Allosteric modulation of the CXCR4:CXCL12 axis by targeting receptor nanoclustering via the TMV-TMVI domain
Figures
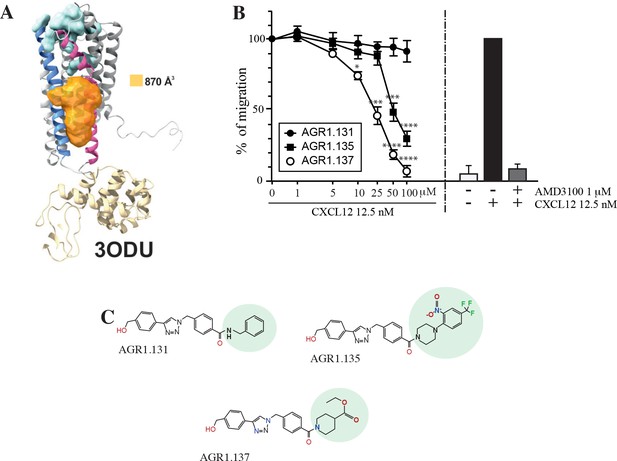
Screening for small compound antagonists acting at the oligomerization site of CXCR4.
(A) Cartoon and surface representation of CXCR4 and the cleft identified between TMV and TMVI. The protein structure is shown in gray, with TMV and TMVI colored in blue and pink, respectively. In green, residues involved in CXCL12 binding. The cavity identified by SurfNet is shown in orange. Volume for the cavity is measured and shown in Å3. T4 lysozyme inserted between TMV and TMVI in the crystallized version of CXCR4 (PDB: 3ODU) is also shown in yellow. (B) Dose-response curve of the selected antagonists in Jurkat cell migration experiments in response to 12.5 nM CXCL12. Data are shown as percentage of migrating cells (mean ± SD; n=5; *≤0.05, *** p≤0.001, **** p≤0.0001). (C) Chemical structure of the selected compounds (AGR1.131, AGR1.135 and AGR1.137). The differences between the lateral chains of the three compounds are shaded in green.
-
Figure 1—source data 1
Compounds with minimal interaction energy in the area of interest.
Structure of the compounds employed in the study. The compounds studied in detail (AGR1.131, AGR1.135 and AGR1.137) are highlighted in green boxes. The percentage of inhibition of cell migration in response to CXCL12 is also shown.
- https://cdn.elifesciences.org/articles/93968/elife-93968-fig1-data1-v1.docx
-
Figure 1—source data 2
Compound synthesis and characterization.
- https://cdn.elifesciences.org/articles/93968/elife-93968-fig1-data2-v1.docx
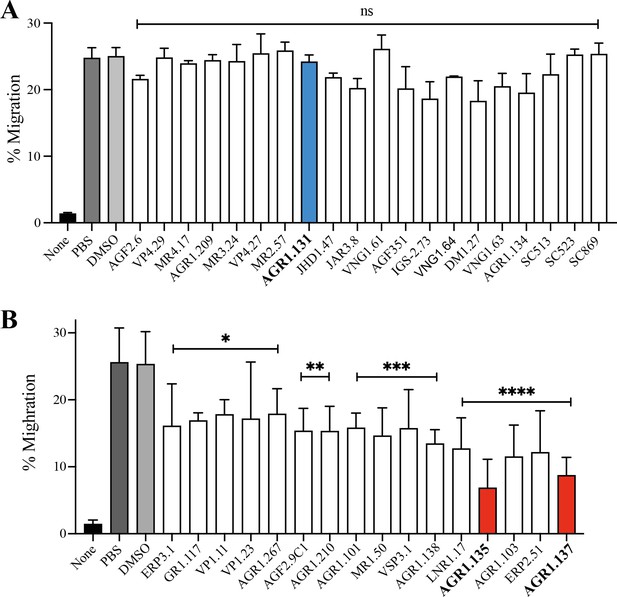
Small compound-mediated inhibition of CXCL12-induced cell migration.
(A, B) Migration of Jurkat cells, untreated or treated with the selected small compounds (50 μM) or DMSO (vehicle) in the previous in silico screening, in response to 12.5 nM CXCL12. Data shown as percentage of migrating cells (mean ± SD; n = 5; *p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤0.0001, ns : not significant).
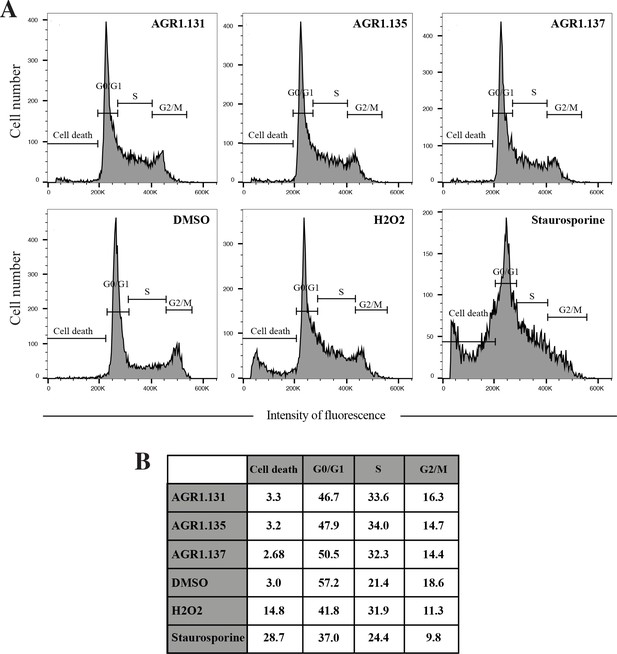
Effect of small compounds on Jurkat cell cycle.
(A) Cell cycle analysis using propidium iodide incorporation and flow cytometry of Jurkat cells treated with AGR1.131, AGR1.135 and AGR1.137 (2 hours, 37ºC) or with DMSO (vehicle), H2O2 (10%) and staurosporine (10 mM) as controls. The percentage of cells in each cycle stage is shown. (B) Summary of the percentage of treated cells in each cell cycle phase.
