Purging viral latency by a bifunctional HSV-vectored therapeutic vaccine in chronically SIV-infected macaques
Figures
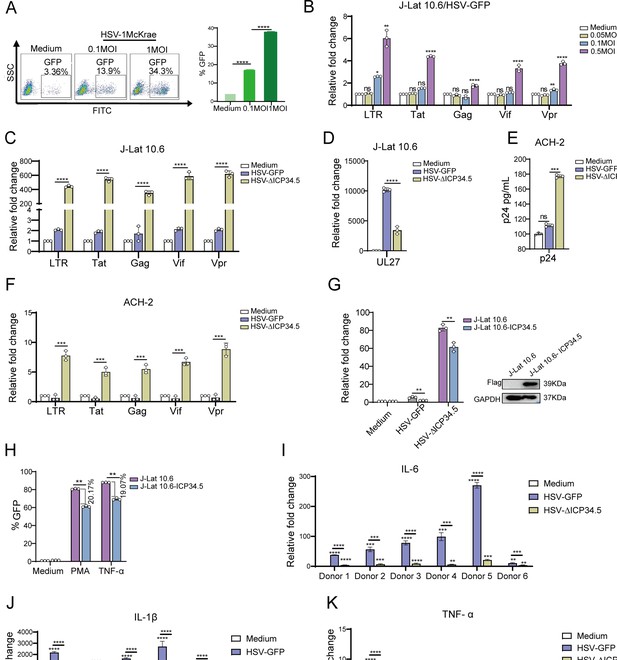
The modified HSV-ΔICP34.5-based constructs reactivated human immunodeficiency virus (HIV) latency more efficiently than wild-type herpes simplex virus (HSV) counterparts.
(A) J-Lat 10.6 cells (1×106) were infected with varying multiplicities of infection (MOIs) of wild-type HSV type I (HSV-1) Mckrae strain for 30 hr. The proportion of green fluorescent protein (GFP+) cells, indicating activated latent cells, is shown in the pseudocolor plot (left) and the corresponding bar chart (right). (B) J-Lat 10.6 cells (1×106) were infected with varying MOIs of HSV-1 17 strain containing GFP (HSV-GFP) for 30 hr, and the mRNA levels of HIV-1 transcripts driven by the 5’ LTR, as well as Tat, Gag, Vpr, and Vif, are presented in the histogram. (C) J-Lat 10.6 cells (1×106) were infected with HSV-GFP or HSV-ΔICP34.5 at an MOI of 0.1 for 30 hr. The mRNA levels of HIV-1 transcripts driven by the 5’ LTR, as well as Tat, Gag, Vpr, Vif, and (D) HSV-1 UL27 are presented in the histogram. (E) ACH-2 cells (1×106) were infected with HSV-GFP or HSV-ΔICP34.5 at an MOI of 0.1 for 30 hr. The p24 protein level was detected using an HIV-1 p24 ELISA kit, and the mRNA levels of HIV-1 transcripts driven by the 5’ LTR, as well as Tat, Gag, Vpr, and Vif, are shown in the histogram (F). (G) J-Lat 10.6 and J-Lat 10.6-ICP34.5 cells were infected with HSV-GFP or HSV-ΔICP34.5, and the mRNA levels of HIV-1 Tat were shown with the histogram (left). Blotting showed that J-Lat 10.6 cells stably expressing HSV ICP34.5 (J-Lat 10.6-ICP34.5) can appropriately express ICP34.5 protein using Flag-tag antibodies (right). (H) J-Lat 10.6 and J-Lat 10.6-ICP34.5 cells were respectively stimulated with phorbol 12-myristate 13-acetate (PMA) (10 ng/mL) and TNF-α (10 ng/mL), and the expression level of GFP+ cells is displayed with the corresponding bar chart. (I–K) Primary CD4+ T cells from people living with HIV (PLWH) were infected with HSV-GFP or HSV-∆ICP34.5. The inflammatory response was assessed by evaluating mRNA levels of IL-6, IL-1β, and TNF-α using qPCR. Data shown are mean ± SD. Three independent experiments were repeated. **p<0.01, ***p<0.001, ****p<0.0001. ns: no significance.
