Rab11 suppresses neuronal stress signaling by localizing dual leucine zipper kinase to axon terminals for protein turnover
Figures
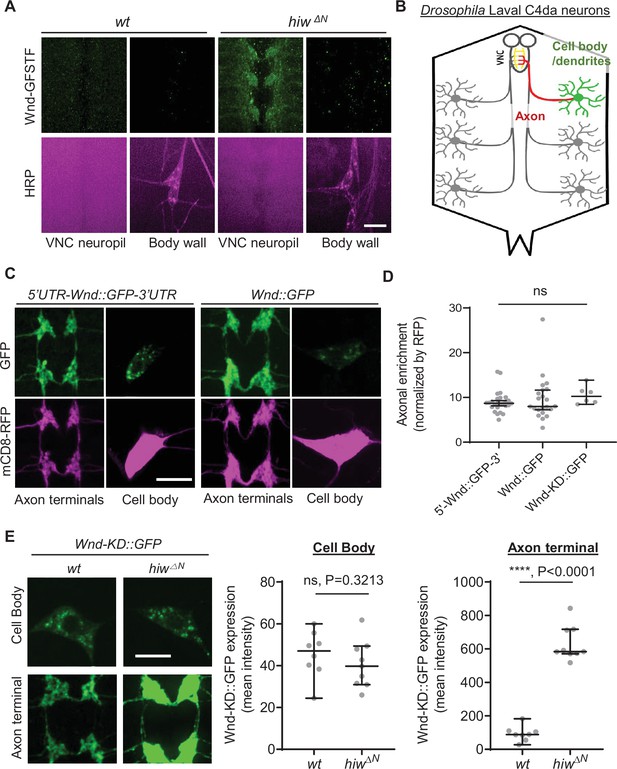
Wallenda (Wnd) protein turnover by Highwire (Hiw) is restricted in the axon terminals of Drosophila sensory neurons.
Wnd is highly enriched in axon terminals (A–D). (A) A representative image of Mi{MIC}wnd GFSTF in the neuropil of larval ventral nerve cords (top) and C4da neuron cell body (bottom) from wild-type male (w1118) or hemizygous male hiw mutant (hiw△N) were shown. Wnd-GFP (Wnd-GFSTF, green) and anti-HRP staining (magenta). Scale bar = 10 µm. (B) Schematic of Drosophila larval C4da neurons. C4da cell bodies and dendrites are located in the body wall (green) while their axons (red) terminate in the ventral nerve cord (VNC) collectively forming the ladder-like structure of axon terminals (yellow). (C) The Wnd::GFP (with or without wnd-UTRs, green) along with mCD8::RFP (magenta) were expressed in the larval C4da neurons in the presence of a dual leucine zipper kinase (DLK) inhibitor, DLKi. The expression levels of transgenes were shown in C4da cell body and the axon terminals. (D) The axonal enrichment index was expressed as median ± 95% CI. Sample numbers were 5’-Wnd::GFP-3’ (w1118; UAS-wnd-5’UTR-Wnd::GFP-wnd-3’UTR/+;ppk-GAL4, UAS-mCD8::RFP/+) (n=26), Wnd::GFP (w1118; UAS-Wnd::GFP-SV40 3’UTR /+; ppk-GAL4, UAS-mCD8::RFP/+) (n=22), Wnd-KD::GFP (w1118; UAS-Wnd-KD::GFP/+; ppk-GAL4, UAS-mCD8::RFP/+) (n=6). All samples were treated with DLKi; One-way ANOVA (F(2,51) = 0.5176) followed by post hoc Tukey’s multiple comparison test. (E) The Wnd-KD::GFP transgene along with mCD8::RFP was expressed in C4da neurons with the ppk-Gal4 driver, in wild-type (wt) (w1118;UAS-Wnd-KD::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, sample n=8) or in hiw mutant (hiw△N; UAS-Wnd-KD::GFP/+;ppk-Gal4,UAS-mCD8::RFP/+, sample n=9) larvae. Representative images of Wnd-KD::GFP in C4da cell bodies and axon terminals, stained for anti-GFP (green). The average Wnd-KD::GFP intensity from cell bodies and axon terminals were quantified and expressed as median ± 95% CI; (Cell body: U=25, p=0.3213, Axon terminal: U=0, p<0.0001, two-tailed Mann-Whitney test). Scale bar = 10 µm.
-
Figure 1—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig1-data1-v1.xlsx
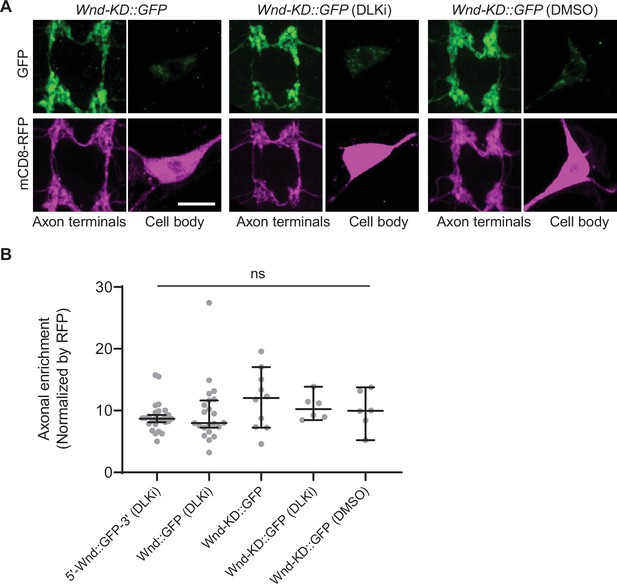
The axon terminal enrichment of Wallenda (Wnd) is independent of Wnd kinase activity.
(A) A Wnd kinase-dead (Wnd-KD::GFP, green) along with mCD8::RFP (magenta) were expressed in the larval C4da neurons in either regular food, food containing DMSO (vehicle control) or DLKi. The expression levels of transgenes were shown in C4da cell body and the axon terminals. Scale bar = 10 µm. (B) The axonal enrichment index was expressed as median± 95%CI. Genotypes and sample numbers were 5’-Wnd::GFP-3’ (w1118; UAS-wnd-5’UTR-Wnd::GFP-wnd-3’UTR/+;ppk-GAL4, UAS-mCD8::RFP/+) with DLKi (n=26), Wnd::GFP (w1118; UAS-Wnd::GFP-SV40 3’UTR /+; ppk-GAL4, UAS-mCD8::RFP/+) with DLKi (n=22), Wnd-KD::GFP (w1118; UAS-Wnd-KD::GFP/+; ppk-GAL4, UAS-mCD8::RFP/+) (n=10), with DLKi (n=6), with DMSO (n=6); One-way ANOVA (F(4,65) = 1.017) followed by post hoc Tukey’s multiple comparison test.