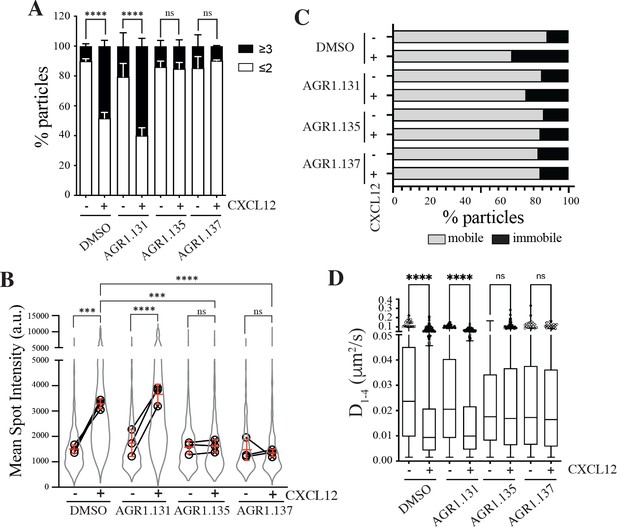
AGR1.135 and AGR1.137 alter CXCL12-mediated CXCR4 dynamics and nanoclustering.
Single-particle tracking analysis of JK−/− cells transiently transfected with CXCR4 (JK−/−-X4) treated with DMSO (control), AGR1.131, AGR1.135 or AGR1.137 on fibronectin (FN)- or FN +CXCL12-coated coverslips (DMSO: 581 particles in 59 cells on FN; 1365 in 63 cells on FN +CXCL12; AGR1.131: 1019 particles in 71 cells on FN; 1291 in 69 cells on FN +CXCL12; AGR1.135: 862 particles in 70 cells on FN; 1003 in 77 cells on FN +CXCL12; AGR1.137: 477 particles in 66 cells on FN; 566 in 64 cells on FN +CXCL12) n=3. (A) Frequency of CXCR4-AcGFP particles containing monomers plus dimers (≤2) or nanoclusters (≥3),± SEM, calculated from mean spot intensity values of each particle as compared with the value of monomeric CD86-AcGFP (n=3; n.s., not significant; ****p≤0.0001). (B) Intensity distribution (arbitrary units, a.u.) from individual CXCR4-AcGFP trajectories on unstimulated and CXCL12-stimulated JK−/−-X4 cells pretreated or not with the indicated compounds or vehicle (DMSO). Graph shows the distribution of all trajectories (gray outline), with the mean value of each experiment (black circles) and the mean of all experiments ± SD (red lines) (n=3; n.s., not significant; ***p≤0.001, ****p≤0.0001). (C) Percentage of mobile and immobile CXCR4-AcGFP particles at the membrane of cells treated as indicated. (D) Diffusion coefficients (D1–4) of mobile single particle trajectories at the membrane of cells treated as indicated, represented by box-and-whiskers plots (Tukey method) with median (black line) indicated. (n.s., not significant, ****p≤0.0001).
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with DMSO and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) at steady state (FN). The video was acquired and displayed at 10.2 frames/s.
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with DMSO and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) in response to CXCL12. The video was acquired and displayed at 10.2 frames/s.
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with AGR1.131 and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) at steady state (FN). The video was acquired and displayed at 10.2 frames/s.
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with AGR1.131 and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) in response to CXCL12. The video was acquired and displayed at 10.2 frames/s.
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with AGR1.135 and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) at steady state (FN). The video was acquired and displayed at 10.2 frames/s.
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with AGR1.135 and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) in response to CXCL12. The video was acquired and displayed at 10.2 frames/s.
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with AGR1.137 and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) at steady state (FN). The video was acquired and displayed at 10.2 frames/s.
Related to Figure 2.
Representative video of CXCR4-AcGFP on live JKX4-/- cells treated with AGR1.137 and captured by SPT-TIRF, showing the diffusion of CXCR4 particles (monomers, dimers, and nanoclusters) in response to CXCL12. The video was acquired and displayed at 10.2 frames/s.
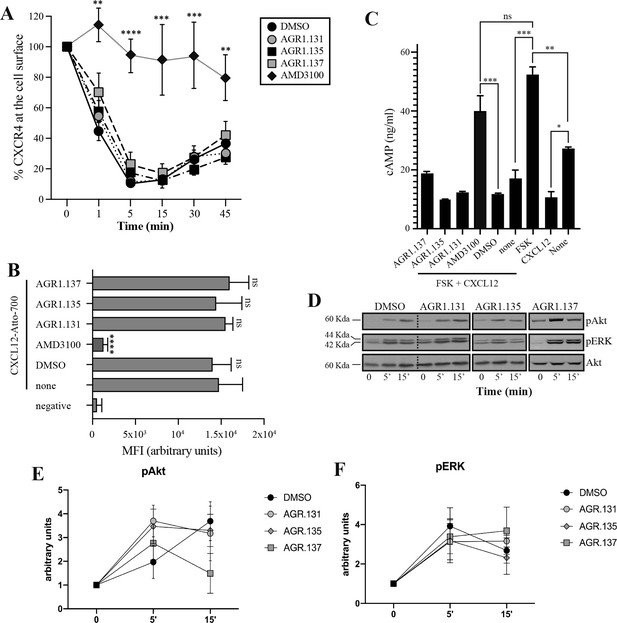
AGR1.135 and AGR1.137 treatments do not block CXCL12-mediated signaling pathways.
