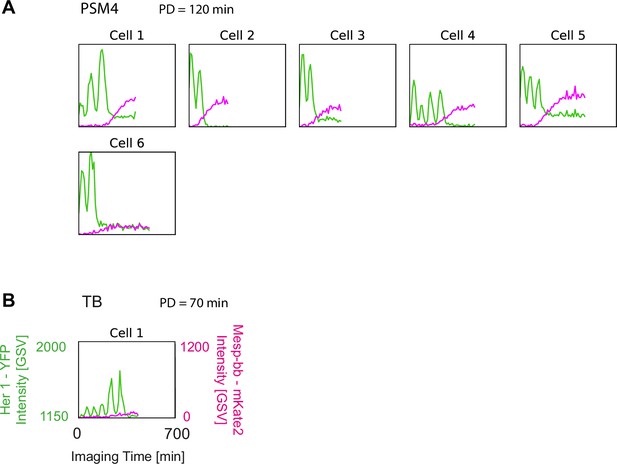
Cells isolated one per well reproduce autonomous behaviour.
(A, B) Mesp-ba-mKate2 and Her1-YFP intensity traces in PSM4 (A) and TB (B) cells isolated one per well in a 24-well plate (N=3 embryos, n=6 PSM4 cells, n=1 TB cell). Time of imaging start post-dissociation (PD) given. Oscillations, intensity increase and arrest in concert with Mesp-ba-mKate2 signal-onset in the isolated cells observed.
Cell-autonomous Her1-YFP dynamics do not depend on Tg(mesp-ba-mKate2).
(A) Her1-YFP intensity traces in cultured PSM4 cells from embryos carrying only Tg(her1-YFP) (N=3 embryos; n=64 cells) that were cultured in parallel to PSM4 from Tg(her1-YFP;me- sp-ba-mKate2) embryos (experiments 449 and 450 in Figure 1—figure supplement 3). (B) Number of Her1-YFP peaks. (C) Her1-YFP intensity traces aligned by the first peak show on average lengthening period, increasing peak intensity (I+) and constant trough intensity (I-) over successive cycles.
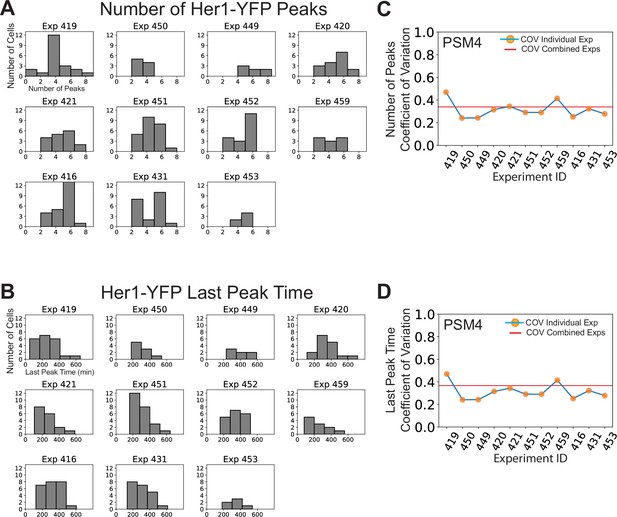
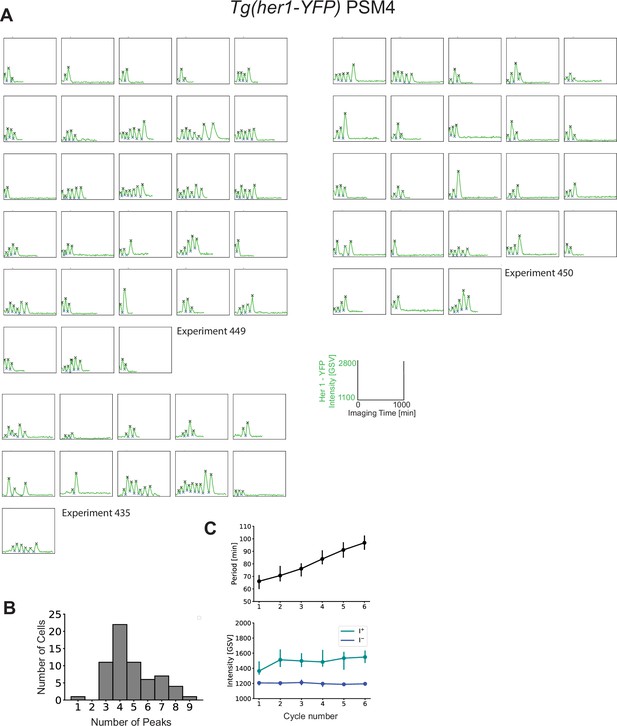
Variability in cell-autonomous timing of oscillatory arrest is not due solely to inter-experimental differences.