-
Figure 1—figure supplement 1—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig1-figsupp1-data1-v1.xlsx
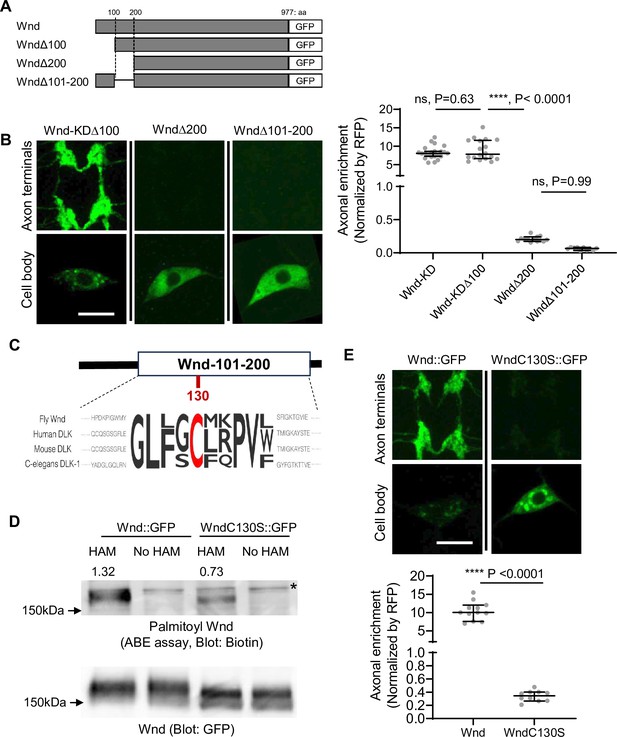
Wallenda (Wnd) palmitoylation is essential for Wnd axonal localization.
(A) Schematic of Wnd deletion mutants. Deletion of the N-terminal 1-100aa of Wnd (Wnd∆100), Deletion of the N-terminal 1–200 aa of Wnd (Wnd∆200), and Deletion of the N-terminal 101–200 aa of Wnd (Wnd∆101–200). (B) The transgenes of serially deleted Wnd (green) along with mCD8::mRFP were expressed in the larval C4da neurons. Note that Wnd∆100 was further mutated to remove kinase activity (Wnd-KD∆100). The expression levels of Wnd::GFP proteins in C4da cell body and the axon terminals were shown (left). Scale bars = 10 µm. The axonal enrichment of each Wnd transgenes was measured using mCD8::RFP (not shown in figure) as a normalization control and expressed as median ± 95% CI. One-way ANOVA (F(3,59) = 103.8) followed by post hoc Tukey’s multiple comparison test. The p-values from Tukey’s test are indicated in the graph. Genotypes and sample numbers were Wnd-KD (UAS-Wnd-KD::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=22), Wnd-KD∆100 (UAS-Wnd-KD∆100::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=18), Wnd∆200 (UAS-Wnd∆200::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=13), and Wnd∆101–200 (UAS-Wnd∆101–200::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=10). (C) Wnd-C130 is an evolutionarily conserved palmitoylation site. The protein sequence alignment of DLK proteins from the indicated species is shown. The conserved Cysteine residue is shown in red. (D) GFP-tagged Wnd proteins were expressed in S2 cells before being pulled down by GFP nanobody beads. The Acyl Biotin Exchange assay was performed on the beads coupled Wnd proteins. Wnd-C130S::GFP is a palmitoylation defective version of Wnd. ‘No HAM’ serves as a negative control since Biotin exchange does not occur in the absence of HAM. Biotin blot shows protein palmitoylation levels while total Wnd shows Wnd protein levels in the pulldown. The numbers indicate the total intensity of the palmitoylation blot that were normalized by GFP blot from Wnd::GFP and Wnd-C130S::GFP. (E) A wild-type Wnd::GFP transgene (Wnd::GFP) and a palmitoylation-defective Wnd transgene (Wnd-C130S::GFP) along with mCD8::RFP were expressed in the larval C4da neurons in the presence of DLKi. GFP immunostaining from Wnd::GFP proteins in C4da cell body and the axon terminals were shown (top). Scale bar = 10 µm. The axonal enrichment of the Wnd transgenes was measured using mCD8::RFP (not shown in figure) as a normalization control and expressed as median ± 95% CI (U=0, p<0.0001, two-tailed Mann-Whitney test). Genotypes and samples numbers were Wnd::GFP (UAS-Wnd::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=12) and Wnd-C130S::GFP (UAS-WndC130S::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=10).
-
Figure 2—source data 1
The original western blots for Figure 2D.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig2-data1-v1.zip
-
Figure 2—source data 2
The original western blots for Figure 2D with relevant bands labeled.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig2-data2-v1.zip
-
Figure 2—source data 3
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig2-data3-v1.xlsx
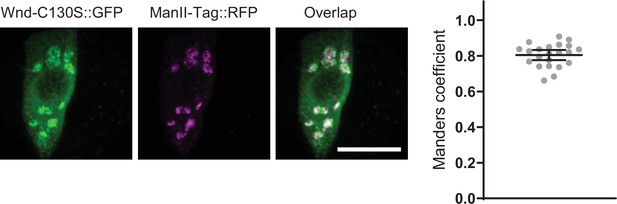
Wallenda (Wnd) palmitoylation is not required for Wnd localization in somatic Golgi complex.
Wnd-C130S::GFP (green) along with a Golgi marker, ManII-Tag::RFP (magenta) were expressed in the larval C4da neurons. The expression patterns of the two transgenes in C4da cell bodies were shown (left). Scale bar = 10 µm. The Manders’ overlap coefficient between Wnd transgenes and ManII-Tag::RFP was measured and expressed as median ± 95% CI (right). The Genotype and sample numbers were Wnd-C130S::GFP (w1118; UAS-Wnd-C130S-KD::GFP/+;ppk-GAL4/UAS-ManII-Tag-RFP, n=22).
-
Figure 2—figure supplement 1—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig2-figsupp1-data1-v1.xlsx
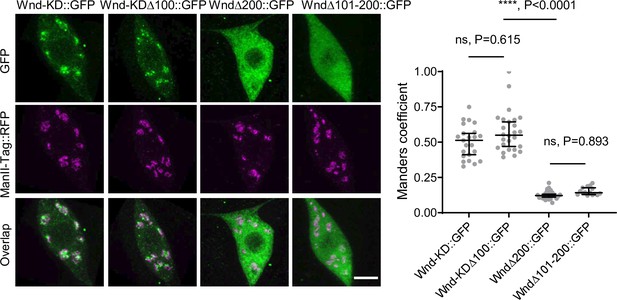
Somatic Golgi localization is necessary for Wallenda (Wnd) localization in axon terminals.
