Formation of a giant unilocular vacuole via macropinocytosis-like process confers anoikis resistance
Figures
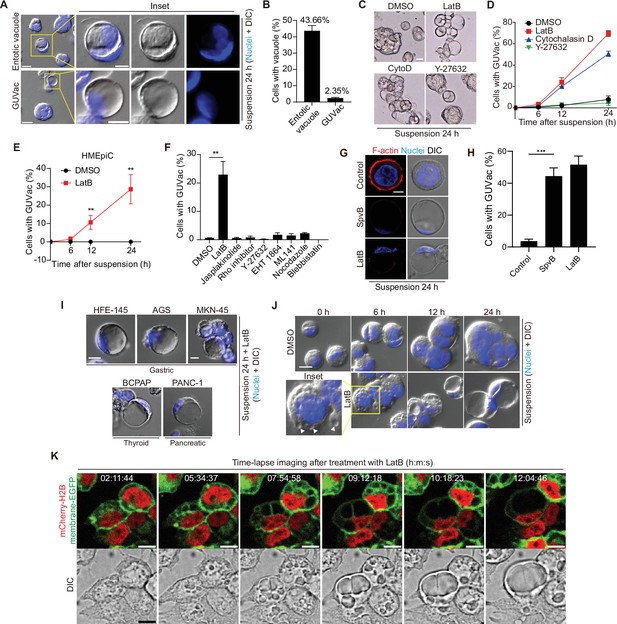
Giant unilocular vacuole (GUVac) formation induced by F-actin disruption in matrix-deprived mammary epithelial cells.
(A) Representative differential interference contrast (DIC) microscopy images of suspended MCF-10A cells showing entotic vacuole or GUVac formation after culture for 24 hr. Nuclei were stained with Hoechst 33342. Scale bars: 20 μm (main panels) or 10 μm (inset). (B) Percentage of suspended MCF-10A cells showing entotic vacuole or GUVac formation after 24 hr (n = 772). (C) Representative bright-field microscopy images of MCF-10A cells suspended with F-actin cytoskeleton inhibitors. Scale bar: 15 μm (D) Percentage of suspended MCF-10A cells showing GUVac formation after exposure to the indicated drugs for the indicated times. dimethyl sulfoxide (DMSO): 0 hr (n = 512), 6 hr (n = 723), 12 hr (n = 690), and 24 hr (n = 690). latrunculin B (LatB): 6 hr (n = 634), 12 hr (n = 693), and 24 hr (n = 428). Cytochalasin D: 6 hr (n = 613), 12 hr (n = 618), and 24 hr (n = 464). Y-27632: 6 hr (n = 448), 12 hr (n = 601), and 24 hr (n = 660). (E) Percentage of suspended human primary mammary epithelial cells (HMEpiCs) showing GUVac formation after incubation with DMSO or LatB for the indicated times. DMSO: 6 hr (n = 722), 12 hr (n = 647), 24 hr (n = 417). LatB: 6 hr (n = 1225), 12 hr (n = 1505), and 24 hr (n = 1335). (F) Percentage of suspended MCF-10A cells showing GUVac formation after incubation with the indicated drugs for 18 hr. DMSO (n = 1087), LatB (n = 1634), jasplakinolide (n = 2578), Rho inhibitor (n = 839), Y-27632 (n = 1322), EHT 1864 (n = 905), ML 141 (n = 997), nocodazole (n = 1342), and blebbistatin (n = 520). (G) Representative DIC microscopy images with phalloidin staining show disruption of the actin cytoskeleton and the GUVac formation in SpvB-expressing cells after suspension culture for 24 hr. Scale bar: 10 μm (H) Percentage of GUVac formation in control or SpvB-expressing MCF-10A cells. (I) Representative DIC microscopy images after suspension culture of indicated cell lines for 24 hr with LatB. Scale bar: 10 μm (J) Representative DIC images of suspended MCF-10A cells treated with DMSO or LatB for the indicated times. White arrowheads indicate the accumulation of vacuoles. Nuclei were stained with Hoechst 33342. Scale bar, 20 μm. (K) Representative fluorescence and DIC time-lapse images of MCF-10A cells expressing mCherry-H2B and membrane-targeted EGFP obtained at the indicated times after the onset of LatB treatment. Times are presented as hour:minute:second (h:m:s). Scale bars, 10 μm. All quantitative data are means ± SD. The n values represent the total number of cells quantified for two (D) or three (B, E, F, H) independent experiments. **p<0.01 (two-tailed unpaired t-test) for the indicated comparison (F, H) or versus the corresponding value for DMSO (E).