PSM4 cell data in Figure 1 was pooled from 11 different experiments, each with one embryo. (A) The distribution of the numbers of Her1-YFP peaks generated by cells is shown for each experiment. (B) The distribution of the time of the Her1-YFP last peak in cells for each experiment. (C, D) Coefficient of Variation (COV) for each individual experiment (orange points, blue line) and mean COV for the combined set of experiments (red line) for both numbers of peaks (C) and last peak time (D).
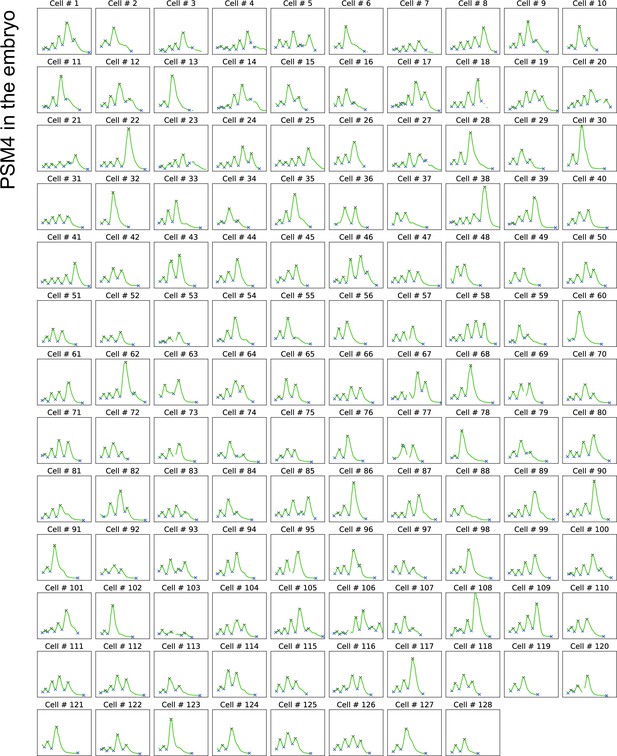
Her1-YFP intensity traces from PSM4 cells tracked in the embryo until somite formation.
Cells were selected within PSM4 of 15 somite-staged Tg(her1-YFP;h2b-mCherry) embryos, then tracked until somite formation as shown in Figure 1G and H (N=2 embryos, n=128 cells). Her1-YFP intensity traces with peaks and troughs (X).
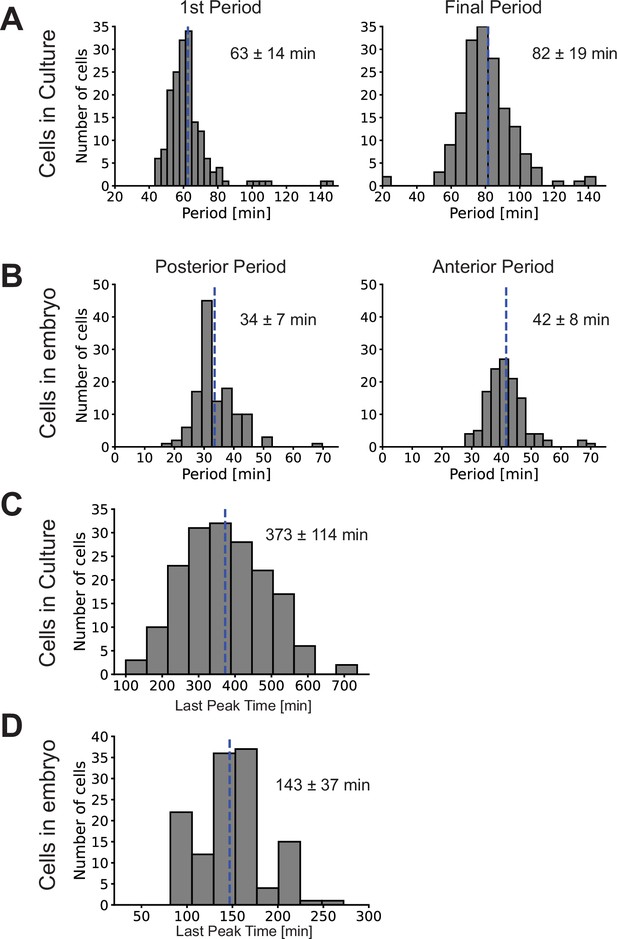
Lengthening of period and cell-autonomous transient dynamics in culture.