Wnd transgenes (green) along with a Golgi marker, ManII-Tag::RFP (magenta) were expressed in the larval C4da neurons. The expression patterns of the two transgenes in C4da cell bodies were shown (left). Scale bar = 5 µm. The Manders’ overlap coefficient between Wnd transgenes and ManII-Tag::RFP was measured and expressed as median ± 95% CI (right). Genotypes and sample numbers were Wnd-KD::GFP (w1118; UAS-Wnd-KD::GFP/+;ppk-GAL4/UAS-ManII-Tag-RFP, n=23), Wnd-KD∆100::GFP (w1118; UAS-Wnd-∆100-KD::GFP/+;ppk-GAL4/UAS-ManII-Tag-RFP, n=26), Wnd-∆200::GFP (w1118;UAS-Wnd-∆200::GFP/+;ppk-GAL4/UAS-ManII-Tag-RFP, n=28) and Wnd-∆101–200::GFP (w1118;UAS-Wnd∆101–200::GFP/+;ppk-GAL4/UAS-ManII-Tag-RFP, n=15). One-way ANOVA (F(3,88) = 129.8) followed by post hoc Tukey’s multiple comparison test. The p-values from Tukey’s test are indicated in the graph.
-
Figure 2—figure supplement 2—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig2-figsupp2-data1-v1.xlsx
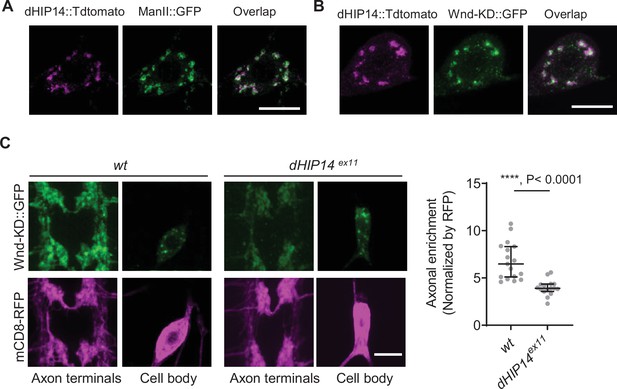
dHIP14 is required for Wallenda (Wnd) localization in axon terminals.
(A) A Golgi marker, ManII::GFP (green) was expressed in the larval C4da neurons along with dHIP14::Tdtomato (magenta). The image was taken from a C4da cell body. Scale bar = 10 µm. (B) Wnd-KD::GFP (green) was expressed in the larval C4da neurons along with dHIP14::Tdtomato (magenta). The image was taken from a C4da cell body. Scale bar = 10 µm. (C) Wnd-KD::GFP (green) along with mCD8::RFP (magenta) were expressed in the larval C4da neurons from wild-type (wt) or homozygous dHIP14 mutants (dHIP14ex11). Scale bar = 10 µm. The axonal enrichment of Wnd-KD::GFP was measured and expressed as a median ± 95% CI (Mann-Whitney U=13, p<0.0001, two-tailed Mann-Whitney test). Genotypes and sample numbers were wt (UAS-Wnd-KD::GFP/ppk-Gal4, U-mCD8::RFP;+/+, n=17) and dHIP14ex11(UAS-Wnd-KD::GFP/ppk-Gal4, U-mCD8::RFP;dHIP14ex11, n=16).
-
Figure 2—figure supplement 3—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig2-figsupp3-data1-v1.xlsx
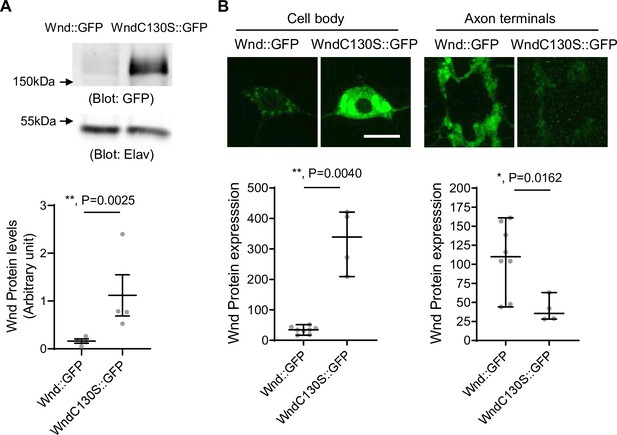
Increased protein expression levels of palmitoylation-defective Wallenda (Wnd).
(A) Wnd::GFP and Wnd-C130S::GFP were expressed in the larval nervous system using a pan-neuronal driver, Elav-GAL4 in the presence of DLKi. Larval brain lysates were subjected to western blot analysis using a GFP antibody. Elav blot was used as a loading control. The GFP blots were normalized by Elav blots and expressed as mean ± SEM (n=4, t=9.409, df = 3, p=0.0025, two-tailed ratio paired t-test). Genotypes were Wnd::GFP (elav-Gal4/+; UAS-Wnd::GFP/+;) and WndC130S::GFP (elav-Gal4/+; UAS-WndC130S::GFP/+;). (B) Wnd transgenes (green) along with mCD8::RFP were expressed in the larval C4da neurons to directly compare the expression levels of Wnd::GFP and Wnd-C130S::GFP in C4da cell bodies and axon terminals. The larvae were treated with DLKi. Scale bar = 10 µm. The average fluorescent intensities from GFP staining of each larva sample (C4da cell bodies and axon terminals) were measured and expressed as median ± 95% CI (Cell body: Mann-Whitney U=0, p=0.0040, two-tailed Mann-Whitney test, Axon terminal: Mann-Whitney U=2, p=0.0162, two-tailed Mann-Whitney test,). Genotypes and sample numbers were Wnd::GFP (UAS-Wnd::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=8) and Wnd-C130S::GFP (UAS-WndC130S::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+, n=4).
-
Figure 3—source data 1
The original western blots for Figure 3A.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig3-data1-v1.zip
-
Figure 3—source data 2
The original western blots for Figure 3A with relevant bands labeled.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig3-data2-v1.zip
-
Figure 3—source data 3
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig3-data3-v1.xlsx

Palmitoylation-defective Wallenda (Wnd) caused exacerbated stress responses.