-
Figure 1—source data 1
Quantification data corresponding to Figure 1B.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Quantification data corresponding to Figure 1E.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Quantification data corresponding to Figure 1F.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig1-data3-v1.xlsx
-
Figure 1—source data 4
Quantification data corresponding to Figure 1H.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig1-data4-v1.xlsx
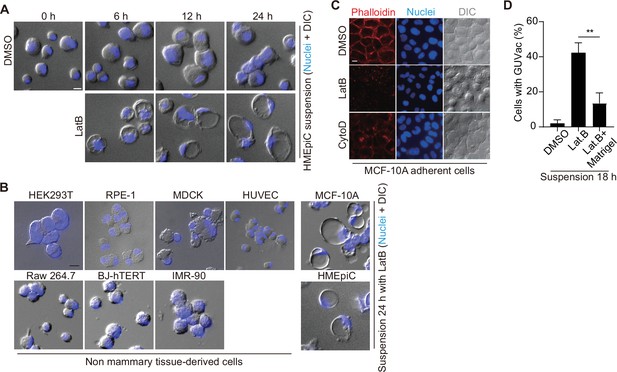
GUVac develops specifically in suspended cells that originate from secretory-related tissues.
(A) Representative differential interference contrast (DIC) images of suspended human primary mammary epithelial cells (HMEpiCs) treated with dimethyl sulfoxide (DMSO) or latrunculin B (LatB) for 0, 6, 12, or 24 hr. Nuclei were stained with Hoechst 33342. Scale bar, 10 μm. (B) Representative DIC images of suspended cells of the indicated cell lines after treatment with LatB for 24 hr. Nuclei were stained with Hoechst 33342. Scale bar, 10 μm. (C) Representative fluorescence images for F-actin (phalloidin staining) or nuclei (Hoechst 33342 staining) as well as DIC images of adherent MCF-10A cells treated with DMSO, LatB, or cytochalasin D (CytoD) for 12 hr. Scale bar, 10 μm. (D) Percentage of suspended MCF-10A cells showing giant unilocular vacuole (GUVac) formation after incubation for 18 hr with DMSO (n = 523), LatB (n = 571), or LatB plus 2.5% Matrigel (n = 587). Data are means ± SD for the indicated numbers of cells examined in three independent experiments. **p<0.01 (two-tailed unpaired t-test).
-
Figure 1—figure supplement 1—source data 1
Quantification data corresponding to Figure 1—figure supplement 1D.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig1-figsupp1-data1-v1.xlsx
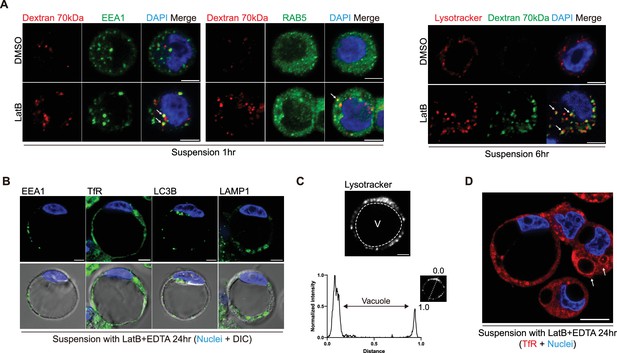
GUVac membranes do not consistently align with particular organelle markers.
(A) Representative fluorescence images with dextran 70 kDa and indicated organelle markers in suspended MCF-10A cells treated with dimethyl sulfoxide (DMSO) or latrunculin B (LatB) for 1 or 6 hr. Nuclei were stained with DAPI. Scale bar, 5 μm. Co-localization sites of two fluorescence signals are marked by arrowheads. (B) Representative fluorescence images stained with indicated organelle markers in suspended MCF-10A cells after treatment with LatB and EDTA for 24 hr. Nuclei were stained with DAPI. Scale bar, 5 μm. (C) Representative images of suspended cell with giant unilocular vacuole (GUVac) stained with LysoTracker. The profile on the bottom represents the distribution of fluorescence intensity along the indicated line. Scale bar, 5 μm. V, vacuole. (D) Representative fluorescence images for Transferrin receptor of suspended MCF-10A cells treated with LatB and EDTA for 24 hr. Scale bar, 10 μm.