(A) Cell surface expression of CXCR4 in Jurkat cells after stimulation with CXCL12 (12.5 nM) at different time points and analyzed by flow cytometry using an anti-CXCR4 antibody in nonpermeabilized cells. Results show mean ± SEM of the percentage of CXCR4 expression at the cell surface (n=4; n.s. not significant; **p≤0.01, ***p≤0.001, **** p≤0.0001). (B) CXCL12-Atto-700 binding on untreated or Jurkat cells pre-treated with DMSO (vehicle), AGR1.131, AGR1.135, AGR1.137 or with AMD3100 as a control, followed by flow cytometry analysis. Results are expressed as mean fluorescence intensity (MFI) values (arbitrary units). Negative corresponds to basal cell fluorescence in the absence of CXCL12-Atto-700 (mean ± SD, n=3 n.s. not significant; ****p≤0.0001). (C) Cells untreated or pretreated with the small compounds (50 μM, 30 min 37 °C) or AMD3100 (1 μM, 30 min 37 °C) were stimulated with CXCL12 (50 nM, 5 min, 37 °C) followed by forskolin (10 μM, 10 min, 37 °C). Cells were then lysed and cAMP levels were determined (mean ± SD; n=3; n.s. not significant; *p≤0.05, **p≤0.01, ***p≤0.001). (D) Western blot analysis of phospho (p) Akt and pERK in Jurkat cells pre-treated with DMSO, AGR1.131, AGR1.135 or AGR1.137, in response to CXCL12. As a loading control, membranes were re-blotted with an anti-Akt antibody. Representative experiments are shown (n=4). (E, F) Densitometry evaluation of the specific bands in D (mean ± SD; n=4).
-
Figure 3—source data 1
PDF file containing original western blots for Figure 3D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/93968/elife-93968-fig3-data1-v1.zip
-
Figure 3—source data 2
Original files for western blot analysis displayed in Figure 3D.
- https://cdn.elifesciences.org/articles/93968/elife-93968-fig3-data2-v1.zip
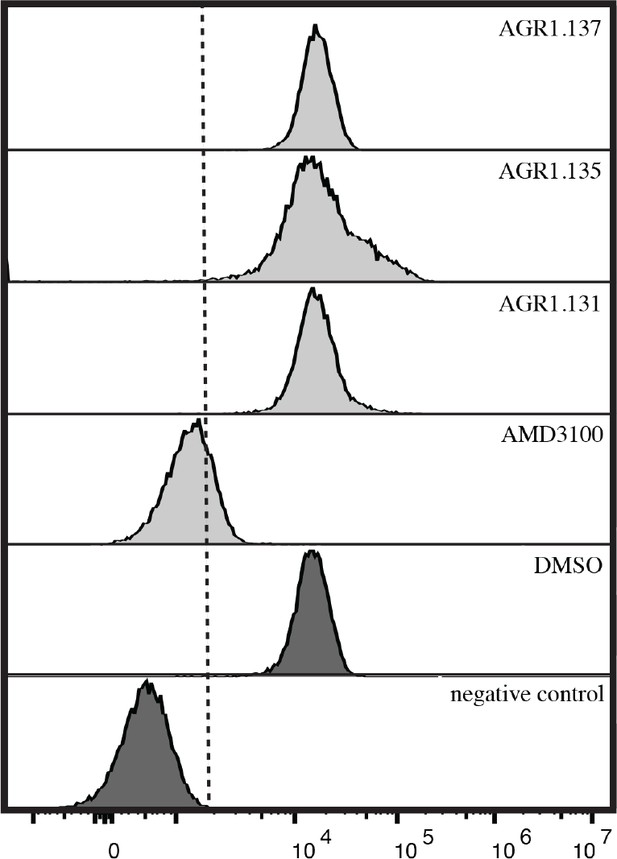
Flow cytometry analysis of the binding of CXCL12-ATTO700 to JK cells.
Cells were pre-incubated for 30 min at 37ºC with the indicated antagonists, followed by incubation with CXCL12- ATTO700 (30 min 37ºC). CXCL12 binding was determined in flow cytometry. A representative experiment of 4 performed is shown.
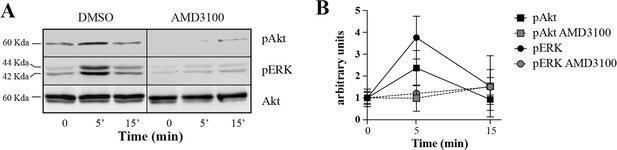
Western blot analysis of AMD3100-treated JK cells.
(A) Cells were pre-incubated for 30 min at 37ºC with AMD3100 or DMSO (vehicle) as control, stimulated with CXCL12 and analyzed by western blot using specific antibodies. (B) Densitometry analysis of the western blot images in (A) using Image J software.
-
Figure 3—figure supplement 2—source data 1
PDF file containing original western blots for Figure 3—figure supplement 2, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/93968/elife-93968-fig3-figsupp2-data1-v1.zip
-
Figure 3—figure supplement 2—source data 2
Original files for western blot analysis displayed in Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/93968/elife-93968-fig3-figsupp2-data2-v1.zip
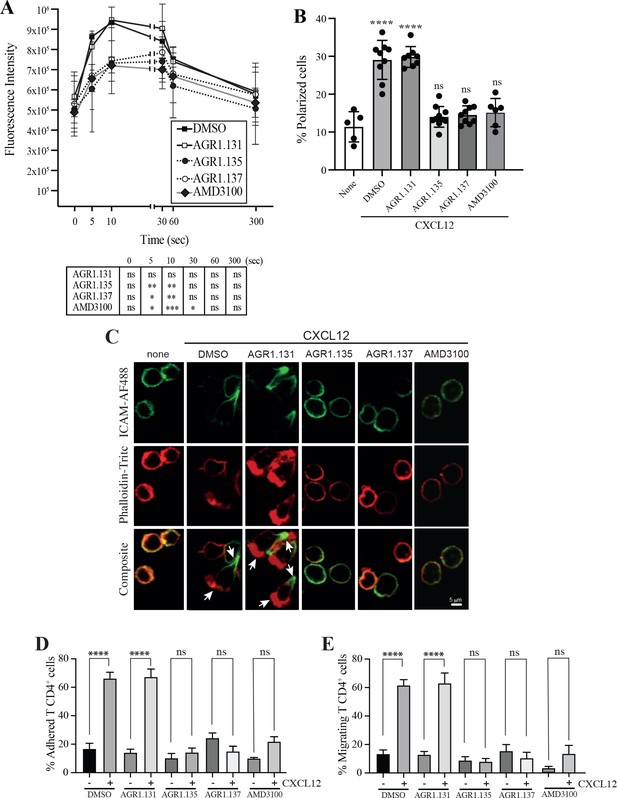
AGR1.135 and AGR1.137 treatment alter CXCL12-mediated actin polymerization.