-
Figure 1—source data 1
PDF file containing original western blots for Figure 1G, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig1-data1-v1.pdf
-
Figure 1—source data 2
Original files for western blot analysis displayed in Figure 1G.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig1-data2-v1.zip
-
Figure 1—source data 3
Source data for Figure 1A–K.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig1-data3-v1.xlsx
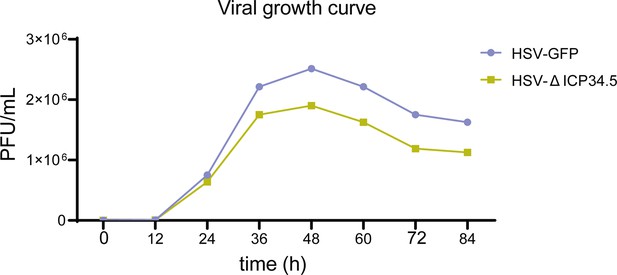
Viral growth curve.
Vero cells were infected with herpes simplex virus (HSV)-green fluorescent protein (GFP) and HSV-∆ICP34.5 at a multiplicity of infection (MOI) of 0.01. Cells and supernatants were collected at various time points, and viral titers were determined by plaque assay.
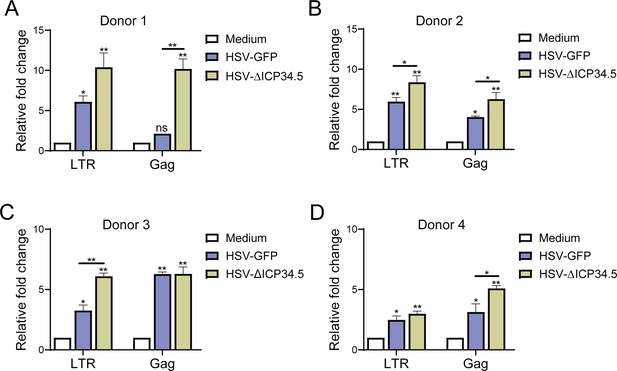
The reactivation effect of herpes simplex virus type I (HSV-1) on the latent human immunodeficiency virus (HIV) reservoir in primary CD4+ T cells from people living with HIV (PLWH).
CD4+ T cells from four PLWH donors (A–D) were infected with HSV-green fluorescent protein (GFP) and HSV-∆ICP34.5. LTR‑initiation transcripts and HIV-1 Gag mRNA levels were detected by quantitative PCR. Experiments were performed in three independent biological replicates. *p<0.05, **p<0.01. ns: no significance.
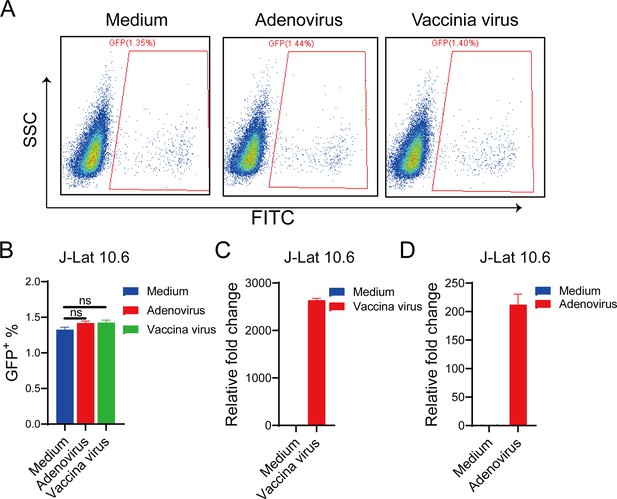
Adenovirus and vaccinia virus cannot reactivate human immunodeficiency virus (HIV) latency in J-Lat 10.6 cells.
J-Lat 10.6 cells were infected with adenovirus and vaccinia virus for 30 hr. The proportion of green fluorescent protein (GFP+) cells, indicating activated latent cells, is shown in the pseudocolor plot (A) and the corresponding bar chart (B). The bar charts represent the mRNA levels of adenovirus L1 gene (C) and vaccinia virus tk gene (D). ns: no significance.

Deletion of ICP0 diminished the reactivation effect of human immunodeficiency virus (HIV) latency by herpes simplex virus type I (HSV-1).
The J-Lat 10.6 cells were infected with HSV-green fluorescent protein (GFP), HSV-∆ICP34.5 and HSV-∆ICP0. LTR‑initiation transcripts were detected by quantitative PCR. Three independent experiments were repeated. ****p<0.0001.

ICP0 promotes human immunodeficiency virus (HIV) reactivation in J-Lat10.6 cells.