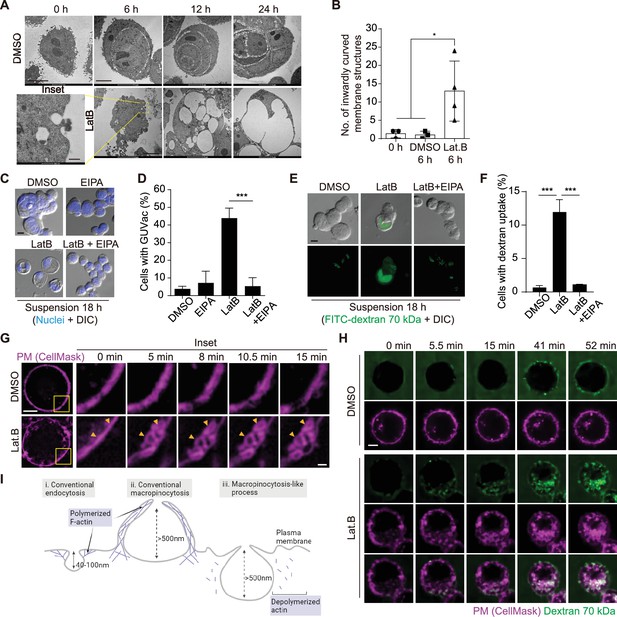
Macropinocytosis-like process contributes to giant unilocular vacuole (GUVac) formation.
(A) Representative transmission electron microscopy (TEM) images of suspended MCF-10A cells treated with dimethyl sulfoxide (DMSO) or latrunculin B (LatB) for the indicated times. Scale bars, 5 µm (main panels) or 500 nm (inset). (B) Number of inwardly curved plasma membrane structures with a diameter of >500 nm detected by TEM in individual suspended MCF-10A cells treated with DMSO or LatB for 6 hr. Each data point represents an individual cell analyzed. (C) Representative differential interference contrast (DIC) images of suspended MCF-10A cells treated with 5-(N-ethyl-N-isopropyl) amiloride (EIPA) or LatB for 18 hr. Nuclei were stained with Hoechst 33342. Scale bar, 20 μm. (D) Percentage of cells showing GUVac formation in experiments as in (C). DMSO (n = 1489), EIPA (n = 1256), LatB (n = 1482), and LatB + EIPA (n = 1159). (E) Representative DIC and FITC-dextran fluorescence images of suspended MCF-10A cells treated with LatB or EIPA for 18 hr. The cells were exposed to FITC–dextran (70 kDa) at 1 mg/ml. Scale bar, 20 μm. (F) Percentage of cells showing FITC-dextran uptake in experiments as in (E). DMSO (n = 797), LatB (n = 1534), and LatB + EIPA (n = 821). (G) Representative super-resolution structured illumination microscopy (SIM) time-lapse images of suspended MCF-10A cells showing the plasma membrane labeled by CellMask obtained at the indicated times after DMSO or LatB treatment. Arrowheads indicate plasma membrane invagination and vesicle formation. Scale bars, 5 μm (main panels) or 1 μm (inset). (H) Representative spinning disk confocal time-lapse images of suspended MCF-10A cells showing CellMask-labeled plasma membrane and 70 kDa dextran obtained at the indicated times after DMSO or LatB treatment. Scale bar, 5 μm. (I) Schematic showing the differences among conventional endocytosis, conventional macropinocytosis, and unconventional macropinocytosis-like process with regard to the size of endocytic cups and reliance on actin polymerization. Schematic was created using Biorender. All quantitative data are means ± SD. The n values represent the total number of cells examined in three independent experiments (D, F). *p<0.05, ****p<0.0001 by one-way ANOVA (B) or one-way ANOVA with Tukey’s multiple comparisons (D, F).
-
Figure 2—source data 1
Quantification data corresponding to Figure 2B.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Quantification data corresponding to Figure 2D.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Quantification data corresponding to Figure 2F.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig2-data3-v1.xlsx
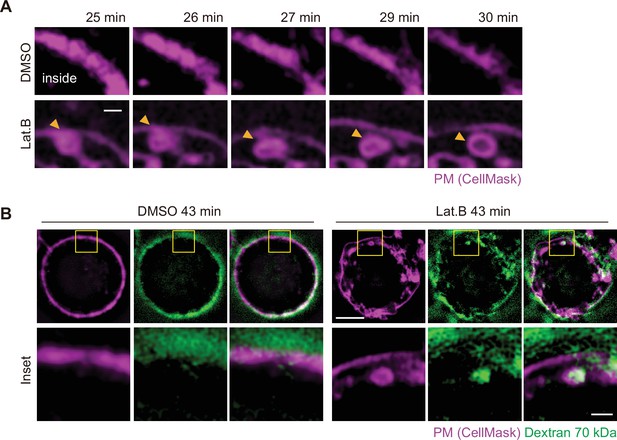
Colocalization of macropinosomes with the invaginated plasma membrane during early vacuole formation.