(A) puc-LacZ was expressed along with 2XUAS-Wnd::GFP and 2XUAS-Wnd-C130S::GFP in the larval C4da neurons. The average fluorescent intensities from puc-LacZ staining of each larva sample in C4da cell body were measured and expressed as median ± 95% CI (Mann-Whitney U=56, p<0.0001, two-tailed Mann-Whitney test). Genotypes and sample numbers were 2XUAS-Wnd::GFP (2XUAS-Wnd::GFP/ppk-Gal4,UAS-mCD8::RFP;puc-LacZ/+, n=28) and 2XUAS-Wnd-C130S::GFP (2XUAS-Wnd-C130S::GFP/ ppk-Gal4,UAS-mCD8::RFP;puc-LacZ/+, n=28). Scale bar = 10 µm. (B) 2XUAS-Wnd::GFP and 2XUAS-Wnd-C130S::GFP were expressed under an eye-specific driver, GMR-GAL4. Adult fly lethality was scored and analyzed using the Chi-square (Chi-square, df = 41.57,1, z=6.448, p<0.0001, two-sided contingency test). Genotypes and sample numbers were Wnd::GFP (2XUAS-Wnd::GFP/GMR-Gal4, n=188), Wnd-C130S::GFP (2XUAS-Wnd-C130S::GFP/GMR-Gal4, n=195). (C) Representative images of fly eyes from Wnd::GFP (2XUAS-Wnd::GFP/+), Wnd-C130S::GFP (2XUAS-Wnd-C130S::GFP/+), Wnd::GFP under GMR-GAL4 (2XUAS-Wnd::GFP/GMR-Gal4), Wnd-C130S::GFP under GMR-GAL4 (2XUAS-Wnd-C130S::GFP/GMR-Gal4).
-
Figure 4—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig4-data1-v1.xlsx
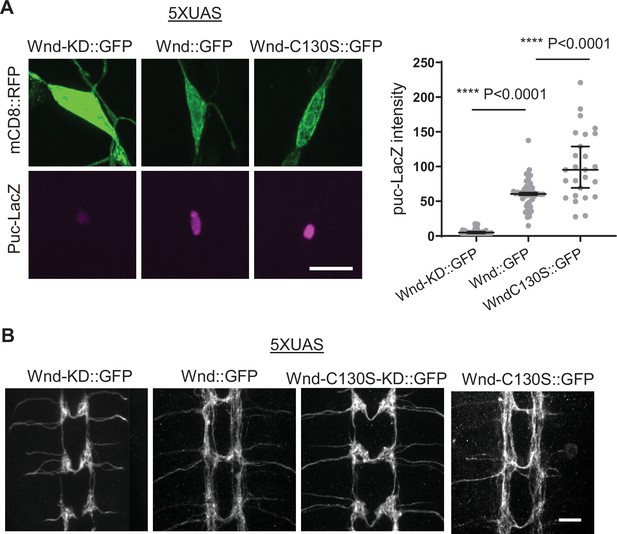
Wallenda (Wnd)-C130S::GFP retains Wnd signaling capacity.
(A) puc-LacZ was expressed along with 5XUAS-Wnd-KD::GFP, 5XUAS-Wnd::GFP, and 5XUAS-Wnd-C130S::GFP in the larval C4da neurons. The average fluorescent intensities from puc-LacZ staining of each larva sample in the C4da cell body were measured and expressed as median ± 95% CI (One-way ANOVA, F (2, 114)=108.3). Genotypes and sample numbers were 5XUAS-Wnd-KD::GFP (;ppk-Gal4,UAS-mCD8::RFP/5XUAS-Wnd-KD::GFP;puc-LacZ/+, n=42), 5XUAS-Wnd::GFP (;ppk-Gal4,UAS-mCD8::RFP/5XUAS-Wnd::GFP;puc-LacZ/+, n=49), and 5XUAS-Wnd-C130S::GFP (;ppk-Gal4,UAS-mCD8::RFP/5XUAS-Wnd-C130S::GFP;puc-LacZ/+, n=26) Scale bar = 10 µm. (B) Wnd transgenes along with mCD8::RFP were expressed in the larval C4da neurons using ppk-GAL4. The axon terminal morphology was visualized by mCD8::RFP. Wnd-KD::GFP (UAS-Wnd-KD::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+), Wnd::GFP(UAS-Wnd::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+), WndC130S::GFP (UAS-WndC130S::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+), and WndC130S-KD::GFP (UAS-WndC130S-KD::GFP/+;ppk-Gal4, UAS-mCD8::RFP/+). Scale bar = 10 µm.
-
Figure 4—figure supplement 1—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig4-figsupp1-data1-v1.xlsx
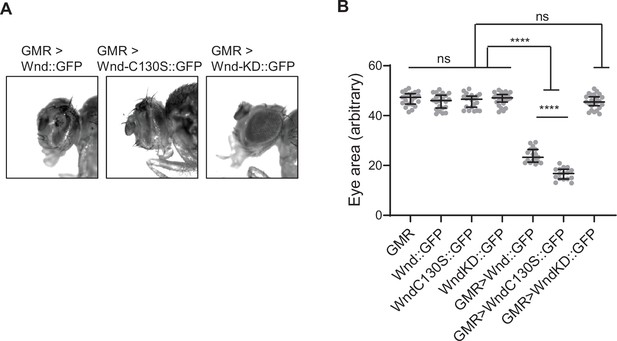
Wallenda (Wnd)-C130S::GFP exhibits higher toxicity levels than Wnd::GFP.
(A) Representative images of fly eyes from Wnd::GFP under GMR-GAL4 (GMR >Wnd::GFP), Wnd-C130S::GFP under GMR-GAL4 (GMR >Wnd-C130::GFP), and Wnd-KD::GFP under GMR-GAL4 (GMR >Wnd-KD::GFP). Flies were reared at 18 ºC. (B) Eye area was quantified and expressed as median ± 95% CI. One-way ANOVA (F(6, 136)=362.0) followed by post hoc Tukey’s multiple comparison test (****p<0.0001). Genotypes and sample numbers were GMR (GMR-GAL4/+, n=21), Wnd::GFP (UAS-Wnd::GFP/+, n=22), Wnd-C130S::GFP (UAS-Wnd-C130S::GFP/+, n=20), Wnd-KD::GFP (UAS-Wnd-KD::GFP/+, n=24), GMR >Wnd::GFP (GMR-GAL4/UAS-Wnd::GFP, n=17), GMR >Wnd-C130S::GFP (GMR-GAL4/UAS-Wnd-C130S::GFP, n=16), and GMR >Wnd- KD::GFP (GMR-GAL4/UAS-Wnd-KD::GFP, n=23). Note more severe eye phenotypes in GMR >Wnd-C130S::GFP as compared to GMR >Wnd::GFP.