(A) Actin polymerization in response to CXCL12 as determined by F-actin (phalloidin-TRITC) staining in Jurkat cells treated with DMSO (vehicle) or the indicated modulators. Statistical significance of the different time points in comparison with the control (DMSO) is shown in the table (mean ± SD; n=3; n.s. not significant; *p≤0.05, **p≤0.01, ***p≤0.001). (B) Percentage of polarized T cell blasts adhered to fibronectin and treated or not with CXCL12 in the presence of the indicated antagonists, as analyzed by immunostaining with anti-ICAM3-Alexa fluor 488 and phalloidin-TRITC. More than 500 cells were analyzed in each condition. Data are presented as percentage of polarized cells (mean ± SD; n=3; n.s. not significant; ****p<0.0001). (C) Representative images of T cell blasts adhered to fibronectin and treated or not with CXCL12 in the presence of the indicated antagonists, as analyzed by immunostaining with anti-ICAM3-Alexa fluor 488 and phalloidin-TRITC as in B. (D) CD4+ T cells pretreated with AGR1.131, AGR1.135 or AGR1.137 were perfused in flow chambers coated with ICAM-1-containing lipid bilayers, alone or CXCL12-coated, and analyzed for cell contacts with the substrate. Data are presented as percentage of adhered cells (mean ± SD; n=3; n.s. not significant; ****p≤0.0001). (E) Cells in D were analyzed for cell migration. Data are presented as percentage of migrating cells (mean ± SD; n=3; n.s. not significant; ****p≤0.0001).
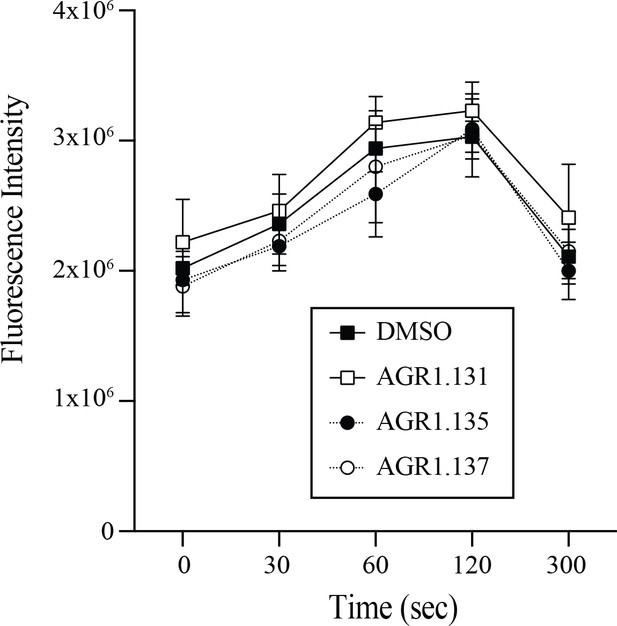
AGR1.135 and AGR1.137 do not alter anti-CD3-induced actin polimerization.
F-actin (Phalloidin-TRITC) staining of Jurkat cells treated with the indicated compounds or DMSO (vehicle), adhered to fibronectin and stimulated with anti-CD3 antibody, as indicated (mean ± SD; n = 3).
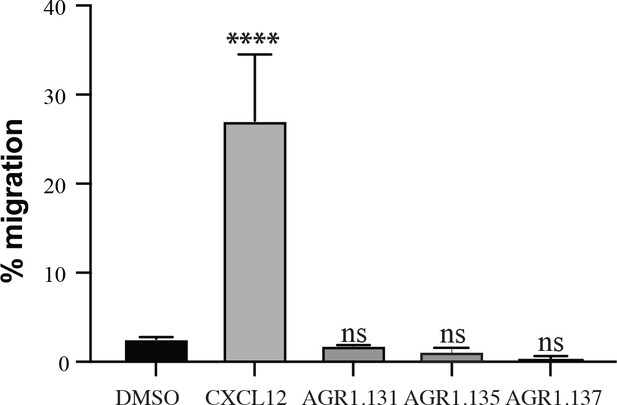
AGR1.131, AGR1.135 and AGR1.137 are not CXCR4 agonists.
Migration of Jurkat cells in response to 12.5 nM CXCL12, 50 μM AGR1.131, 50 μM AGR1.135 or 50 μM AGR1.137. Data shown as percentage of migrating cells (mean ± SD; n = 5; n.s. not significant, **** p ≤0.0001).
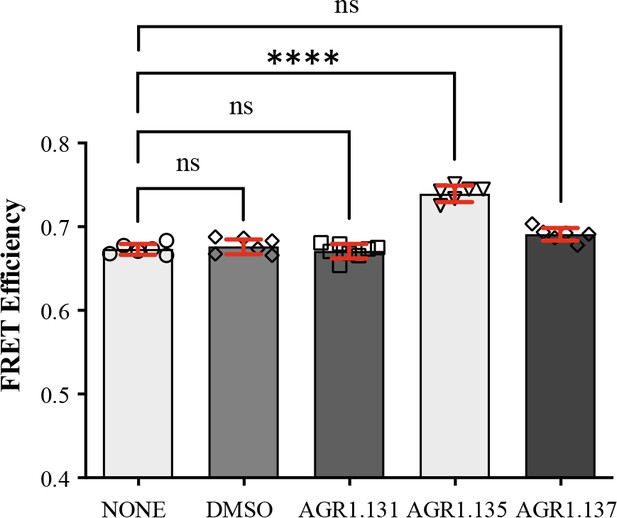
FRET analysis of CXCR4 in the presence of AGR1.131, AGR1.135 and AGR1.137.