(A) Representative super-resolution structured illumination microscopy (SIM) time-lapse images of suspended MCF-10A cells showing CellMask-labeled plasma membrane obtained at the indicated times after dimethyl sulfoxide (DMSO) or latrunculin B (LatB) treatment. Arrowheads indicate vesicle formation at the plasma membrane. Scale bar, 1 μm. (B) Representative super-resolution SIM time-lapse images of suspended MCF-10A cells showing CellMask-labeled plasma membrane and 70 kDa dextran obtained at 43 min after DMSO or LatB treatment. Scale bars, 5 μm (main panels) or 1 μm (inset).
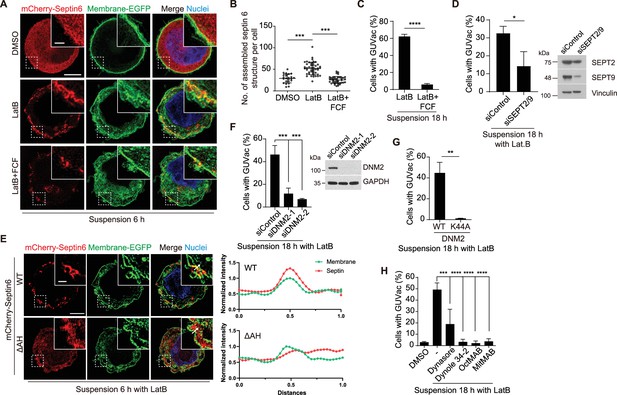
Recruitment of septin to the fluctuating plasma membrane drives macropinocytosis-like phenomenon.
(A) Representative super-resolution structured illumination microscopy (SIM) fluorescence images (maximum intensity projection) of MCF-10A cells expressing membrane-targeted EGFP and mCherry–septin 6 treated with indicated drugs for 6 hr. Scale bars, 5 μm (main panels) or 1 μm (inset). (B) Number of assembled mCherry-septin 6 structures (filaments and collar-like) per cell, quantified in experiments as in (A). Dimethyl sulfoxide (DMSO) (n = 24), latrunculin B (LatB) (n = 47), LatB + forchlorfenuron (FCF) (n = 41). Each data point represents an individual cell analyzed from two independent experiments. (C) Percentage of suspended MCF-10A cells showing giant unilocular vacuole (GUVac) formation after incubation with the indicated drugs for 18 hr. LatB (n = 783), LatB + FCF (n = 1016). (D) Percentage of GUVac formation in 18 hr suspended MCF-10A cells after co-depletion of Septin 2 and 9 or control knockdown. siControl (n = 1270) and siSEPT2/9 (n = 511). (E) Representative super-resolution SIM fluorescence images (maximum intensity projection) of MCF-10A cells expressing membrane-targeted EGFP and either mCherry-septin 6 WT or a mutant lacking the AH domain (ΔAH), which were suspended in the presence of LatB for 6 hr. Scale bars, 5 μm (main panels) or 1 μm (inset). The profiles on the right side represent the normalized intensity of each fluorescence signal along the indicated arrow from the inset of the merge panel. (F) Percentage of cells showing GUVac formation for MCF-10A cells that had been transfected with control or dynamin 2 siRNAs and treated with LatB in suspension culture for 18 hr (left panel). siControl (n = 746), siDNM2-1 (n = 822), and siDNM2-2 (n = 712). Immunoblot analysis of dynamin 2 in MCF-10A cells transfected with control or two independent dynamin 2 siRNAs (right panel). (G) Percentage of cells showing GUVac formation for MCF-10A cells expressing WT or dominant negative mutant (K44A) forms of dynamin 2 that had been suspended in the presence of LatB for 18. WT (n = 607) and K44A (n = 737). (H) Percentage of MCF-10A cells showing GUVac formation after suspension in the presence of the indicated inhibitors and incubation for 18 h. DMSO (n = 1423), LatB (n = 1151), LatB + Dynasore (n = 1407), LatB + Dynole 34-2 (n = 947), LatB + OctMAB (n = 699), and LatB + MitMAB (n = 1404). All quantitative data are means ± SD. The n values represent the total number of cells examined in two (B) or three independent experiments (C, D, F–H). **p<0.01, ***p<0.001, ****p<0.0001 by one-way ANOVA with Tukey’s multiple comparisons (B, F, H) or two-tailed unpaired t-test (C, D, G).
