DNAH3 deficiency causes flagellar inner dynein arm loss and male infertility in humans and mice
Figures
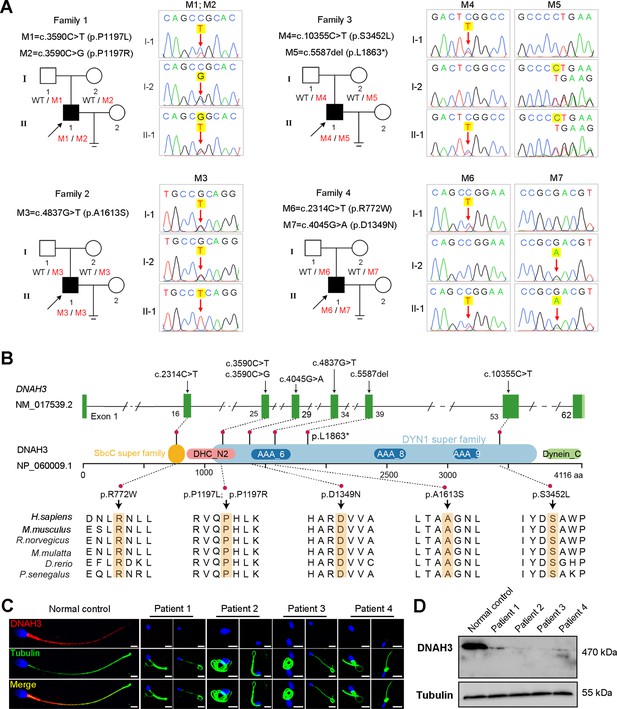
Identification of biallelic pathogenic variants in DNAH3 from four unrelated infertile families.
(A) Pedigrees of four families affected by DNAH3 variants (M1–M7). Black arrows indicate the probands in these families. (B) Location of the variants and conservation of affected amino acids in DNAH3. Black arrows indicate the position of the variants. (C) Immunofluorescence staining of DNAH3 in sperm from the patients and normal control. Red, DNAH3; green, α-Tubulin; blue, DAPI; scale bars, 5 μm. (D) Western blotting analysis of DNAH3 expressed in spermatozoa from the patients and normal control.
-
Figure 1—source data 1
Summary of whole exome sequencing and the candidate variants identified.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Primers for Sanger sequencing.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig1-data2-v1.docx
-
Figure 1—source data 3
PDF file containing original western blotting for Figure 1D.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig1-data3-v1.zip
-
Figure 1—source data 4
Original files for western blotting analysis displayed in Figure 1D.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig1-data4-v1.zip
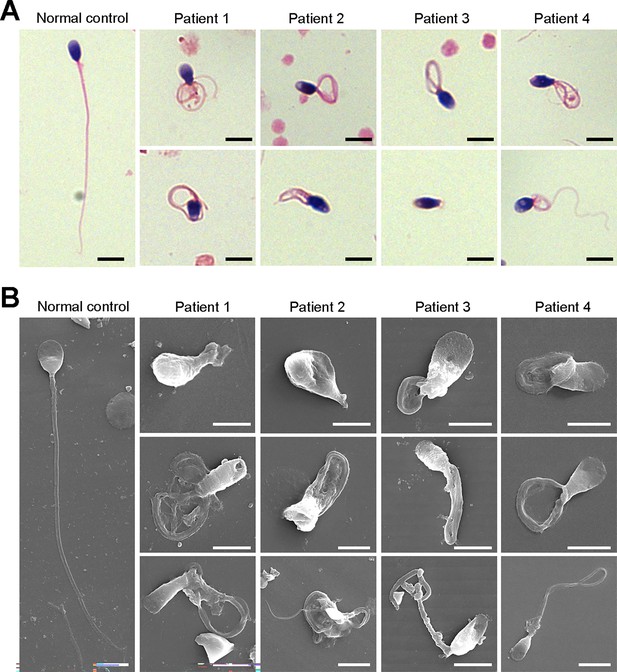
Defects in sperm morphology of the patients harboring DNAH3 variants.