FRET efficiency in HEK-293 cells transiently transfected with CXCR4-YFP/ CXCR4-CFP (ratio 15:9), in the presence of 50 μM AGR1.131, 50 μM AGR1.135, 50 μM AGR1.137 or vehicle (DMSO). Data shows FRET efficiency (a.u.) (mean ± SD; n=6; n.s. not significant, **** p≤0.0001).
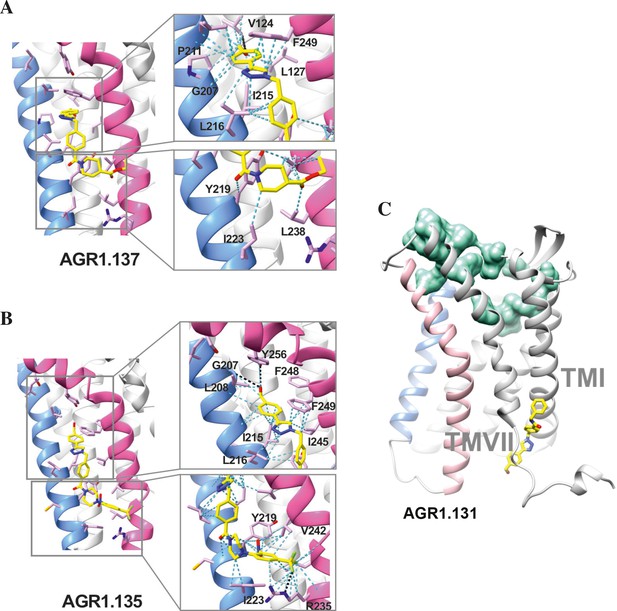
Ribbon and sticks representation of the CXCR4 modulators bound to the receptor.
In the ribbon and sticks representations, the TMV helix is colored in blue and TMVI in pink. Figures show the binding of ligands AGR1.135 (A) and AGR1.137 (B) Zoom images showing the residues involved in the interaction of both compounds are also shown (right panels).(C) Binding of AGR1.131 represented as sticks with carbon atoms in yellow, oxygen in red, nitrogen in blue and fluorine in green. CXCR4 residues involved in CXCL12 engagement and initial signal transmission are represented as green spheres.
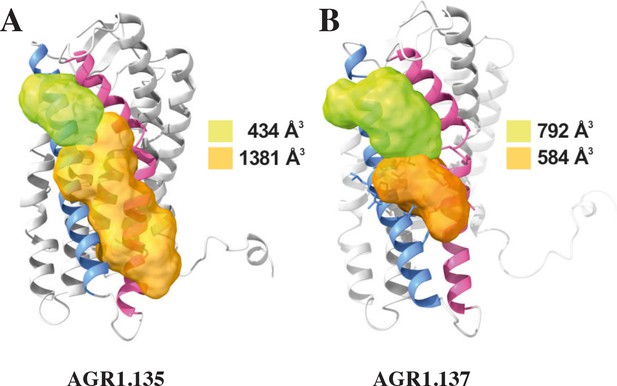
Cartoon and surface representation of CXCR4 and the clefts identified between TMV and TMVI.
The protein structure is shown in gray, with TMV and TMVI colored in blue and pink, respectively. Cavities associated to AGR1.135 (A) and AGR1.137 (B) binding were identified by SurfNet software are shown in orange and green. Volumes for each cavity are measured (Å3).
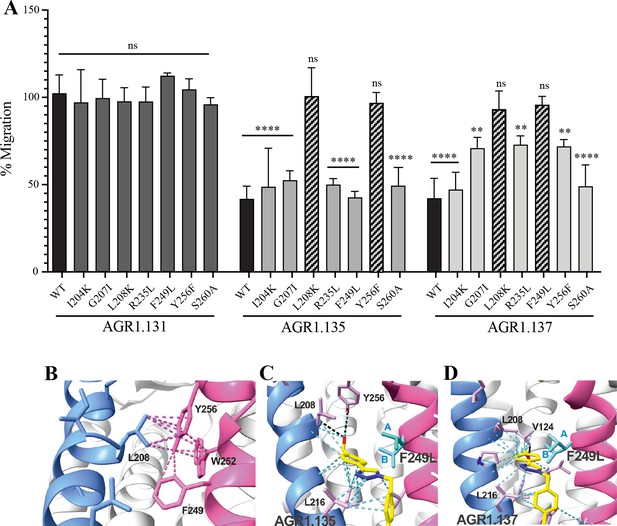
The antagonistic behavior of AGR1.135 and AGR1.137 depends on specific residues of CXCR4.
(A) CXCL12-induced migration of AGR1.131- AGR1.135- or AGR1.137- pretreated JK−/− cells transiently transfected with CXCR4wt or the different mutants described. Data are shown as the mean percentage ± SD of input cells that migrate (n=4; n.s. not significant, **p≤0.01, ****p≤0.0001). (B) Cartoon representation of Y256 and its intramolecular interactions in the CXCR4 X-ray solved structure 3ODU. (C, D) Cartoon representation of the interaction of CXCR4 F249L mutant with AGR1.135 and AGR1.137, respectively. The two most probable conformations of leucine rotamers are represented in cyan. Van der Waals interactions are depicted in cyan dashed lines and hydrogen bonds in black dashed lines.