-
Figure 3—source data 1
Quantification data corresponding to Figure 3B.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Quantification data corresponding to Figure 3C.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data2-v1.xlsx
-
Figure 3—source data 3
Quantification data corresponding to Figure 3D.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Quantification data corresponding to Figure 3F.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data4-v1.xlsx
-
Figure 3—source data 5
Quantification data corresponding to Figure 3G.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data5-v1.xlsx
-
Figure 3—source data 6
Quantification data corresponding to Figure 3H.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data6-v1.xlsx
-
Figure 3—source data 7
Uncropped blot images with sample labeling used in Figure 3D.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data7-v1.zip
-
Figure 3—source data 8
Original blot images used in Figure 3D.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data8-v1.zip
-
Figure 3—source data 9
Uncropped blot images with sample labeling used in Figure 3F.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data9-v1.zip
-
Figure 3—source data 10
Original blot images used in Figure 3F.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig3-data10-v1.zip
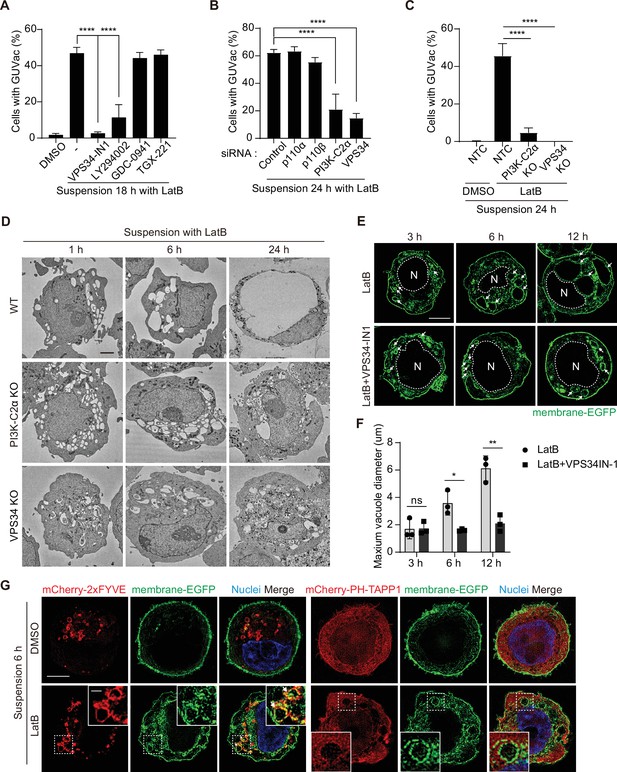
PI(3)P is required for vacuole fusion during giant unilocular vacuole (GUVac) formation.
(A) Percentage of suspended MCF-10A cells showing GUVac formation after treatment with the indicated drugs for 18 hr. Dimethyl sulfoxide (DMSO) (n = 1122), latrunculin B (LatB) (n = 1386), LatB + VPS34-IN1 (n = 989), LatB + LY294002 (n = 876), LatB + GDC-0941 (n = 787), and LatB + TGX-221 (n = 801). (B) Percentage of cells showing GUVac formation for MCF-10A cells transfected with the indicated siRNAs and suspended in the presence of LatB for 24 hr. siRNAs: control (n = 456), p110α (n = 718), p110β (n = 709), PI3K-C2α (n = 421), and VPS34 (n = 295). (C) Percentage of cells showing GUVac formation for nontargeting control (NTC), PI3K-C2α KO, or VPS34 KO MCF-10A cells suspended in the presence of DMSO or LatB for 24 hr. NTC/DMSO (n = 746), NTC/LatB (n = 730), PI3K-C2α KO/LatB (n = 824), and VPS34 KO/LatB (n = 939). (D) Representative transmission electron microscopy (TEM) images of suspended WT, PI3K-C2α KO, or VPS34 KO MCF-10A cells treated with LatB for the indicated times. Scale bars, 2 µm. (E) Representative super-resolution structured illumination microscopy (SIM) fluorescence images of MCF-10A cells expressing membrane-targeted EGFP that had been suspended in the presence of the indicated drugs for the indicated times. The nucleus is denoted by the letter ‘N’ and outlined with a dashed line. Arrows indicate vacuoles. Scale bars, 5 μm. (F) Maximum diameter of vacuoles in each cell for experiments as in (E). LatB: 3 hr (n = 29), 6 hr (n = 39), and 12 hr (n = 36). LatB + VPS34-IN1: 3 hr (n = 47), 6 hr (n = 43), and 12 hr (n = 39). (G) Representative super-resolution SIM fluorescence images of MCF-10A cells expressing membrane-targeted EGFP and either mCherry-2xFYVE or mCherry–PH-TAPP1 that had been suspended in the presence of DMSO or LatB for 6 hr. All quantitative data are means ± SD. The n values represent the total number of cells examined in three independent experiments (A–C, F). *p<0.05, **p<0.01, ****p<0.0001; ns, not significant by one-way ANOVA with Tukey’s multiple comparisons (A–C) or two-tailed unpaired t-test (F).