(A, B) Abnormal sperm morphology was observed through Papanicolaou staining (A), and SEM analysis (B) compared to normal control. Scale bars, 5 μm.
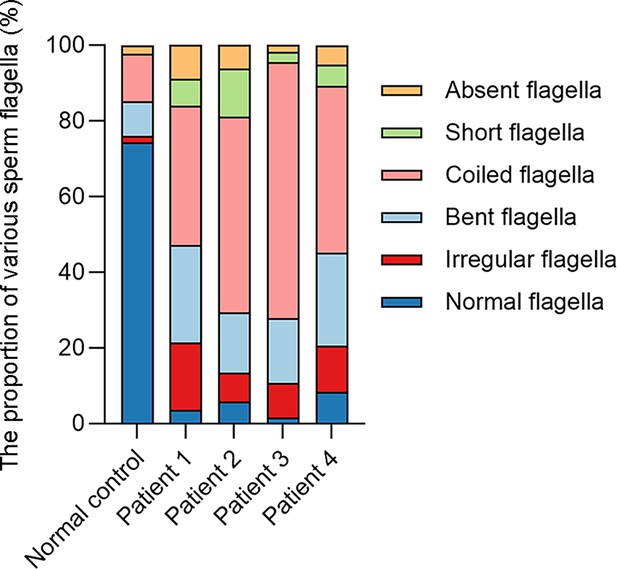
The histogram of various flagellar morphology in the normal control and patients.
-
Figure 2—figure supplement 1—source data 1
The percentage distribution of various flagellar morphology in the normal control and patients.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig2-figsupp1-data1-v1.xlsx
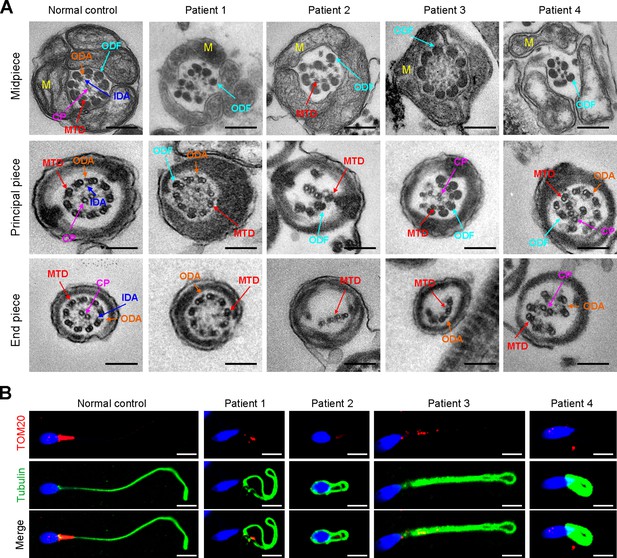
Ultrastructural and mitochondrial defects in sperm from infertile men with DNAH3 variants.
(A) TEM analysis of sperm obtained from a normal control and patients harboring DNAH3 variants. Cross-sections of the midpiece, principal piece and endpiece of sperm from normal control showed the typical ‘‘9+2’’ microtubule structure, and an IDA and an ODA were displayed on the A-tube of each microtubule doublet. Cross-sections of the midpiece, principal piece and endpiece of sperm from the patients displayed absent or disordered CPs, MTDs and ODFs, as well as an evident missing of the IDAs in different pieces of the flagella. M, mitochondria sheath; ODF, outer dense fiber; MTD, microtubule doublets; CP, central pair; IDA, inner dynein arms; ODA, outer dynein arms. Scale bars, 200 nm. (B) Immunofluorescence staining of TOM20 in sperm from the patients and normal control. Red, TOM20; green, α-Tubulin; blue, DAPI; scale bars, 5 μm.