J-Lat10.6 cells (2×10⁶) were electroporated with 2 µg of either pVAX-empty or pVAX-ICP0 plasmid. Green fluorescent protein (GFP) expression was analyzed by flow cytometry 24 hr post-transfection to assess HIV reactivation in the latent J-Lat10.6 cell line.
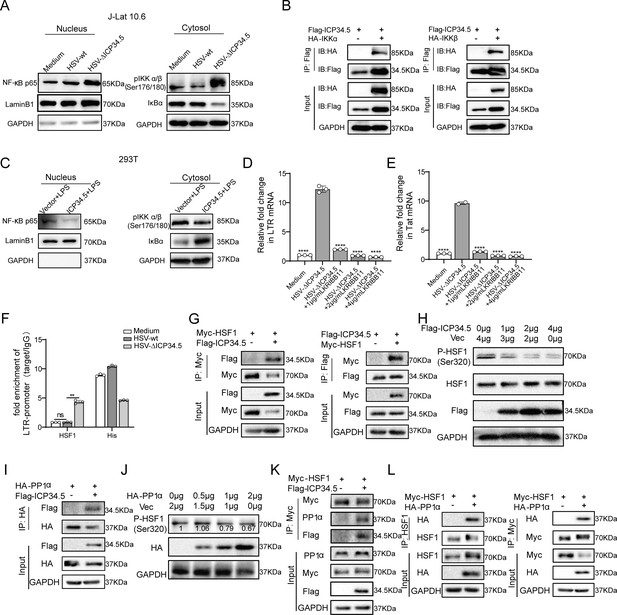
The modified herpes simplex virus (HSV)-based constructs effectively reactivated human immunodeficiency virus (HIV) latency by modulating the NF-κB pathway and HSF1 pathway.
(A) J-Lat 10.6 cells were infected with HSV-green fluorescent protein (GFP) and HSV-ΔICP34.5 at a multiplicity of infection (MOI) of 0.1. Cytoplasmic and nuclear protein fractions were analyzed for NF‑κB p65, phosphorylated IKKα/β (p-IKKα/β), and IkBα. GAPDH and Lamin B1 served as loading controls for cytoplasmic and nuclear fractions, respectively. (B) 293T cells were co-transfected with Flag-ICP34.5 and either IKKα (left) or IKKβ (right), and the interactions were examined by co-immunoprecipitation (Co-IP) assays. (C) 293T cells were transfected with Flag-ICP34.5 or an empty vector (Vec) for 24 hr and then treated with lipopolysaccharide (LPS; 1 μg/mL) for 8 hr. Cytoplasmic and nuclear protein fractions were subsequently analyzed by Western blot (WB). (D–E) J-Lat 10.6 cells were infected with HSV-ΔICP34.5 and treated with increasing concentrations of KRIBB11 concentrations. The mRNA levels of Tat and of HIV‑1 transcripts driven by the 5′ LTR were quantified by qPCR. (F) J-Lat 10.6 cells were infected with HSV-wt or HSV-ΔICP34.5 at an MOI of 0.1 for 36 hr. Chromatin immunoprecipitation followed by qPCR (ChIP–qPCR) was performed to assess HSF1 binding to LTR. Normal IgG and an anti‑histone H3 antibody served as negative and positive controls, respectively. (G) 293T cells were co-transfected with Flag-ICP34.5 and Myc-HSF1, and the interactions were examined by Co-IP assays. (H) The immunoblot depicts the alterations in protein levels in 293T cells transfected with either empty vector or ICP34.5 for 6 hr, followed by treatment with 10 μM MG132 for 24 h. (I) 293T cells transfected with Flag-ICP34.5 and HA-PP1α, analyzed by Co-IP assays. (J) The immunoblot depicts the alterations in protein levels in 293T cells transfected with either empty vector or HA-PP1α for 6 hr, followed by 24 hr of treatment with 10 μM MG132. (K) 293T cells transfected with Myc-HSF1 with or without Flag-ICP34.5, analyzed by Co-IP assays. (L) 293T cells transfected with Myc-HSF1 and HA-PP1α, analyzed through Co-IP assays. Data shown are mean ± SD. **p<0.01, ****p<0.0001. ns: no significance.