-
Figure 4—figure supplement 2—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig4-figsupp2-data1-v1.xlsx
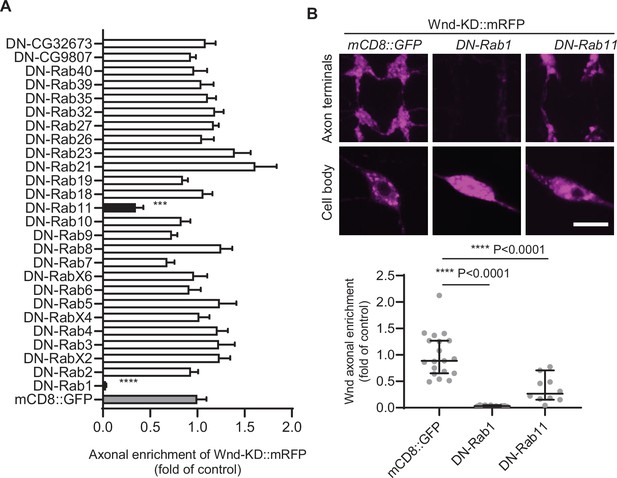
Screening of dominant negative Rab proteins for Wallenda (Wnd) axonal enrichment.
(A) YFP-tagged dominant negative-Rab (DN-Rab) transgenes along with Wnd-KD::mRFP were expressed in the larval C4da neurons. The axonal enrichment of Wnd-KD::mRFP was measured by Wnd-KD::mRFP expression levels from C4da cell bodies and axon terminals and expressed as mean ± sem of a fold change of control (mCD8::GFP) (n≥10). One-way ANOVA (F(27,284) = 7.146) followed by post hoc Dunnett test. A mCD8::GFP transgene was used as a transgene control (UAS-Wnd-KD::mRFP/+;ppk-Gal4, UAS-mCD8::GFP/+). Note that YFP::DN-Rab1 and YFP::DN-Rab11 reduced Wnd axonal enrichment by more than 50% compared to the control (gray bar). (B) Representative images of Wnd-KD::mRFP expression in the axon terminals and cell bodies from the C4da neurons that express mCD8::GFP, DN-Rab1, and DN-Rab11 were shown. The Wnd axonal enrichment was expressed as median ± 95% CI (Mann-Whitney U=0, p<0.0001 for DN-Rab1, U=13, p<0.0001 for DN-Rab11, two-tailed Mann-Whitney test). Genotypes and sample numbers were mCD8::GFP (UAS-Wnd-KD::mRFP/+;ppk-Gal4, UAS-mCD8::GFP/+, n=19) DN-Rab1 (UAS-Wnd-KD::mRFP/+;ppk-Gal4 /UAST YFP.Rab1 S25N, n=12) and DN-Rab11 (UAS-Wnd-KD::mRFP/ UASp-YFP.Rab11 S25N; ppk-Gal4/+, n=10) Scale bar = 10 µm.
-
Figure 5—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig5-data1-v1.xlsx
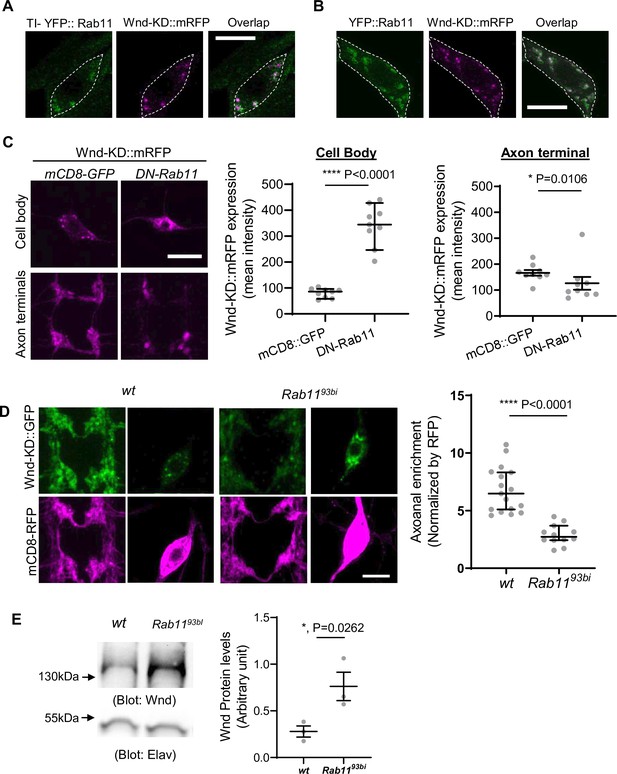
Rab11 is necessary for Wallenda (Wnd) localization in axon terminals and Wnd protein turnover.
(A) and (B). Wnd localizes at the Rab11-positive endosomes in C4da cell bodies. Wnd-KD::mRFP was expressed in C4da neurons of TI-YFP::Rab11 larvae (A). TI-YFP::Rab11 is endogenously YFP-(Yellow fluorescent protein) tagged Rab11 allele. Wnd-KD::mRFP was expressed in C4da neurons along with YFP::Rab11 (B). Wnd-KD::mRFP (magenta) and YFP::Rab11 (green) were visualized in C4da cell bodies using an RFP and GFP antibodies, respectively. Genotypes and sample numbers were TI-YFP::Rab11 (UAS-Wnd-KD::mRFP/+;TI{TI}Rab11[EYFP]/ppk-Gal4, n=28) and YFP::Rab11 (UAS-Wnd-KD::mRFP/UAS-YFP::Rab11;ppk-Gal4/+;, n=30). Scale bar = 10 µm. (C) Wnd transgenes (magenta) along with mCD8::GFP or YFP::DN-Rab11 were expressed in the larval C4da neurons to directly compare the expression levels of Wnd-KD::mRFP in C4da cell bodies and axon terminals. Scale bar = 10 µm. The average fluorescent intensities from mRFP staining of each larva sample were measured and expressed as median ± 95% CI (Cell body: Mann-Whitney U=0, p<0.0001, two-tailed Mann-Whitney test; Axon terminal: Mann-Whitney U=12, p=0.0160, two-tailed Mann-Whitney test). Genotypes and sample numbers were wt (UAS-Wnd-KD::mRFP/+;ppk-Gal4, UAS-mCD8::GFP/+, n=9) and YFP::DN-Rab11 (UAS-Wnd-KD::mRFP/UASp-YFP.Rab11.S25N;ppk-Gal4, UAS-mCD8::RFP/+, n=9). (D) Wnd-KD::GFP (green) along with mCD8::RFP (magenta) were expressed in the larval C4da neurons from wild-type (wt) or from homozygous Rab11 mutants (Rab1193bi). Images from C4da axon terminals and cell bodies were shown. Scale bar = 10 µm. The axonal enrichment of Wnd-KD::GFP was measured using mCD8::RFP as a normalization control and expressed as median ± 95% CI (Mann-Whitney U=0, p<0.0001, two-tailed Mann-Whitney test). Genotypes and sample numbers were wt (UAS-Wnd-KD::GFP/ppk-Gal4, U-mCD8::RFP, n=17) and Rab1193bi (UAS-Wnd-KD::GFP/ppk-Gal4, U-mCD8::RFP; Rab1193bi, n=12). (E) A western blot was performed on the larval brain lysates from wild-type (wt, w1118) or homozygous Rab1193bi using Wnd antibody to measure total Wnd protein levels. Elav blot was used as a loading control. The Wnd blots were normalized by Elav blots and expressed as mean ± SEM (n=3, t=6.06, df = 2, p=0.0262, two-tailed ratio paired t-test).