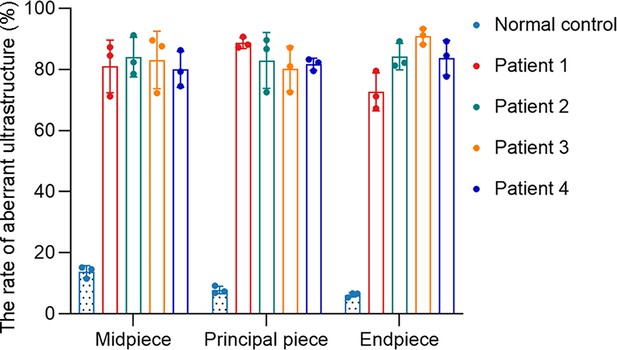
The percentage of aberrant ultrastructure in different cross-sections of sperm from the normal control and patients.
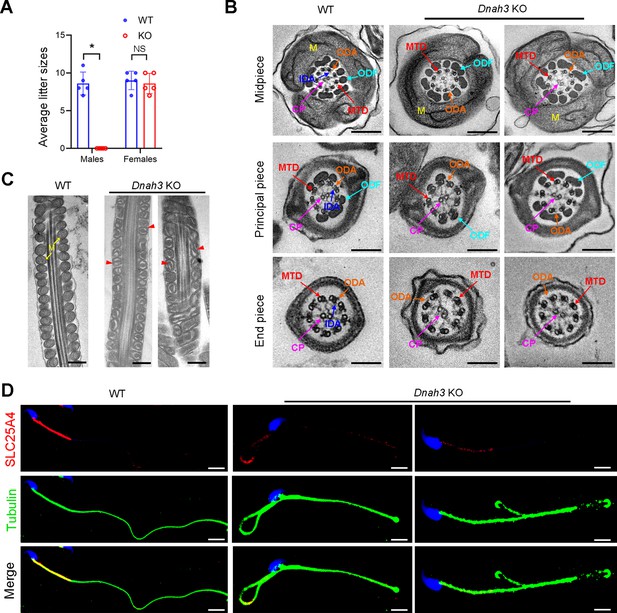
Dnah3 KO male mice are infertile.
(A) Fertility of Dnah3 KO mice. The KO male mice were infertile (n=five biologically independent WT mice or KO mice; Student’s t test; *, p<0.05; NS, no significance; error bars, s.e.m.). (B) TEM analysis of the cross-sections of spermatozoa from Dnah3 KO mice revealed an obvious absence of IDAs in different pieces of the flagella compared to WT mice. M, mitochondrion sheath; ODF, outer dense fiber; MTD, microtubule doublet; CP, central pair; IDA, inner dynein arm; ODA, outer dynein arm. Scale bars, 200 nm. (C) Disrupted mitochondria were observed in spermatozoa tail from Dnah3 KO mice by TEM analysis. The yellow arrows indicate the normal mitochondria. The red arrowheads indicate the dilated intermembrane spaces and dissolved mitochondrial material. M, mitochondrion sheath. Scale bars, 200 nm. (D) Immunofluorescence staining of SLC25A4 indicated impaired mitochondrial formation in Dnah3 KO mice compared to WT mice. Red, SLC25A4; green, α-Tubulin; blue, DAPI; scale bars, 5 µm.
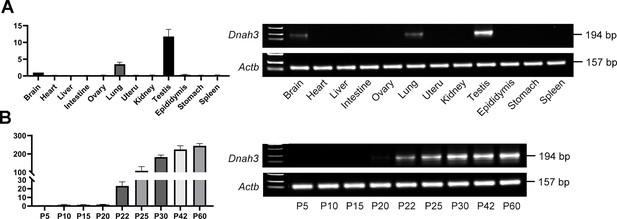
The expression of DNAH3 in mouse testis.
(A) qPCR analysis revealed that Dnah3 was highly expressed in the mouse testis. (B) qPCR analysis showed that Dnah3 expression was significantly elevated beginning on postnatal Day 12, peaked at postnatal Day 30, and maintained a stable expression level thereafter.