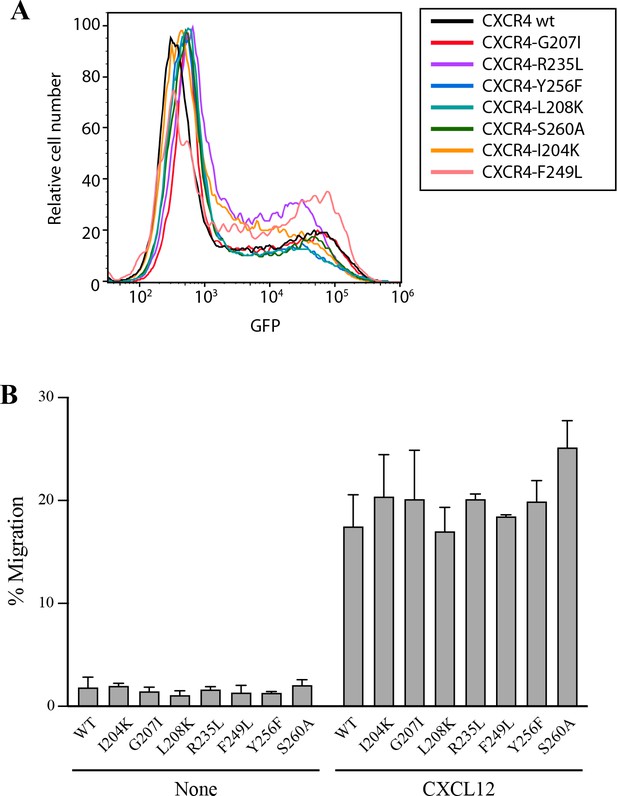
Expression and function of CXCR4 and CXCR4 mutants coupled to AcGFP in transfected JK-/- cells.
(A) Flow cytometry analysis of cells transiently transfected with the indicated CXCR4 mutants or wt CXCR4, coupled to AcGFP and their expression was determined 24 hours after transfection by GFP detection in flow cytometry. A representative experiment of 4 performed is shown. (B) CXCL12-induced migration of CXCR4-defficient Jurkat cells (JK-/-) transiently transfected with CXCR4wt or its mutants, CXCR4I204K, CXCR4G207I, CXCR4L208K, CXCR4R235L, CXCR4F249L, CXCR4Y256F and CXCR4S260A. Data are shown as the mean ± SD percentage of input cells that migrate (n = 3).
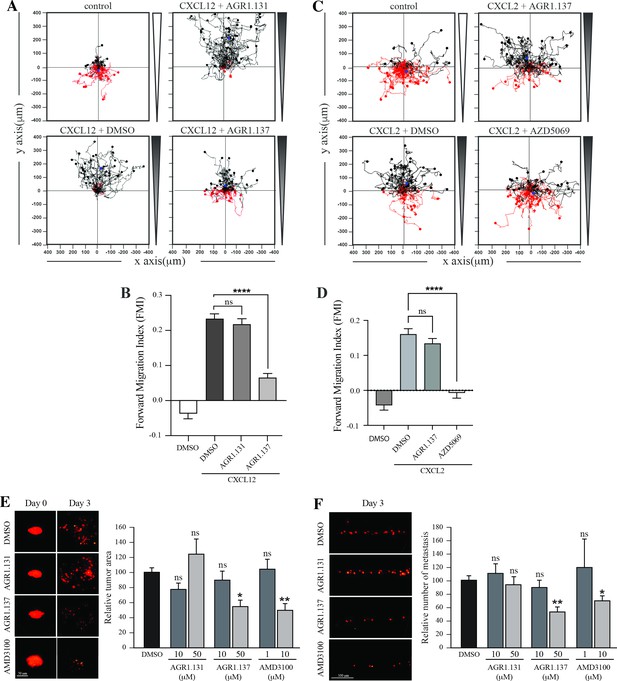
AGR1.137 reduces tumorigenesis and metastasis in a zebrafish model.
(A–D) Migration of HeLa cells treated with vehicle (DMSO) or with the selected small compounds or inhibitor as indicated, on μ-chambers in response to a CXCL12 (A, B) or CXCL2 (C, D) gradient (n=2, in duplicate, with at least 50 cells tracked in each condition). Panels (A) and (C) show representative spider plots with the trajectories of tracked cells migrating along the gradient (black) or moving in the opposite direction (red). Black and red dots in the plots represent the final position of each single tracked cell. (B, D) Quantification of the Forward Migration Index of experiments performed in (A) and (C) (mean ± SD; n=3; n.s. not significant, ****p≤0.0001). (E) Representative fluorescent images of DiI-labeled HeLa cells in zebrafish larvae treated with vehicle (DMSO), AGR1.131 50 μM, AGR1.137 50 μM or AMD3100 10 μM at 0 or 3 days post-implantation and treatment. Quantitation of the relative tumor size at day 3 compared with that of day 0 normalized to the relative tumor size in the DMSO control group, is shown for each experimental group (mean ± SD; n=20; n.s. not significant, **p≤0.01). (F) Representative fluorescent images of the caudal hematopoietic plexus of larvae from the same groups as shown in E at 3 days postimplantation. Quantitation of the relative amount of metastasized cells in each group relative to DMSO-controls is shown (mean ± SD; n=20, n.s. not significant, * p≤0.05, *** p≤0.001).
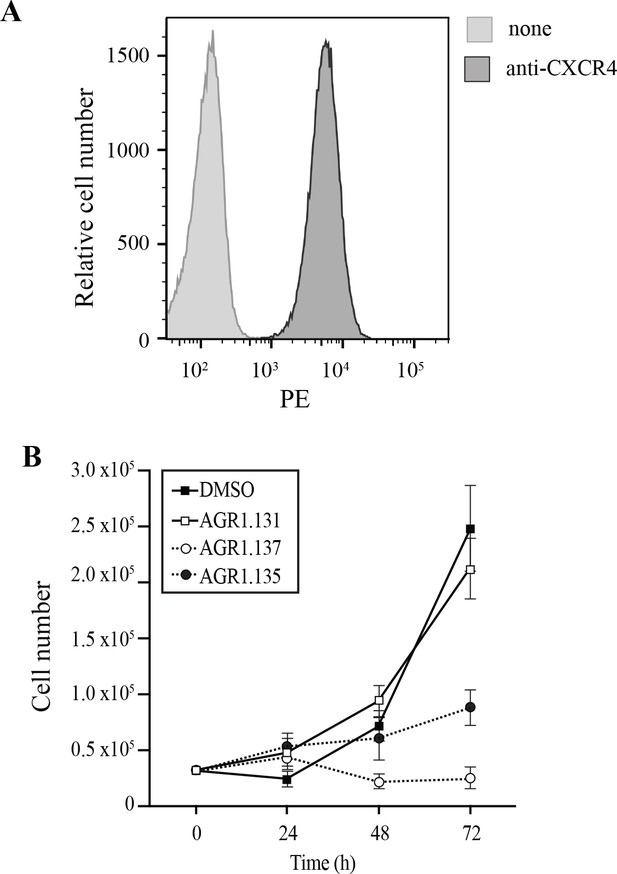
Characterization of HeLa cells.