-
Figure 4—source data 1
Quantification data corresponding to Figure 4A.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Quantification data corresponding to Figure 4B.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Quantification data corresponding to Figure 4F.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-data3-v1.xlsx
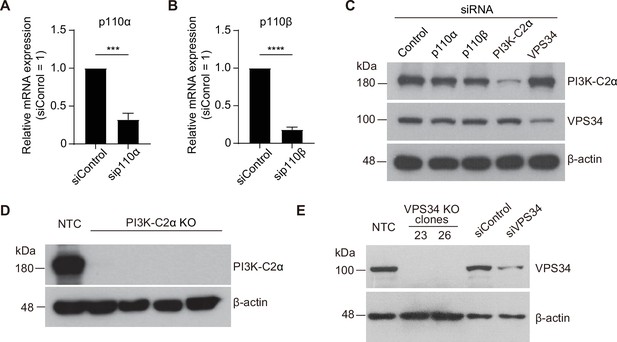
Verification of PI3K knockdown or knockout in MCF-10A cell.
(A) Quantitative RT-PCR analysis of relative p110α mRNA abundance in MCF-10A cells transfected with control or p110α siRNAs. Data are means ± SD from three independent experiments. ***p<0.001 (two-tailed unpaired t-test). (B) Quantitative RT-PCR analysis of relative p110β mRNA abundance in MCF-10A cells transfected with control or p110β siRNAs. Data are means ± SD from three independent experiments. ****p<0.0001 (two-tailed unpaired t-test). (C) Immunoblot analysis of PI3K-C2α and VPS34 in MCF-10A cells transfected with the indicated siRNAs. β-actin was examined as a loading control. (D) Immunoblot analysis of PI3K-C2α in nontargeting control (NTC) and PI3K-C2α KO MCF-10A cell clones. (E) Immunoblot analysis of VPS34 in VPS34 KO MCF-10A clones and siRNA KD cells.
-
Figure 4—figure supplement 1—source data 1
Uncropped blot images with sample labeling used in Figure 4—figure supplement 1C.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original blot images used in Figure 4—figure supplement 1C.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-figsupp1-data2-v1.zip
-
Figure 4—figure supplement 1—source data 3
Uncropped blot images with sample labeling used in Figure 4—figure supplement 1D.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-figsupp1-data3-v1.zip
-
Figure 4—figure supplement 1—source data 4
Original blot images used in Figure 4—figure supplement 1D.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-figsupp1-data4-v1.zip
-
Figure 4—figure supplement 1—source data 5
Uncropped blot images with sample labeling used in Figure 4—figure supplement 1E.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-figsupp1-data5-v1.zip
-
Figure 4—figure supplement 1—source data 6
Original blot images used in Figure 4—figure supplement 1E.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig4-figsupp1-data6-v1.zip
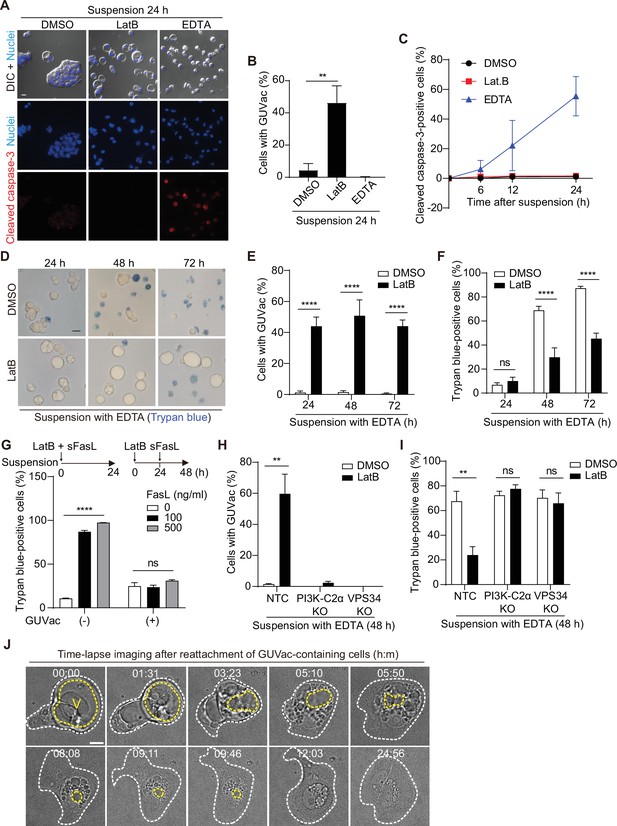
Giant unilocular vacuole (GUVac) formation promotes cell survival in altered actin and matrix environments.