-
Figure 2—source data 1
PDF file containing original western blots for Figure 2A–C and G–L, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig2-data1-v1.pdf
-
Figure 2—source data 2
Original files for western blot analysis displayed in Figure 2A–C and G–L.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig2-data2-v1.zip
-
Figure 2—source data 3
Source data for Figure 2D–F.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig2-data3-v1.xlsx
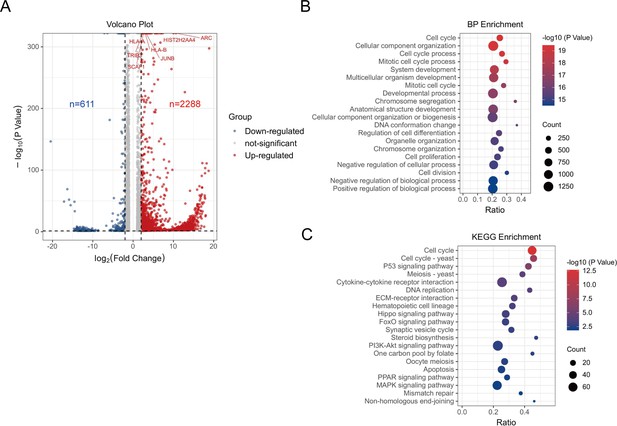
RNA-seq analysis for HSV-ΔICP34.5-induced signaling pathways.
J-Lat 10.6 cells were either infected or not infected with HSV-green fluorescent protein (GFP) and HSV-ΔICP34.5, followed by extraction of total cellular RNA for sequencing and analysis. (A) Volcano plot showing differentially expressed genes between the two groups, with blue indicating downregulation and red indicating upregulation. (B) Bubble plot illustrating biological function enrichment analysis. (C) Bubble plot demonstrating enrichment of differential genes in signaling pathways. Circle size represents the number of genes enriched in that pathway, with colors ranging from blue to red indicating increasing -log10 (p-value) values.
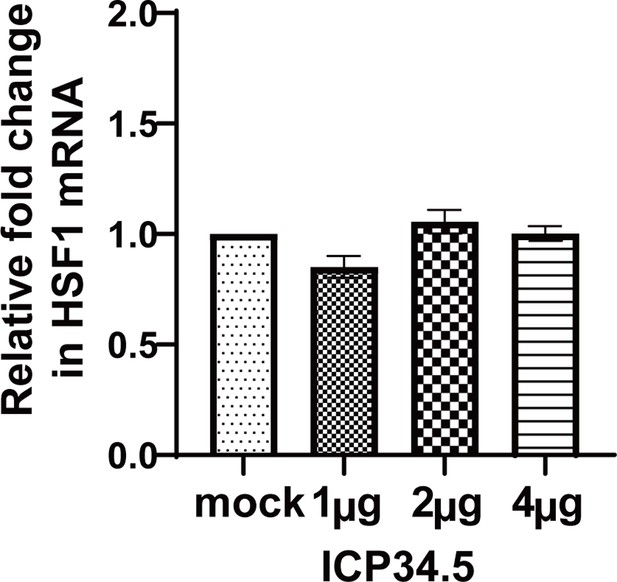
The overexpression of ICP34.5 have no influence on the level of HSF1 expression.
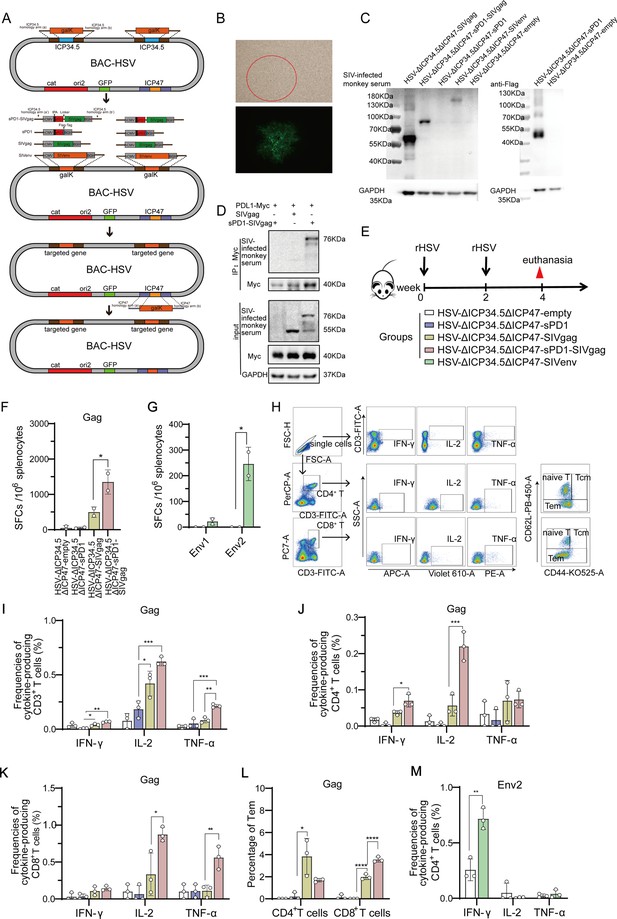
Recombinant herpes simplex virus type I (HSV-1) vector-based simian immunodeficiency virus (SIV) vaccines induce specific T cell immune responses in mice.