-
Figure 6—source data 1
The original western blots for Figure 6E.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig6-data1-v1.zip
-
Figure 6—source data 2
The original western blots for Figure 6E with relevant bands labeled.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig6-data2-v1.zip
-
Figure 6—source data 3
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig6-data3-v1.xlsx
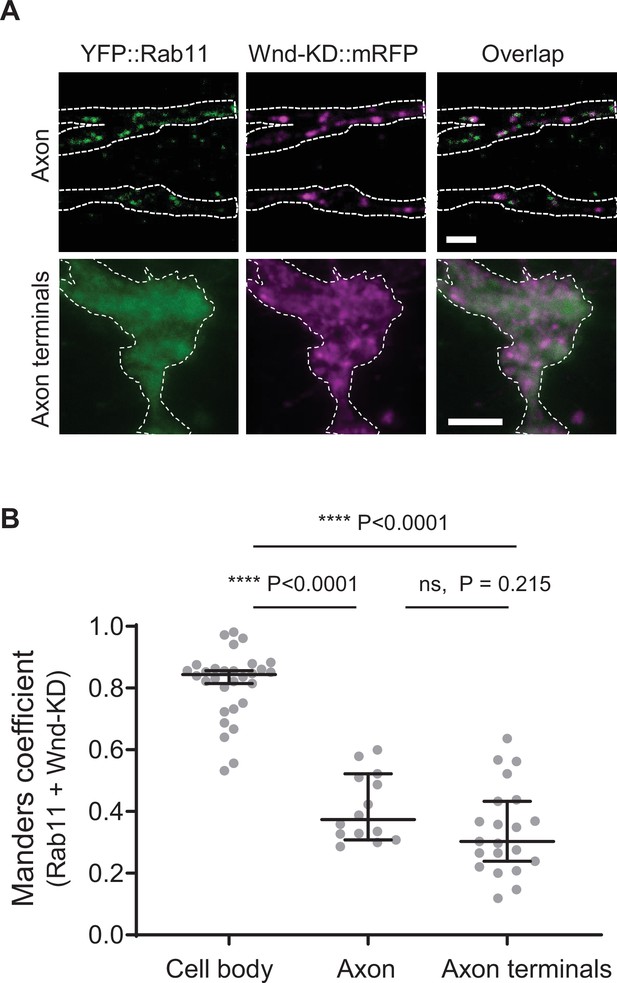
Wallenda (Wnd) does not localize in the Rab11-positive endosomes in axons and axon terminals.
(A) Wnd-KD::mRFP and YFP::Rab11 were expressed in the larval C4da neurons using ppk-GAL4. Wnd-KD::mRFP (magenta) and YFP::Rab11 (green) were visualized in the segmental nerve/axons (top) and in C4da axon terminals (bottom) using RFP and GFP antibodies, respectively. Scale bar = 10 µm. (B) The Manders’ overlap coefficient between Wnd-KD::mRFP and YFP::Rab11 was measured in cell bodies, axons (segmental nerve), and axon terminals. They are expressed as median ± 95% CI. One-way ANOVA (F(2, 62)=109.8) followed by post hoc Tukey’s multiple comparison test. Sample numbers were; Cell body n=30, axons n=14, neuropils n=21.
-
Figure 6—figure supplement 1—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig6-figsupp1-data1-v1.xlsx
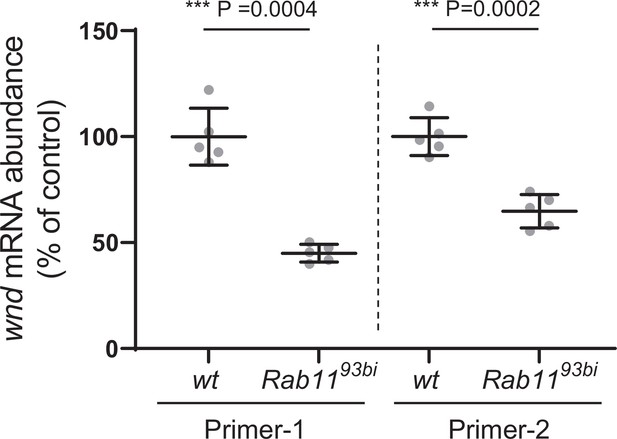
Rab11 mutations did not increase Wallenda (wnd) mRNA abundance.
Total RNAs extracted from the wondering third instar larval brains from a wild-type (wt) control (w1118) and Rab1193bi were subjected to RT-qPCR to measure wnd mRNA abundance. The wnd mRNA levels were normalized by GAPDH1 mRNA levels via the 2ΔΔCT method. Two independent wnd primer sets were used (Primer-1 and Primer-2). The data were expressed as mean ± SEM of five technical replicates. Welch’s t-test (Primer-1: t=8.778, df = 4.772, Primer-2: t=6.576, df = 7.875).