(A–C) MCF-10A cells were suspended in the presence of latrunculin B (LatB) or EDTA for the indicated times and then immunostained for cleaved caspase-3. Nuclei were stained with Hoechst 33342. Representative differential interference contrast (DIC) and fluorescence images at 24 hr (scale bar, 20 μm) (A), the percentage of cells positive for GUVac formation at 24 hr (B), and the time course for the percentage of cells positive for cleaved caspase-3 (C) are shown. Dimethyl sulfoxide (DMSO): 0 hr (n = 1030), 6 hr (n = 1742), 12 hr (n = 1423), and 24 hr (n = 1273). LatB: 6 hr (n = 1479), 12 hr (n = 1508), and 24 hr (n = 1443). EDTA: 6 hr (n = 1976), 12 hr (n = 1578), and 24 hr (n = 1296). (D–F) MCF-10A cells were suspended with EDTA and in the presence of DMSO or LatB for the indicated times and then stained with trypan blue. Representative bright-field images (scale bar, 20 μm) (D) as well as the percentage of cells positive for GUVac formation (E) and the percentage of trypan blue-positive cells (F) are shown. DMSO: 24 hr (n = 1003), 48 hr (n = 797), and 72 hr (n = 943). LatB: 24 hr (n = 1473), 48 hr (n = 2100), and 72 hr (n = 1350). (G) Percentage of trypan blue-positive cells for suspended MCF-10A cells that either were simultaneously treated with both LatB and the indicated concentrations of sFasL for 24 hr (a condition under which cell death signaling precedes GUVac formation) or were treated with LatB for 24 hr and then incubated in the presence of sFasL for 24 hr (a condition under which GUVac formation precedes cell death signaling). GUVac(–): sFasL at 0 ng/ml (n = 1193), 100 ng/ml (n = 1441), or 500 ng/ml (n = 994). GUVac(+): sFasL at 0 ng/ml (n = 1240), 100 ng/ml (n = 1418), or 500 ng/ml (n = 1529). (H, I) Nontargeting control (NTC), PI3K-C2α KO, and VPS34 KO MCF-10A cells were suspended with EDTA in the presence of DMSO or LatB for 48 hr, after which the percentage of cells showing GUVac formation (H) and the percentage of trypan blue-positive cells (I) were determined. NTC: DMSO (n = 927), LatB (n = 1369). PI3K-C2α KO: DMSO (n = 627), LatB (n = 883). VPS34 KO: DMSO (n = 1026), and LatB (n = 885). (J) Time-lapse bright-field images of cells with GUVacs after matrix reattachment. MCF-10A cells were suspended in the presence of LatB for 24 hr, washed to eliminate LatB, and then cultured in an adhesive confocal dish for 14 hr before imaging for the indicated times (hour:minute). The yellow dotted line represents the largest vacuole in each cell, while the white dotted line indicates the spread cell area. Scale bar, 10 μm. All quantitative data are means ± SD. The n values represent the total number of cells examined in three (B, C, G–I) or four (E, F) independent experiments. **p<0.01, ****p<0.0001; ns, not significant (two-tailed unpaired t-test). V, vacuole.