(A) Schematic diagram illustrating the construction of recombinant HSV through the bacterial artificial chromosome (BAC)/galactokinase (galK) selection system. The ICP34.5 gene was replaced with the galK gene via homologous recombination, followed by substituting galK with a target gene expression cassette containing the hCMV promoter and BGH terminator. Finally, the ICP47 gene was deleted. (B) Brightfield (top) and fluorescence (bottom) images of a clone of the rescued recombinant HSV. (C) Vero cells infected with recombinant HSV constructs, with protein expression of targeted genes detected using SIV-infected monkey serum. (D) HeLa cells transfected with Myc-PDL1 and then infected with HSV-ΔICP34.5ΔICP47-empty, HSV-ΔICP34.5ΔICP47-SIVgag, or HSV-ΔICP34.5ΔICP47-sPD1-SIVgag at a multiplicity of infection (MOI) of 0.1 for 24 hr. Cell lysates were subjected to co-immunoprecipitation (Co-IP) analysis. (E) Schematic schedule of mouse vaccination. Twenty-five mice were randomly allocated to five groups: HSV-ΔICP34.5ΔICP47-empty, HSV-ΔICP34.5ΔICP47-sPD1, HSV-ΔICP34.5ΔICP47-SIVgag, HSV-ΔICP34.5ΔICP47-sPD1-SIVgag, and HSV-ΔICP34.5ΔICP47-SIVenv. Mice were injected with the corresponding vaccines at weeks 0 and 2. At week 4, mice were sacrificed, and spleen lymphocytes were collected to evaluate immune response.(F–G) Column graphs showing the number of Gag or Env1, Env2-specific spot-forming cells (SFCs) per 106 spleen lymphocytes, as measured by interferon γ (IFN-γ) ELISpot assay. (H) Pseudocolor plot of flow cytometry illustrating the gating strategy. Column graphs showing the frequencies of IFN-γ, IL-2, and TNF-α production from gag-specific CD3+ T (I), CD4+ T (J), and CD8+ T cells (K). (L) Bar chart showing the proportion of Tem (effector memory T cells) among CD4+ T and CD8+ T cells upon stimulation with the SIV Gag peptide pools.(M) Bar chart showing the frequencies of Env2-specific IFN-γ+ CD4+ T cells. Data were expressed as mean ± SD from five mice samples. Three independent experiments for the animal immunization were repeated. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ns: no significance.
-
Figure 3—source data 1
PDF file containing original western blots for Figure 3C, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig3-data1-v1.pdf
-
Figure 3—source data 2
Original files for western blot analysis displayed in Figure 3C.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig3-data2-v1.zip
-
Figure 3—source data 3
Source data for Figure 3F–G and I–M.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig3-data3-v1.xlsx

The identification of recombinant pBAC-HSV plasmid.
Identification was performed by amplifying the pBAC plasmid using the tHSV-F and tHSV-R primers designed in the homologous arm of ICP34.5. lane1: DNA marker; lane2: pBAC-HSV (negative control); lane3: pBAC-HSV-ΔICP34.5ΔICP47; lane4: pBAC-HSV-sPD1; lane5: pBAC-HSV-SIVgag; lane6: pBAC-HSV-sPD1-SIVgag; pBAC-HSV-SIVenv.