(A) First and final periods (mean ± SD in blue) of Her1-YFP oscillations in PSM4 cells in culture. (B) Posterior- and Anterior-most periods of Her1-YFP in PSM4 cells tracked in the embryo. (C–D) Time of the Her1-YFP last peak in PSM4 cells in culture (C) and in the embryo (D). The mean of the last peak time in culture is more than double that in the embryo, indicating that the transient dynamics are skewed to comparatively longer times in culture.
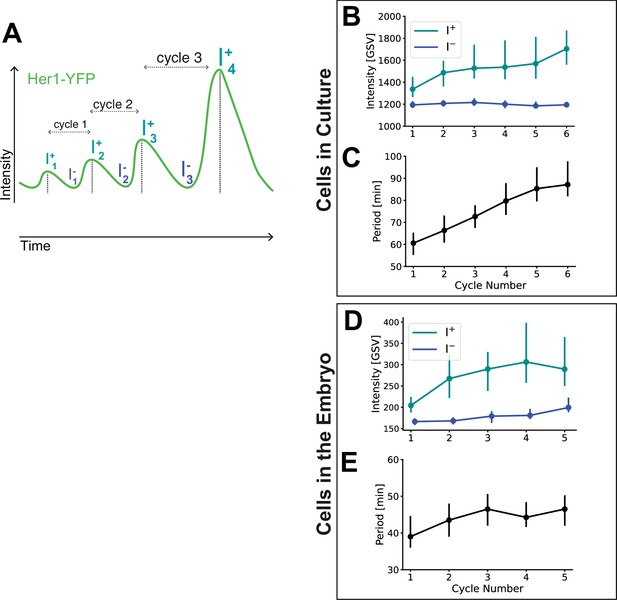
PSM4 periods and peak intensity by cycle.
Diagram of Cycle, Peak Intensities (I+) and Troughs (I-). (B,C) All PSM4 Her1-YFP intensity traces from cells in culture were aligned by the first peak (marked I+ in A). Peak and trough intensity (B) and the period (C) for each cycle are given as median (circle) with 25th and 75th interquartiles (vertical bar). (D, E) All Her1-YFP intensity traces from cells in the embryo were aligned by the first peak. Peak and trough intensity (D) and the period (E) for each cycle shown as in B,C.
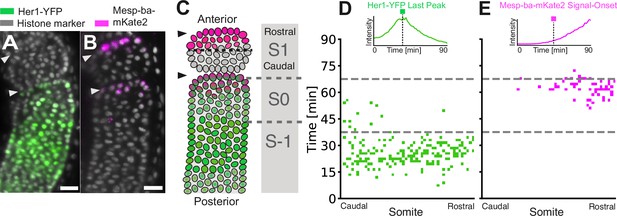
Clock arrest and Mesp-ba-mKate2 signal-onset within the forming somite.