(A) CXCR4 expression on the surface of HeLa cells, analyzed by flow cytometry using a specific anti-CXCR4 antibody. Data show a representative experiment of 6 performed. (B) Effect of the indicated compounds on HeLa cells growth. Cells (0.5x106) were plated on 48 well plates in the presence of 100 mM of the compounds, which were added freshly every 24 hours. Cells were counted at every 24 hours and maintained for 72 h. The mean ± SD (n=3) is shown.
Related to Figure 8A.
Representative video of DMSO-treated HeLa cell migration on fibronectin-coated μ-Slide Chemotaxis chambers in the absence of CXCL12 gradient. Images over time (15 frames/s) are shown. Overlaid trajectories of cells shown in Video were detected and tracked using Fiji software.
Related to Figure 8A.
Representative video of DMSO-treated HeLa cell migration on fibronectin-coated μ-Slide Chemotaxis chambers following a CXCL12 gradient on top. Images over time (15 frames/s) are shown. Overlaid trajectories of cells shown in Video were detected and tracked using Fiji software.
Related to Figure 8A.
Representative video of AGR1.131-treated HeLa cell migration on fibronectin-coated μ-Slide Chemotaxis chambers following a CXCL12 gradient on top. Images over time (15 frames/s) are shown. Overlaid trajectories of cells shown in Video were detected and tracked using Fiji software.
Related to Figure 8A.
Representative video of AGR1.137-treated HeLa cell migration on fibronectin-coated μ-Slide Chemotaxis chambers following a CXCL12 gradient on top. Images over time (15 frames/s) are shown. Overlaid trajectories of cells shown in Video were detected and tracked using Fiji software.
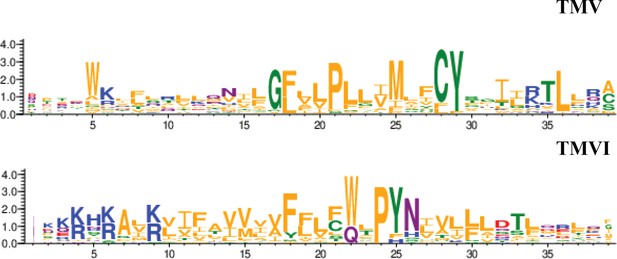
Residue conservation among all GPCR class A chemokine receptors for TMV and TMVI.
Graphs were produced in the WebLogo server after aligning all receptors sequence in the sequence alignment tool included in GPCRdb. Font size for each residue represented in one letter code stands for the degree of conservation among all chemokine receptors. Residues are colored by their chemistry being green for polar, orange for hydrophobic, red for acidic, blue for basic and purple for neutral.
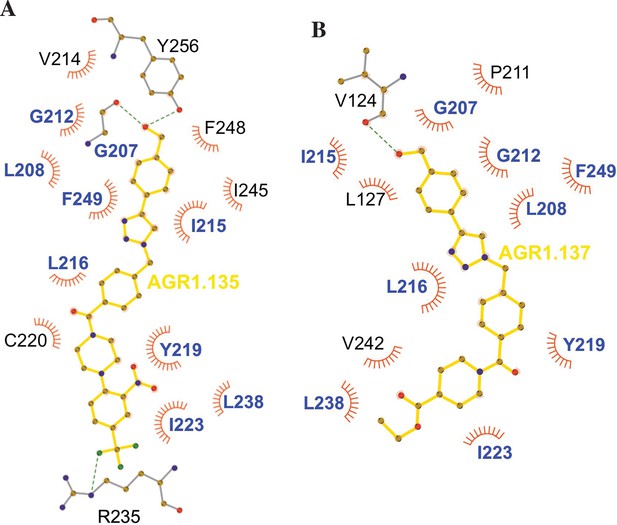
Ligplot representation of antagonists interactions with CXCR4.
Ligplot representations of AGR1.135 (A) and AGR1.137 (B), with hydrogen bonds represented in green dotted lines and non-bonded contacts in red spoke arcs. Residues labeled in blue font are present in the interface of both AGR1.135 and AGR1.137. CXCR4 residues forming a hydrogen bond with the ligands are shown in gray.
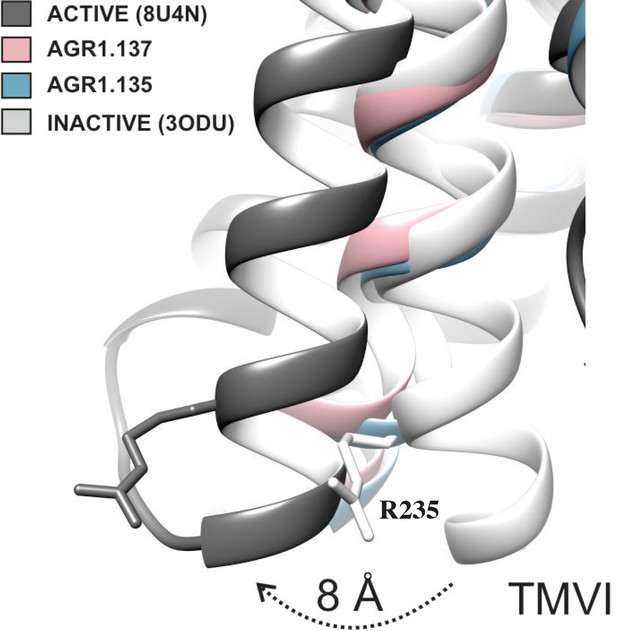
TMVI transition from inactive to active position.
Ribbon representation of TMVI of CXCR4 structures superimposed for inactive (PDB code 3ODU), intermediate active (AGR1.135, AGR1.137) and active (PDB 8U4N) upon interaction with no, negative allosteric modulators and CXCL12-induced Gi-protein interaction, respectively. Arginine 235 is depicted in stick representation as a reference for the movement of the protein.