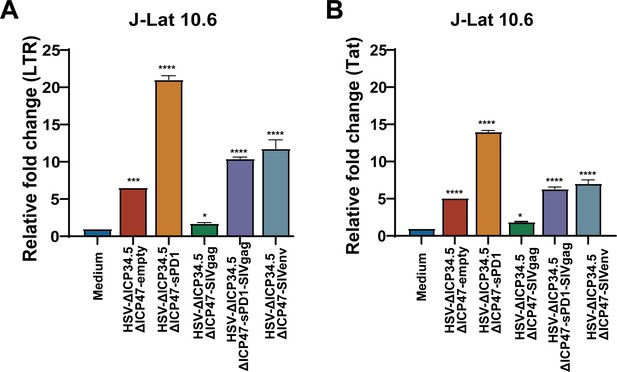
HSV-∆ICP34.5ΔICP47-based recombinant viruses reverse HIV latency in J-Lat 10.6 cells.
J-Lat 10.6 cells (1×106) were infected with HSV-∆ICP34.5ΔICP47-based recombinant viruses. Relative fold-changes in LTR-initiation transcripts (A) and Tat mRNA (B) were detected by qPCR. Data were shown as mean ± SD. Three independent experiments were repeated. *p<0.05, ***p<0.001, ****p<0.0001.
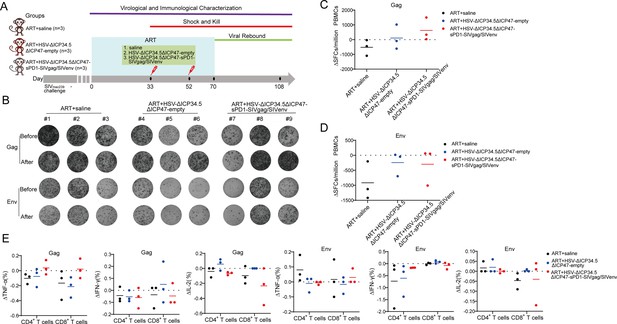
The modified herpes simplex virus (HSV)-based constructs efficiently elicited simian immunodeficiency virus (SIV)-specific immune responses in chronically SIV-infected macaques.
(A) Schematic schedule of the macaque experiment. Nine chronically SIV-infected macaques were assigned into three groups: antiretroviral therapy (ART)+saline group (n=3), ART+HSV-ΔICP34.5ΔICP47-empty group (n=3), and ART+HSV-ΔICP34.5ΔICP47-sPD1-SIVgag/SIVenv group (n=3). All SIV-infected macaques received ART treatment (FTC/6.7 mg/kg/once daily, PMPA/10 mg/kg/once daily) for 33 days. On days 33 and 52, macaques were immunized with saline, HSV-ΔICP34.5ΔICP47-empty, and HSV-ΔICP34.5ΔICP47-sPD1-SIVgag/SIVenv respectively. ART treatment,was interrupted in all macaques on day 70 after the second vaccination. Samples were collected at various time points to monitor virological and immunological parameters. (B) Representative images of Gag or Env-specific spots (2.5×105 cells per well) from each macaque pre-vaccination (before, day 33) and post-vaccination (after, day 70) by ELISpot assay. (C–D) Difference in SIV-specific interferon γ (IFN-γ)-secreting cells (ΔSFCs) between pre- and post-immunization, assessing the immune response induced by HSV-ΔICP34.5ΔICP47-vectored SIV vaccines. (E) Difference in SIV-specific TNF-α/IFN-γ/IL-2-secreting CD4+ T and CD8+ T subsets between pre- and post-immunization, detected by intracellular cytokine staining (ICS) assay.
-
Figure 4—source data 1
Source data for Figure 4C–E.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig4-data1-v1.xlsx
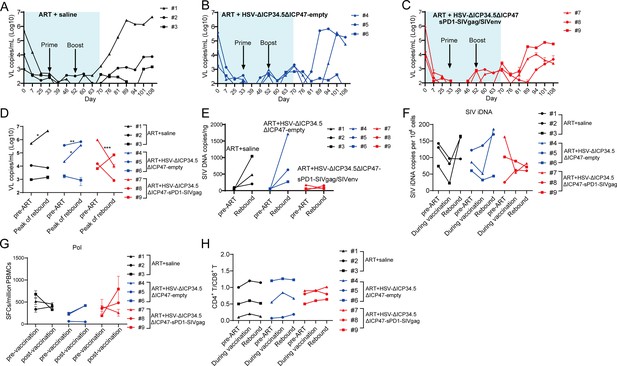
The modified herpes simplex virus (HSV)-based constructs effectively reactivated simian immunodeficiency virus (SIV) latency in vivo in chronically SIV-infected, antiretroviral therapy (ART)-treated macaques.