(A, B) Her1-YFP in a Tg(her1-YFP;h2b-mCherry) embryo (A) and Mesp-ba-mKate2 in a Tg(mesp-ba-mKate2a;h2a-GFP) embryo (B). Representative lateral PSM light-sheet slice. Scale bar is 25 µm. Arrowheads at somite boundaries. (C) Cartoon of the formed somite (S1), the forming somite (SO) and the prospective somite (S-1). (D, E) S1 cells backtracked in a Tg(her1-YFP;h2b-mCherry) embryo (n=233 cells) (D) and a Tg(mesp-ba-mKate2a;h2a-gfp) embryo (n=190 cells) (E). Kymograph of Her1-YFP last peak (D) and Mesp-ba-mKate2 signal-onset time (E) in cells relative to the rostral-caudal somite axis (inset with example traces). Dashed grey line at transitions S-1 to S0 and S0 to S-1.
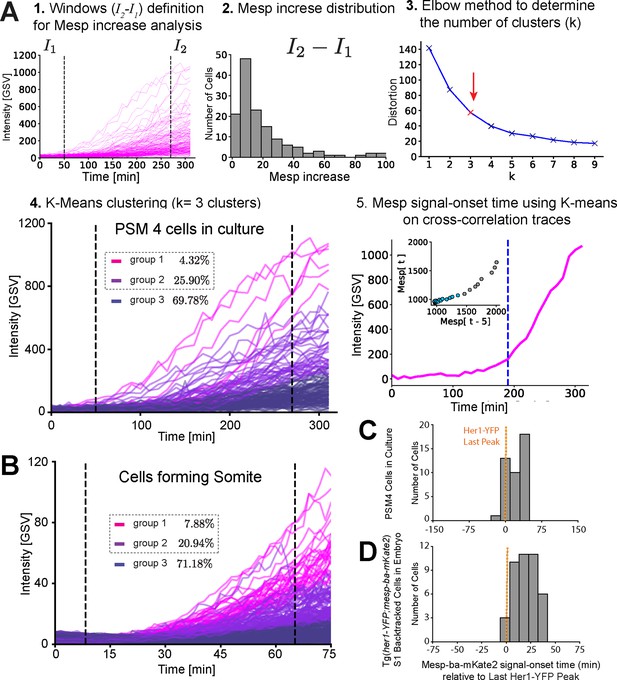
Mesp-ba-mKate2 Signal-Onset defined in individual intensity traces and timing relative to the last Her1-YFP peak.
(A) Mesp-ba-mKate2 signal-onset time defined in intensity traces from PSM4 cells in culture (n=139 PSM4 cells with sufficient trace length). Steps to define signal-onset: (1) An arbitrary window was defined across the intensity traces, then average intensity within each window was calculated. (2) Mesp-ba-mKate2 increase was obtained by subtracting the first intensity window (I1) from the second (I2), shown as a distribution. (3) Because intensity trace profiles varied between cells, we used the elbow method to identify the number of clusters. (4) Mesp-ba-mKate2 increase for each cell was then used to perform K-Means clustering using k=3. Groups 1 and 2 are cells showing an obvious Mesp-ba-mKate2 signal-onset. (5) Signal-onset time was then determined in these groups using K-Means on a lag-plot of the intensity. (B) Mesp-ba-mKate2 signal-onset in cells forming a somite in the embryo (N=2 somites, n=348 pooled cells with sufficient temporal length). (C,D) Time of Mesp-ba-mKate2 signal-onset relative to Her1-YFP Last Peak in PSM4 cells in culture (n=42 cells) (C) and in S1 cells back-tracked in an embryo carrying both mesp-ba-mKate2 and her1-YFP transgenes (n=40 cells)(D).
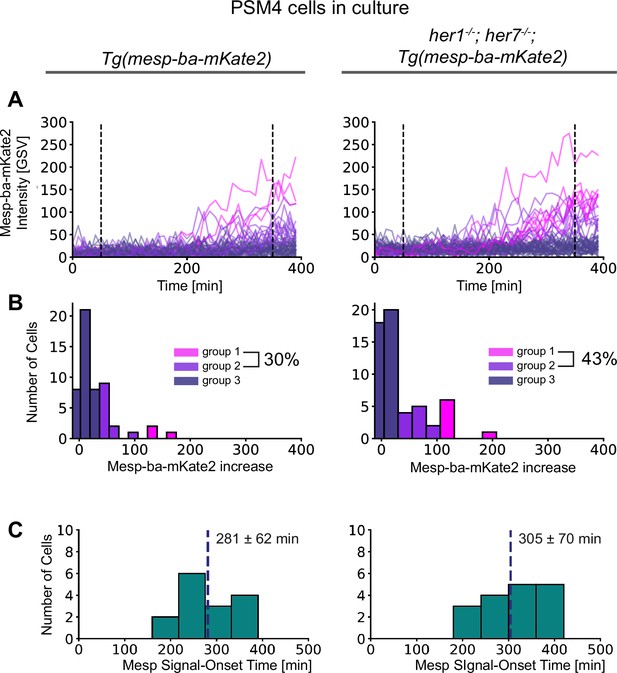
Mesp-ba-mKate2 signal-onset in her1-/-;her7-/- cells with disabled segmentation clock.