-
Figure 6—figure supplement 2—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig6-figsupp2-data1-v1.xlsx
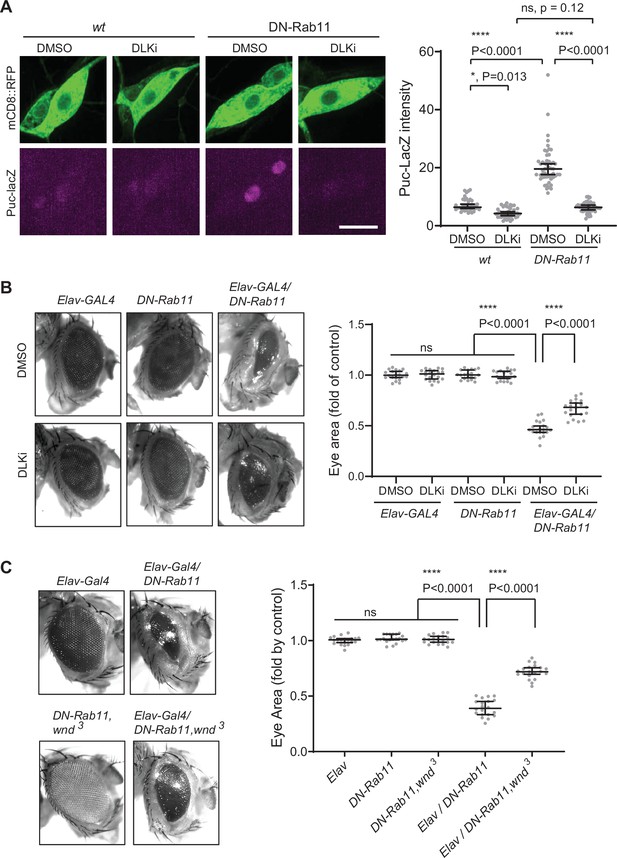
Wallenda (Wnd) mediates stress signaling induced by Rab11 loss-of-function.
(A) puc-LacZ were expressed along with YFP::DN-Rab11 and mCD8::GFP in the larval C4da neurons in the presence of DMSO (vehicle) or dual leucine zipper kinase (DLK) inhibitor (DLKi). The average fluorescent intensities from LacZ staining of each larva sample in C4da cell bodies were measured and quantified as median ± 95% CI. One-way ANOVA (F (3171)=161.8) followed by post hoc Tukey’s multiple comparison test (****p<0.0001). Genotypes and sample numbers were wt (; ppk-Gal4, UAS-mCD8::RFP/+;puc-LacZ/+, in DMSO, n=36, in DLKi n=42) and DN-Rab11 (; ppk-Gal4, UAS-mCD8::RFP/+;puc-LacZ/UASp-YFP.Rab11.S25N, in DMSO, n=51, in DLKi, n=46) Scale bar = 10 µm. (B) Representative images of fly eyes from Elav-GAL4 only, DN-Rab11 only, and DN-Rab11 expression under a pan-neuronal driver, Elav-GAL4 (Elav-GAL4/DN-Rab11). Flies were reared in the presence of DMSO and DLKi (down). Eye area was quantified and expressed as median ± 95% CI. One-way ANOVA (F (5,114)=290.3) followed by post hoc Tukey’s multiple comparison test (****p<0.0001). Genotypes and sample numbers were Elav-Gal4 (Elav-GAL4/+, n=20), DN-Rab11 (;;UASp-YFP.Rab11.S25N/+, n=20), Elav-Gal4/DN-Rab11 (ElavGal4/+;;UASp-YFP.Rab11.S25N /+, n=20). (C) Representative images of fly eyes from Elav-GAL4 only, DN-Rab11 only in heterozygous wnd mutant (DN-Rab11, wnd3), DN-Rab11 expression under a pan-neuronal driver, Elav-GAL4 (Elav-GAL4/DN-Rab11), and DN-Rab11 expression under a pan-neuronal driver in heterozygous wnd mutant (Elav-GAL4/DN-Rab11, wnd3). Eye area was quantified and expressed as median ± 95% CI. One-way ANOVA (F (495)=530.4) followed by post hoc Tukey’s multiple comparison test.
-
Figure 7—source data 1
The numerical source data.
- https://cdn.elifesciences.org/articles/96592/elife-96592-fig7-data1-v1.xlsx
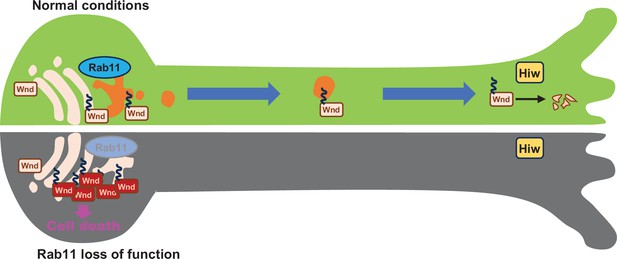
Rab11 suppresses neuronal stress signaling by localizing Dual leucine zipper kinase to axon terminals for protein turnover.
Under normal conditions, the newly synthesized Wallenda (Wnd) proteins are recruited to the Golgi apparatus in the neuronal cell body. Wnd is palmitoylated at somatic Golgi apparatus, which enables it to be sorted into the Recycling Endosomes via Rab11. Wnd is actively transported out of neuronal cell bodies towards axon terminals where Highwire (Hiw)-mediated degradation of Wnd occurs. When Rab11 function is lost, Wnd proteins cannot be transported towards axon terminals. This prevents Wnd protein turnover resulting in higher Wnd protein expression, which in turn triggers the cell death pathway.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-GFP (chicken polyclonal) | Aves Labs | Cat# GFP-1010, RRID: AB_2307313 | IF (1:250), WB (1:1000) |
| Antibody | anti-RFP (rabbit polyclonal) | Rockland | Cat# 600-401-379, RRID: AB_2209751 | IF (1:250) |
| Antibody | anti-LacZ (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 40–1 a | IF (1:5) |
| Antibody | Cy2- conjugated AffiniPure Donkey anti-chicken | Jackson ImmunoResearch | Cat# 703-225-155, RRID:AB_2340370 | IF (1: 500) |
| Antibody | Cy2-conjugated AffiniPure Donkey anti-rabbit | Jackson ImmunoResearch | Cat# 711-225-152, RRID:AB_2340612 | IF (1: 500) |
| Antibody | Cy2-conjugated AffiniPure Donkey anti-mouse | Jackson ImmunoResearch | Cat# 715-225-151, RRID:AB_2340827 | IF (1: 500) |
| Antibody | Cy5-conjugated AffiniPure Donkey anti-rabbit | Jackson ImmunoReesearch | Cat# 711-175-152, RRID:AB_2340607 | IF (1: 500) |
| Antibody | Cy5-conjugated AffiniPure Donkey anti-mouse | Jackson ImmunoResearch | Cat# 715-175-151, RRID:AB_2340820 | IF (1: 500) |
| Antibody | Cy5-conjugated AffiniPure Donkey anti-chic | Jackson ImmunoResearch | Code# 703-175-155, RRID:AB_2340365 | IF (1: 500) |
| Antibody | anti-Biotin (mouse monoclonal) | Jackson ImmunoResearch | Cat# 200-002-211, RRID: AB_2339006 | WB (1:1000) |
| Antibody | anti-Elav (rat monoclonal) | Developmental Studies Hybridoma Bank | Cat# Rat-Elav-7E8A10 anti-elav, RRID: AB_528218 | WB (1:10000) |
| Antibody | anti-Wnd (rabbit polyclonal) | PMID: 16815332 | Gift from Catherine A Collins | WB (1:1000) |
| Chemical compound, drug | GNE-3511 | Sigma-Aldrich | Cat# 5331680001 | DLK inhibitor |
| Chemical compound, drug | Halt protease inhibitor cocktail EDTA- free(100 X) | Thermo Fisher Scientific | Cat# 78437 | |
| Chemical compound, drug | N-ethylmaleimide | Sigma-Aldrich | Cat #04260 | |
| Chemical compound, drug | Hydroxylamine hydrochloride | Sigma-Aldrich | Cat# 55460 | |
| Chemical compound, drug | PEI | Polyscience | Cat# 23966 | |
| Peptide, recombinant protein | EZ-Link BMCC-biotin | Thermo Fisher Scientific | Cat# 21900 | |
| Other | Opti-MEM I | Gibco | Cat# 31985070 | Transfection reagent |
| Other | Drosophila Schneider’s Medium | Thermo Fisher Scientific | Cat# 21720024 | Cell culture medium |
| Other | Fetal bovine serum, Heat Inactivated | Sigma-Aldrich | Cat # F4135 | Cell culture medium |
| Commercial assay or kit | GFP Selector affinity resin | Nano tag | Cat #N0310 | |
| Commercial assay or kit | In-Fusion HD Cloning | Takara Bio USA, Inc | Clontech:639647 | |
| Cell line (D. melanogaster) | Drosophila Schneider 2 (S2)-DRSC | Drosophila Genomics Resource Center | Cat# 181, RRID: CVCL_Z992 | |