(A–C) Viral load (VL) changes in plasma for each animal were monitored throughout the experiment using real-time PCR. The detection limit is 100 copies/mL plasma. The shaded area represented the duration of ART administration. (D) The VL change in plasma between pre-ART and the peak value in the rebound stage after ART discontinuation. (E) Change in total SIV DNA copies between pre-ART and viral rebound after ART discontinuation. (F) SIV integrated DNA (iDNA) copy numbers detected by Alu-PCR at different time points. (G) Change in the number of SIV Pol-specific IFN-γ-secreting cells between pre-immunization (day 33) and post-immunization (day 70), as detected by ELISpot assay. (H) Change in the CD4+ T/ CD8+ T ratio. Data shown are mean ± SD. **p<0.01, ***p<0.0001, ****p<0.0001.
-
Figure 5—source data 1
Source data for Figure 5A–H.
- https://cdn.elifesciences.org/articles/95964/elife-95964-fig5-data1-v1.xlsx
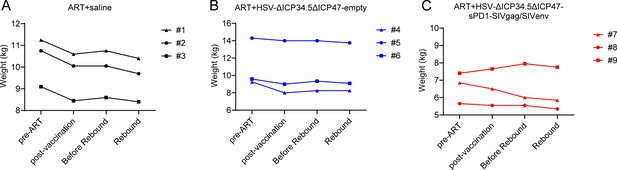
Longitudinal body-weight profiles of macaques throughout the study period.
Body weight was recorded for each macaque (A-C) at four key time points: pre‑ART, post‑vaccination, pre‑rebound, and rebound.
Effect on the cell composition of peripheral blood in the macaques of different groups.
The levels of white blood cells (WBCs) (A), lymphocyte (LYM) (B), neutrophils (NEU) (C), and monocyte (MONO) (D) were monitored by the complete blood count tests.
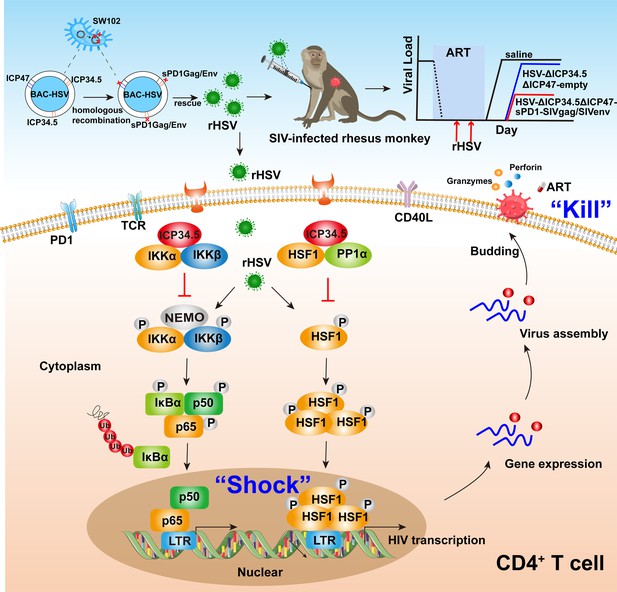
Pattern to illustrate the proof-of-concept strategy based on a bifunctional HSV-ΔICP34.5-vectored therapeutic vaccine for human immunodeficiency virus (HIV) functional cure.
In the present study, the modified HSV-ΔICP34.5-based constructs effectively reactivated HIV/simian immunodeficiency virus (SIV) latency by modulating the IKKα/β-NF-κB pathway and PP1-HSF1 pathway (shock) and simultaneously elicited antigen-specific polyfunctional CD8+ T cells to eliminate cells infected with the reactivated virion (kill). BAC: bacterial artificial chromosome; rHSV: recombinant herpes simplex virus; TCR: T cell receptor; PD1: programmed cell death protein 1; CD40L: CD40 ligand.
Additional files
-
Supplementary file 1
Supplementary figures and tables for this study.
(a) Immunoprecipitation-mass spectrometry (IP-MS) analysis. (b) Baseline information for the experimental macaques in this study. (c) Antibodies for the intracellular cytokine staining (ICS) assay in this study. (d) The sequences of primers.
- https://cdn.elifesciences.org/articles/95964/elife-95964-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95964/elife-95964-mdarchecklist1-v1.docx