PSM4 cells were cultured in parallel from her1-/-;her7-/- mutant embryos carrying Tg(mesp-ba-mKate2) (N=2 embryos, n=75 cells), and control Tg(mesp-ba-mKate2) embryos (N=2 embryos, n=71 cells). (A) Mesp-ba-mKate2 intensity traces of sufficient length were aligned by time (n=52 control cells, n=56 her1-/-;her7-/- cells). (B) Mesp-ba-mKate2 intensity increase clustered into groups (as described in Figure 2—figure supplement 1). Percentage of cells with an obvious Mesp-ba-mKate2 signal-onset detected, groups 1 and 2. (C) Mesp-ba-mKate2 signal-onset times (mean ± SD) determined for group1 and 2 cells.
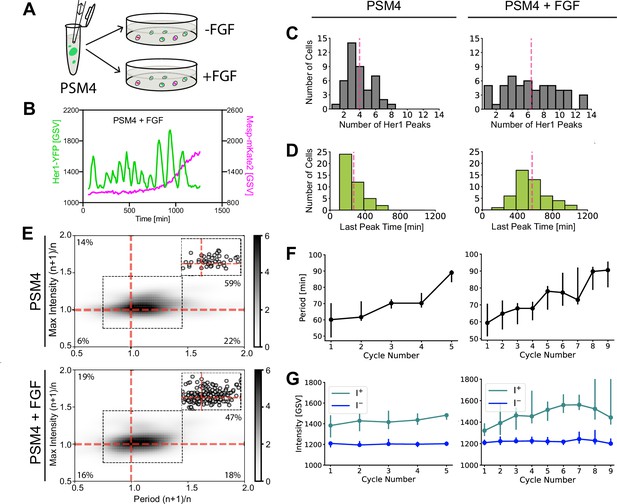
FGF extends cell-autonomous program duration of PSM4 cells in culture.
(A) A pool of dissociated PSM4 cells was split into a control well and one containing FGF-8b, then cultured (N=4 experiments, n=44 PSM4 control cells and n=54 PSM4 cells with added FGF). (B) Representative Her1-YFP and Mesp-mKate2 trace from PSM4 cells cultured with added FGF. (C) Number of Her1-YFP Peaks produced per cell, with pink line at the mean (mean ± SD = 4.0±1.8 peaks in control cells, 6.4±3.5 with FGF). (D) Time of Her1-YFP last peak (min post-dissociation), with pink line at the mean (mean ± SD = 269±116 min post-dissociation in control cells, 568±185 min post-dissociation in cells with FGF). (E) Ratio of successive cycle Her1-YFP periods (Period (n+1)/n) and peak intensity (Max Intensity (n+1)/n). Upper right quadrant indicates successive intensity rise and oscillation slowing. n=51 successive cycle ratios in PSM4 control cells and n=196 in PSM4 cells cultured with FGF, shown as circles in inset and percent of successive cycles is indicated in each quadrant. (F,G) All Her1-YFP intensity traces were aligned by the first peak. The period (F) and intensity of peaks (I+) and troughs (I-) (G) is given for each cycle as a median (circle) with 25th and 75th interquartiles (bar).

Her1-YFP and Mesp-ba-mKate2 cell intensity traces in PSM4 cells cultured with FGF.