-
Figure 4—figure supplement 1—source data 1
PDF file containing original gels for Figure 4—figure supplement 1A, B.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original files for gel analysis displayed in Figure 4—figure supplement 1A, B.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig4-figsupp1-data2-v1.zip
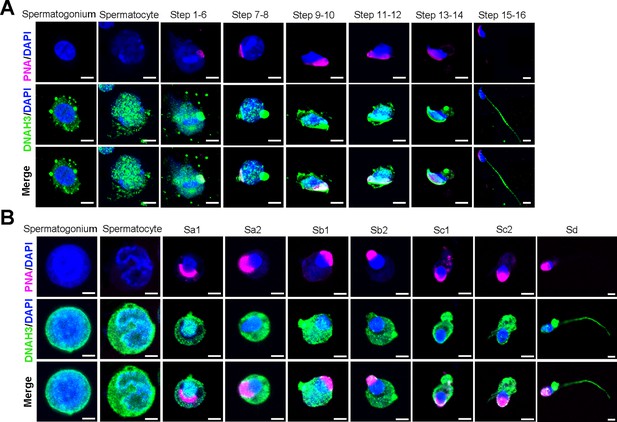
DNAH3 is expressed during spermatogenesis in mice and humans.
(A) Immunofluorescence staining of DNAH3 in isolated mouse germ cells. Pink, PNA; green, DNAH3; blue, DAPI; scale bars, 5 μm. (B) Immunofluorescence staining of DNAH3 in isolated human germ cells. Pink, PNA; green, DNAH3; blue, DAPI; scale bars, 5 μm.
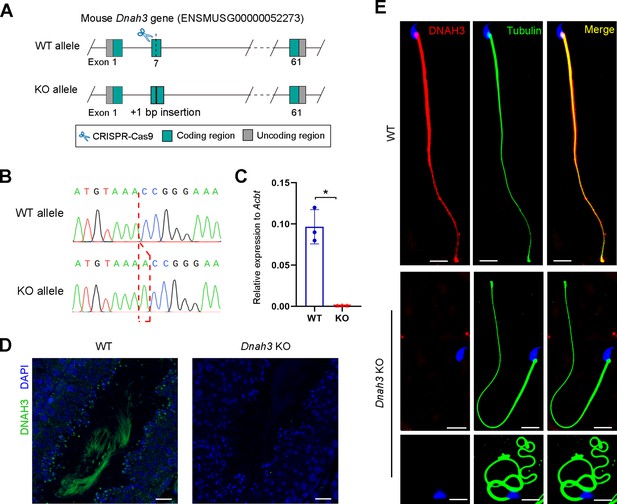
Generation of Dnah3 KO mice.
(A) Schematic illustration of the strategy for the generation of Dnah3 KO mice. (B, C) PCR sequencing (B) and qPCR (C) were used to confirm the genotype and KO efficiency (n=three biologically independent WT mice or KO mice; Student’s t test; *, p<0.05; error bars, s.e.m.). (D) Immunofluorescence staining of DNAH3 in testis of Dnah3 KO mice and WT mice. Green, DNAH3; blue, DAPI; scale bars, 75 μm. (E) Immunofluorescence staining of DNAH3 in spermatozoa isolated from the cauda epididymis of Dnah3 KO mice and WT mice. Red, DNAH3; green, α-Tubulin; blue, DAPI; scale bars, 5 μm.
-
Figure 4—figure supplement 3—source data 1
Primers for Sanger sequencing and qPCR.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig4-figsupp3-data1-v1.docx
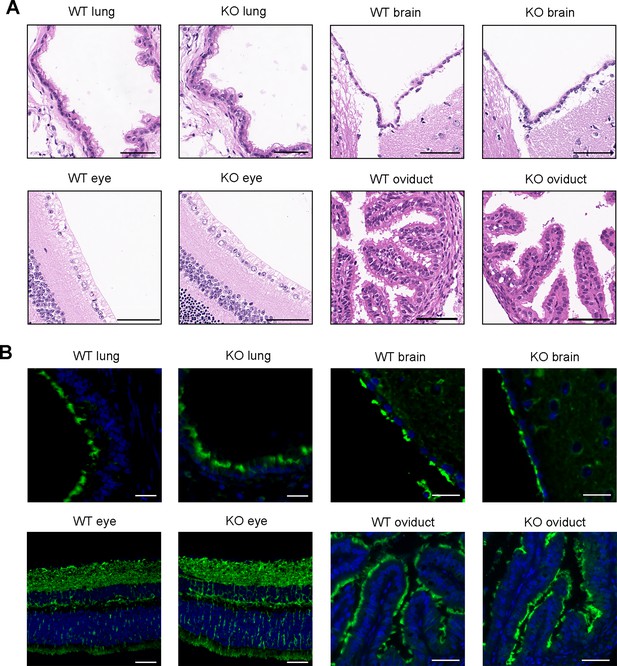
Ciliary development of Dnah3 KO mice.