| Software, algorithm | ImageJ | National Institutes of Health | RRID:SCR_003070 | |
| Software, algorithm | Fiji | National Institutes of Health | RRID:SCR_002285 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_002798 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab1.S25N | Bloomington Drosophila Stock Center | BDSC_9757 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab2.S20N | Bloomington Drosophila Stock Center | BDSC_23640 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.RabX2.S21N | Bloomington Drosophila Stock Center | BDSC_9843 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab3.T35N | Bloomington Drosophila Stock Center | BDSC_9766 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab4.S22N | Bloomington Drosophila Stock Center | BDSC_9769 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.RabX4.T40N | Bloomington Drosophila Stock Center | BDSC_9849 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab5.S43N | Bloomington Drosophila Stock Center | BDSC_9771 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab6.T26N | Bloomington Drosophila Stock Center | BDSC_23249 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.RabX6.S22N | Bloomington Drosophila Stock Center | BDSC_9856 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab7.T22N | Bloomington Drosophila Stock Center | BDSC_9778 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab8.T22N | Bloomington Drosophila Stock Center | BDSC_9780 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab9.S26N | Bloomington Drosophila Stock Center | BDSC_23642 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab10.T23N | Bloomington Drosophila Stock Center | BDSC_9786 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab11.S25N | Bloomington Drosophila Stock Center | BDSC_9792 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab11.S25N | Bloomington Drosophila Stock Center | BDSC_23261 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab18.S19N | Bloomington Drosophila Stock Center | BDSC_23238 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab19.T35N | Bloomington Drosophila Stock Center | BDSC_9799 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab21.T27N | Bloomington Drosophila Stock Center | BDSC_23241 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab23.S51N | Bloomington Drosophila Stock Center | BDSC_9804 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab26.T204N | Bloomington Drosophila Stock Center | BDSC_9807 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab27.T25N | Bloomington Drosophila Stock Center | BDSC_23267 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab32.T33N | Bloomington Drosophila Stock Center | BDSC_23281 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab35.S22N | Bloomington Drosophila Stock Center | BDSC_9819 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab39.S23N | Bloomington Drosophila Stock Center | BDSC_23247 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.Rab40.H25N | Bloomington Drosophila Stock Center | BDSC_9829 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.CG9807.T21N | Bloomington Drosophila Stock Center | BDSC_23257 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.CG9807.T21N | Bloomington Drosophila Stock Center | BDSC_23258 | |
| Strain, strain background (Drosophila melanogaster) | UAST-YFP.CG32673.T21N | Bloomington Drosophila Stock Center | BDSC_23254 | |
| Strain, strain background (Drosophila melanogaster) | ppk-Gal4 | Bloomington Drosophila Stock Center | BDSC_32078 | |
| Strain, strain background (Drosophila melanogaster) | ppk-Gal4 | Bloomington Drosophila Stock Center | BDSC_32079 | |
| Strain, strain background (Drosophila melanogaster) | w1118 | Bloomington Drosophila Stock Center | BDSC_3605 | |
| Strain, strain background (Drosophila melanogaster) | TI{TI}Rab11EYFP | Bloomington Drosophila Stock Center | BDSC_62549 | |
| Strain, strain background (Drosophila melanogaster) | UASp-YFP.Rab11 | Bloomington Drosophila Stock Center | BDSC_50782 | |
| Strain, strain background (Drosophila melanogaster)ain | hiw∆N | Bloomington Drosophila Stock Center | BDSC_51637 | |
| Strain, strain background (Drosophila melanogaster) | GMR-Gal4 | Bloomington Drosophila Stock Center | BDSC_1104 | |
| Strain, strain background (Drosophila melanogaster) | Rab1193Bi | Bloomington Drosophila Stock Center | BDSC_4158 | |
| Strain, strain background (Drosophila melanogaster) | wnd3/Tm3B,Sb | Bloomington Drosophila Stock Center | BDSC_51999 | |
| Strain, strain background (Drosophila melanogaster) | UAS-ManII-GFP | Bloomington Drosophila Stock Center | BDSC_65248 | |
| Strain, strain background (Drosophila melanogaster) | ;;UAS-ManII-TagRFP | Bloomington Drosophila Stock Center | BDSC_65249 | |
| Strain, strain background (Drosophila melanogaster) | elav-Gal4;; | Bloomington Drosophila Stock Center | BDSC_458 | |
| Strain, strain background (Drosophila melanogaster) | ;;MI{MIC}wnd[MI00494]/Tm6B | Bloomington Drosophila Stock Center | BDSC_39656 | |
| Strain, strain background (Drosophila melanogaster) | UAS-dHIP14-Tdtomato | PMID: 1803266 | Gift from Steven Stowers | |
| Strain, strain background (Drosophila melanogaster) | yw;;dHIP14ex11/Tm6c | PMID: 18032660 | Gift from Steven Stowers | |
| Strain, strain background (Drosophila melanogaster) | Puc-LacZ | PMID: 9472024 | Gift from Bing Ye |