For each experiment (N=4), dissociated cells from one or two embryos were split between two wells, then cultured +/-FGF-8b. Single oscillating cells that remained the only cell in the field of view, survived >5 hr post-dissociation, did not divide, and showed transient dynamics were analyzed. Her1-YFP and Mesp-ba-mKate2 intensity traces from single cells shown (n=44 control PSM4 cells, n=54 PSM4 cells +FGF).
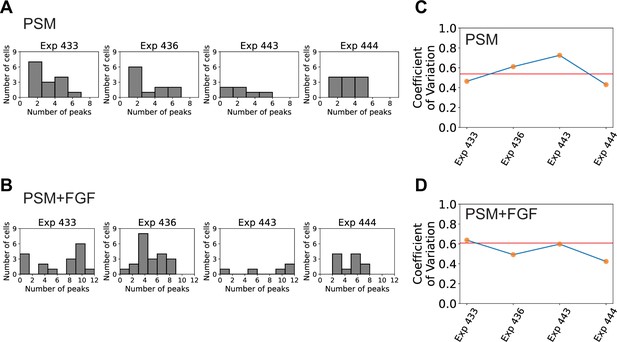
Variability of cell-autonomous Her1-YFP peaks not reduced by the addition of FGF to the culture.
(A, B) PSM4 and PSM4 +FGF cell data was pooled from four different experiments. Distribution of the numbers of Her1-YFP peaks generated by individual cells shown for each experiment. (C, D) Coefficient of Variation (COV) for each individual experiment (orange points, blue line) and mean COV for the combined set of experiments (red line).
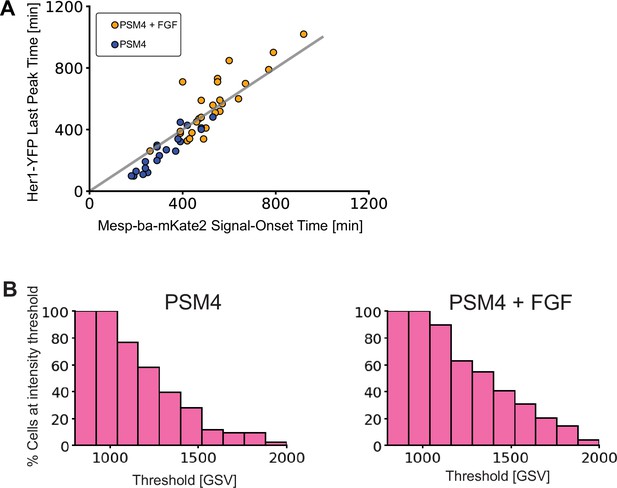
Mesp-ba-mKate2 signal onset and intensity distributions in response to FGF.
(A) Her1-YFP Last peak time correlated with Mesp-ba-mKate2 signal-onset time in PSM4 (blue) and PSM4 +FGF (orange). (B) Mesp-ba-mKate intensity shown by percent of cells reaching a range of intensity thresholds.
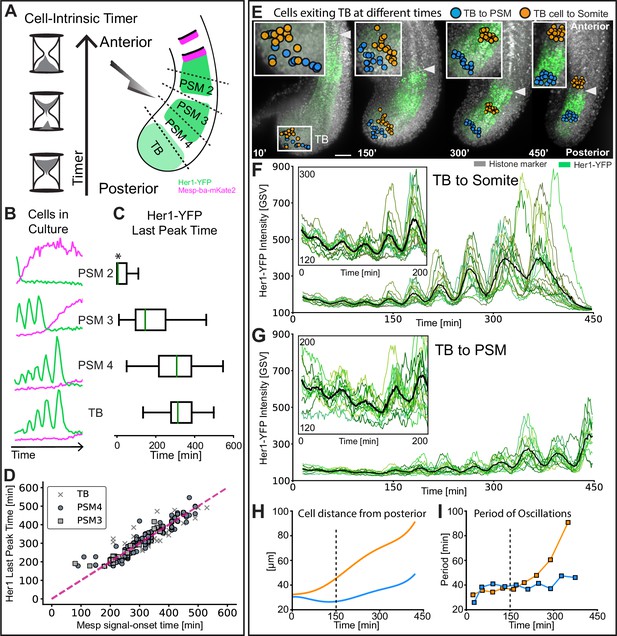
Intrinsic timer initiates upon TB exit and duration shortens in cell flow.