(A) H&E staining of lung, brain, eye, and oviduct from Dnah3 KO mice and WT mice. Scale bars, 100 μm. (B) Analysis of ciliary development in the lung, brain, eye, and oviduct from Dnah3 KO mice and WT mice by using immunofluorescence staining. Green, Ac-Tubulin; blue, DAPI; scale bars, 20 μm.
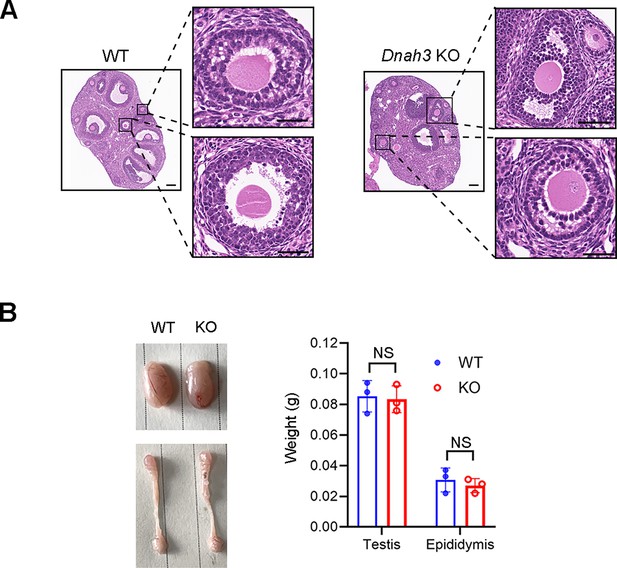
Fertility of Dnah3 KO mice.
(A) H&E staining of ovary tissue sections from 8-week-old Dnah3 KO female mice and WT female mice. Scale bars, 75 μm (n=three biologically independent WT mice or KO mice). (B) Sizes of the testis and epididymis of the 8-week-old Dnah3 KO and WT mice (n=three biologically independent WT mice or KO mice; Student’s t test; NS, no significance; error bars, s.e.m.).
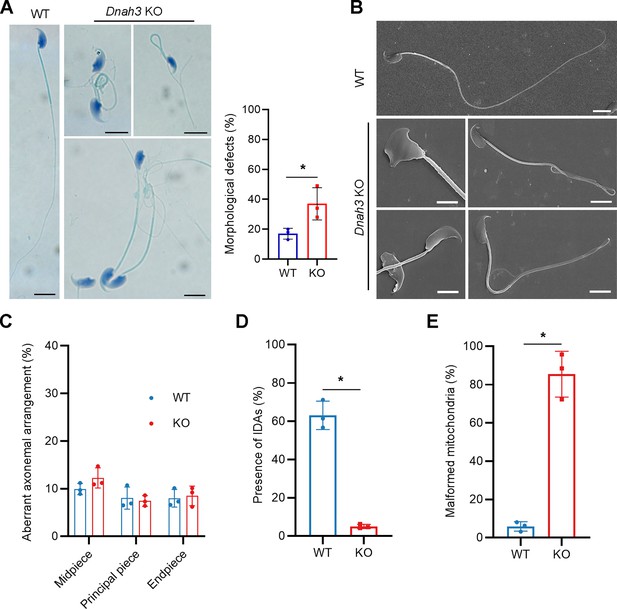
Morphology and ultrastructure of sperm isolated from Dnah3 KO mice.