-
Figure 5—source data 1
Quantification data corresponding to Figure 5B and C.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Quantification data corresponding to Figure 5E and F.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig5-data2-v1.xlsx
-
Figure 5—source data 3
Quantification data corresponding to Figure 5G.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig5-data3-v1.xlsx
-
Figure 5—source data 4
Quantification data corresponding to Figure 5H and I.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig5-data4-v1.xlsx
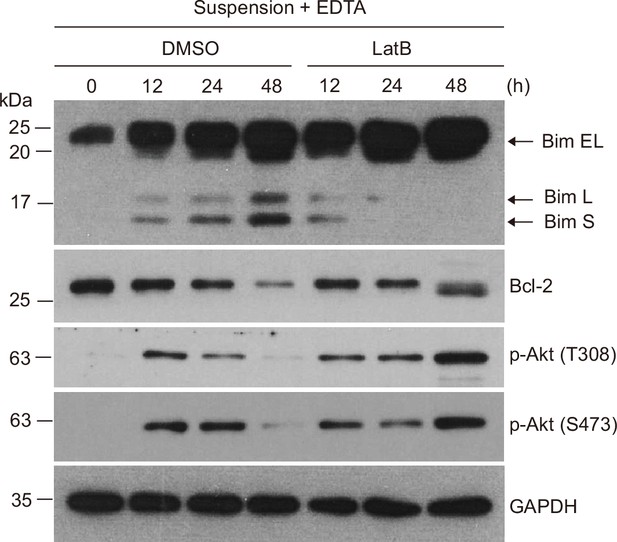
Immunoblot analysis of survival signaling in MCF-10A cells suspended with EDTA and in the presence of dimethyl sulfoxide (DMSO) or latrunculin B (LatB) for the indicated times.
-
Figure 5—figure supplement 1—source data 1
Uncropped blot images with sample labeling used in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Original blot images used in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/96178/elife-96178-fig5-figsupp1-data2-v1.zip
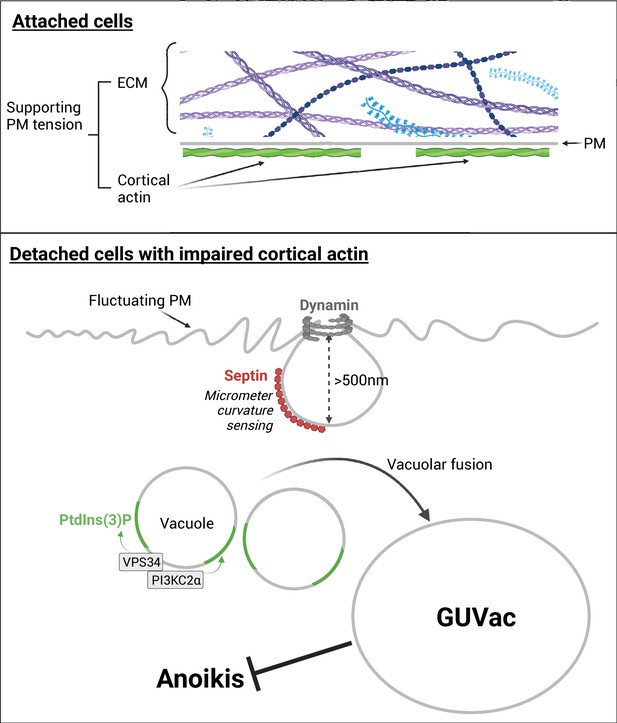
Hypothetical model for the formation of giant unilocular vacuole (GUVac) that enhances the anoikis resistance.
In matrix-attached cells, cortical actin and extracellular matrix (ECM) attachment suppresses plasma membrane (PM) fluctuation and maintains membrane tension. However, in matrix-detached cells with disrupted cortical actin, the PM exhibits micron-scale membrane fluctuations. This promotes the recruitment of septin to the micrometer-sized inwardly curved plasma membrane via its amphipathic helix (AH) domain and the subsequent recruitment further accelerates the PM invagination. As a result of this macropinocytosis-like process, followed by dynamin-mediated pinching off, vacuoles accumulate within the cell and gradually coalesce, a process facilitated by the synthesis of PI(3)P, involving the activity of VPS34 and PI3K-C2α. Eventually, these processes lead to the formation of GUVac. Cells that possess GUVac demonstrate resistance to anoikis. The schematic was generated using Biorender.