(A) Experimental design: Tg(her1-YFP;mesp-ba-mKate2) PSM was dissected into different anteroposterior quarters, dissociated, and then cultured. PSM4 was dissected and cultured in parallel from each embryo to serve as an internal reference. (B) Representative intensity traces for the different anteroposterior quarters. (C) Her1-YFP last peak times as median (green line) with interquartile box and whiskers. Time given as post-dissociation. Data pooled by cell type (N=3 embryos, n=32 PSM2 and 32 PSM4 cells; N=3 embryos, n=65 PSM3 and 41 PSM4 cells; N=3 embryos, n=38 TB and 59 PSM4 cells). PSM2 last peaks that occurred prior to the start of imaging were set to acquisition start time (*). (D) Correlation of Her1-YFP Last Peak and Mesp-ba-mKate2 signal-onset time. (E) Cells backtracked from posterior PSM (blue, n=17) and S1 (orange, n=15) to the TB at 15 somite stage in a representative embryo (7.5 hr imaging, Tg(her1-YFP;h2b-mCherry), N=2). Arrowhead at recently formed somite boundary. Scale bar 100 µm. (F, G) Her1-YFP intensity traces for individual cells (green) and mean intensity (black). Inset zoom of oscillations in the TB. (H) Cell distance from posterior tail tip. (I) Mean Her1-YFP period.
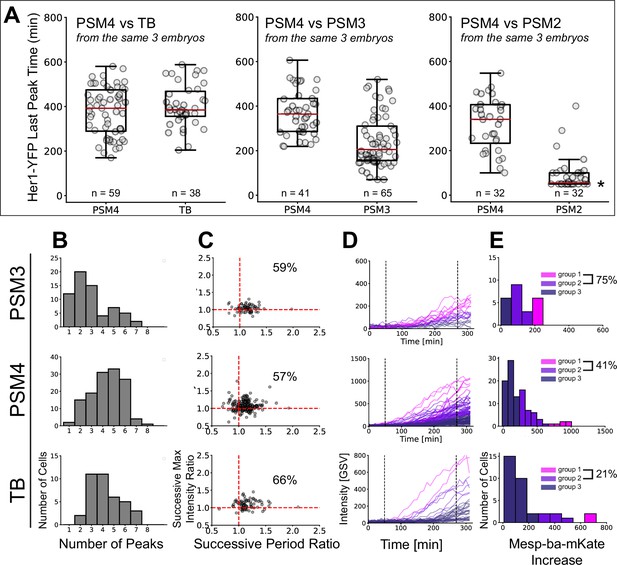
Cell-autonomous Her1-YFP and Mesp-ba-mKate2 dynamics in cells dissected from different anteroposterior positions.
(A) Her1-YFP last peak times in cultured cells dissected from different anteroposterior positions (PSM2, PSM3, PSM4, and TB) in Tg(her1-YFP;mesp-ba-mKate2) embryos. PSM4 was dissected and cultured in parallel to other PSM quarters from the same embryo as an internal reference (N=3 for each comparison). Pooled PSM4 data is shown in Figure 4. Median last peak time (red line) with interquartile box. Many PSM2 cells were in the fall of the last peak when imaging began, thus the last peak time for such cells was set to the time imaging started post-dissociation (*). (B) Number of Her1-YFP peaks (mean ± SD, PSM3 2.95±1.65; PSM4 4.36±1.44; TB 4.26±1.35 peaks) (C) Successive period and intensity ratios. Percentage of successive cycles slowing and increasing intensity. (D, E) Mesp-ba-mKate2 intensity traces and distributions clustered into groups.
Her1-YFP and Mesp-ba-mKate2 intensity traces from PSM2 cells in culture.
Intensity traces from PSM2 cells in culture (N=3 experiments, n=32 cells) and experiment number.

Her1-YFP and Mesp-ba-mKate2 intensity traces from PSM3 cells in culture.
Intensity traces from PSM3 cells in culture (N=3 experiments, n=65 cells) with the peaks and troughs marked (X) and experiment number.