(A, B) Papanicolaou staining (A), and SEM analysis (B) revealed morphological defects in partial spermatozoa from Dnah3 KO mice compared to WT mice. Scale bars in (A), 5 μm; scale bars in (B), 2.5 μm. (n=three biologically independent WT mice or KO mice; Student’s t test; error bars, s.e.m.). (C) The percentage of aberrant axonemal arrangement in different cross-sections of sperm from WT mice and Dnah3 KO mice. (n=three biologically independent WT mice or KO mice; error bars, s.e.m.). (D) The percentage of microtubule doublets that presented IDAs in WT mice and Dnah3 KO mice. (n=three biologically independent WT mice or KO mice; Student’s t test; error bars, s.e.m.). (E) Statistics of malformed mitochondria in the midpiece of sperm from WT mice and Dnah3 KO mice. (n=three biologically independent WT mice or KO mice; Student’s t test; error bars, s.e.m.).
CASA of sperm from WT mice.
Sperm from the epididymis of WT mice were collected, incubated, and recorded under a phase-contrast microscope. A normal quantity and motility of sperm were observed in the WT mice (n=three biologically independent WT mice).
CASA of sperm from Dnah3 KO mice.
Epididymal sperm of Dnah3 KO mice were collected, incubated in HTF medium at 37 °C for 10 min, and recorded under a phase-contrast microscope. The movie showed a significantly reduced motility of sperm from Dnah3 KO (n=three biologically independent Dnah3 mice).
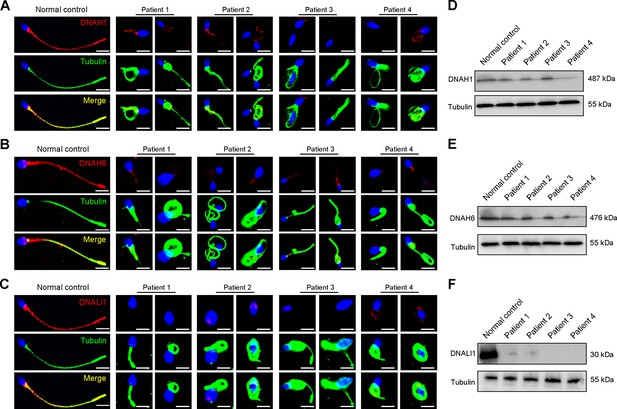
Immunofluorescence staining and western blotting analysis of IDA-associated proteins in spermatozoa obtained from normal control and patients with DNAH3 variants.
(A – C) Immunofluorescence staining of DNAH1 (A), DNAH6 (B) and DNALI1 (C) in spermatozoa from patients and normal controls. Red, DNAH1 in (A), DNAH6 in (B), DNALI1 in (C); green, α-Tubulin; blue, DAPI; scale bars, 5 μm. (D – F) Western blotting analysis of DNAH1(D), DNAH6 (E), DNALI1 (F) in sperm lysates from the patients and normal control.
-
Figure 5—source data 1
PDF file containing original western blotting for Figure 5D–F and Figure 6D–F.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig5-data1-v1.zip
-
Figure 5—source data 2
Original files for western blotting analysis displayed in Figure 5D–F and Figure 6D–F.