AGR1.
135 docking in CXCR4 using the updated protocol for ligand docking. Cartoon representation colored in gray with TMV and TMVI shown in blue and pink, respectively. AGR1.135 is shown in stick representation with carbons in yellow, oxygens in red and nitrogens in blue.

Cartoon representation of Y256 and its intramolecular interactions in the CXCR4 Xray solved structure 3ODU.
TMV helix is colored in blue and TMVI in pink.

AGR1.
135 and AGR1.137 interaction with TMV and TMVI. The model shows the location of the compounds within the TMV-VI cleft, illustrated by a ribbon and stick representation. The CXCR4 segments of TMV and TMVI are represented in blue and pink ribbons respectively, and side chains for some of the residues defining the cavity are shown in sticks. AGR1.135 and AGR1.137 are shown in stick representation with carbon in yellow, nitrogen in blue, oxygen in red, and fluorine in green. Hydrogen bonds are indicated by dashed black lines, while hydrophobic interactions are shown in green. The figure reproduces the panels A, B of Fig. 5 in the revised manuscript.

Evolution of the tumors and metastasis along the time in the absence of any treatment.
HeLa cells were labeled with 8 µg/mL Fast-DiI oil and then implanted in the dorsal perivitelline space of 2-days old zebrafish embryos. Tumors were imaged within 2 hours of implantation and re-imaged each 24 h for three days. Changes in tumor size was evaluated as tumor area at day 1, 2 and 3 divided by tumor area at day 0, and metastasis was evaluated as the number of cells disseminated to the caudal hematopoietic plexus at day 1, 2 and 3 divided by the number of cells at day 3.
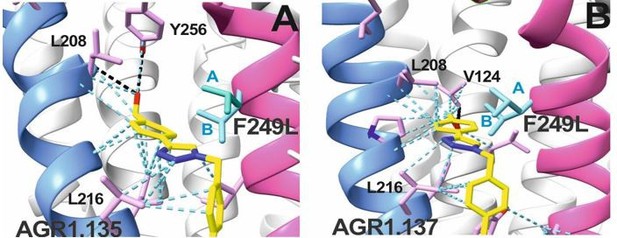
Cartoon representation of the interaction of CXCR4 F249L mutant with AGR1.
135 (A) and AGR1.137 (B). The two most probable conformations of Leucine rotamers are represented in cyan A and B conformations. Van der Waals interactions are depicted in blue cyan dashed lines, hydrogen bonds in black dashed lines. CXCR4 segments of TMV and TMVI are colored in blue and pink, respectively
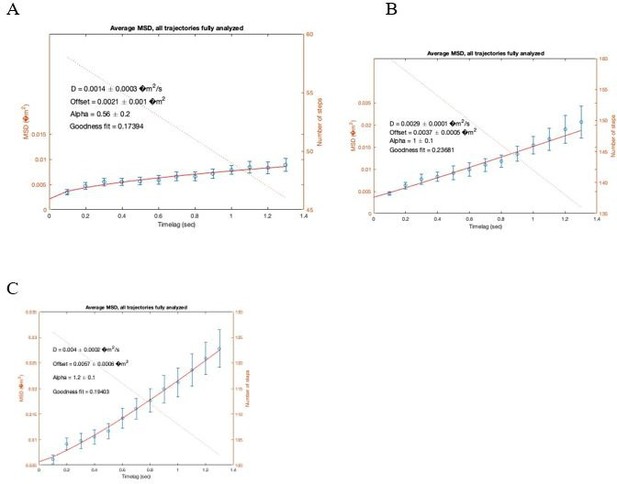
Representative MSD plots from individual trajectories of CXCR4-AcGFP showing different types of motion: (A) confined, (B) Brownian/Free, (C) direct transport of CXCR4-AcGFP particles diffusing at the cell membrane detected by SPT-TIRF in resting JKCD4 cells.

Effects of the negative allosteric modulators on the Types of Motion of CXCR4.
Percentage of single trajectories with different types of motion, classified by MSS (DMSO: 58 particles in 59 cells on FN; 314 in 63 cells on FN+CXCL12; AGR1.131: 102 particles in 71 cells on FN; 258in 69 cells on FN+CXCL12; AGR1.135: 86 particles in 70 cells on FN; 120 in 77 cells on FN+CXCL12; AGR1.137: 47 particles in 66 cells on FN; 74 in 64 cells on FN+CXCL12) n = 3.
Tables
Oligonucleotides used to generate the CXCR4 mutants.
| Mutation | Forward | Reverse |
|---|---|---|
| CXCR4 G207I | cac atc atg gtt att ctt atc ctg cct | agg cag gat aag aat aac cat gat gtg |
| CXCR4 R235L | ggc cac cag aag tta aag gcc ctc aag | CTT GAG GGC CTT TAA CTT CTG GTG GCC |
| CXCR4 Y256F | TGG CTG CCT TAC TTT ATT GGG ATC AGC | GCT GAT CCC AAT AAA GTA AGG CAG CCA |
| CXCR4 S260A | TACTACATTGGGATCgccATCGACTCCTTCATC | GATGAAGGAGTCGATggcGATCCCAATGTAGTA |
| CXCR4 L208K | CACATCATGGTTGGCAAGATCCTGCCTGGTATT | AATACCAGGCAGGATCTTGCCAACCATGATGTG |
| CXCR4 I204K | TTCCAGTTTCAGCACAAGATGGTTGGCCTTATC | GATAAGGCCAACCATCTTGTGCTGAAACTGGAA |
| CXCR4 G212F | GCCCTTATCCTGCCTTTCATTGTCATCCTGTCC | GGACAGGATGACAATGAAAGGCAGGATAAGGCC |
| CXCR4 F249L | GTCATCCTCATCCTGGCTTTCTGCCTGTTGGCTGCCT | AGGCAGCCAACAGGCCAGGAAAGCCAGGATGAGGATGAC |