- https://cdn.elifesciences.org/articles/96755/elife-96755-fig5-data2-v1.zip
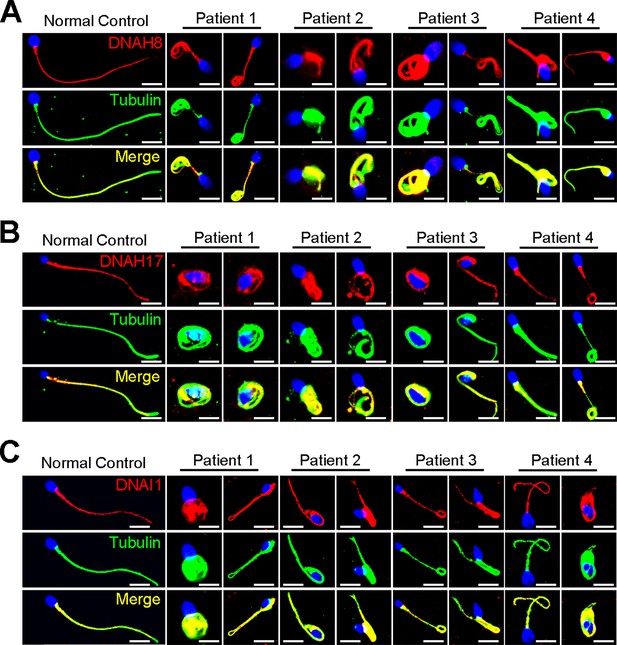
Immunofluorescence staining of ODA-associated proteins in spermatozoa obtained from variants within DNAH3 patients.
(A – C) The expression of DNAH8 (A), DNAH17 (B) and DNAI1 (C) in spermatozoa of the patients was comparable to that in normal controls. Red, DNAH8 in (A), DNAH17 in (B), DNAI1 in (C); green, α-Tubulin; blue, DAPI; scale bars, 5 μm.
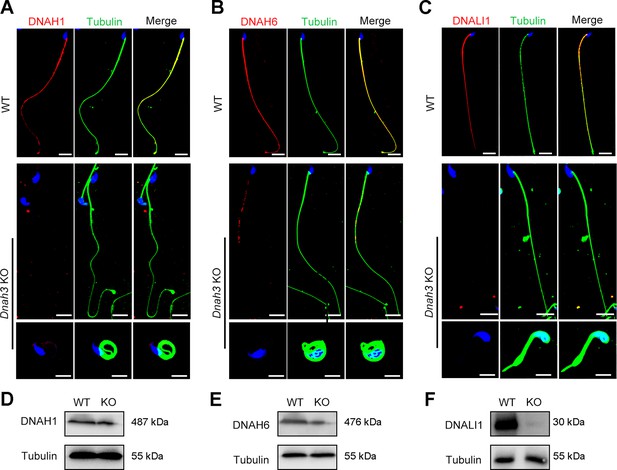
Immunofluorescence staining and western blotting analysis of IDA-associated proteins in spermatozoa from WT and Dnah3 KO mice.
(A – C) Immunofluorescence staining of DNAH1 (A), DNAH6 (B) and DNALI1 (C) in spermatozoa from Dnah3 KO and WT mice. Red, DNAH1 in (A), DNAH6 in (B), DNALI1 in (C); green, α-Tubulin; blue, DAPI; scale bars, 5 μm. (D – F) Western blotting analysis of DNAH1(D), DNAH6 (E) and DNALI1 (F) in spermatozoa lysates from Dnah3 KO and WT mice.
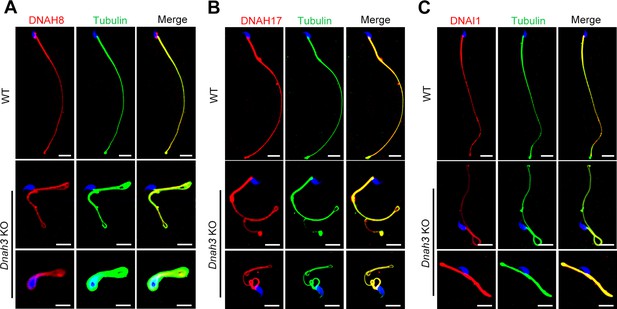
Immunofluorescence staining of ODA-associated proteins in spermatozoa of Dnah3 KO and WT mice.
(A – C) The expression of DNAH8 (A), DNAH17 (B) and DNAI1 (C) in spermatozoa from Dnah3 KO mice was comparable to that in spermatozoa from WT mice. Red, DNAH8 in (A), DNAH17 in (B), DNAI1 in (C); green, α-Tubulin; blue, DAPI; scale bars, 5 μm.
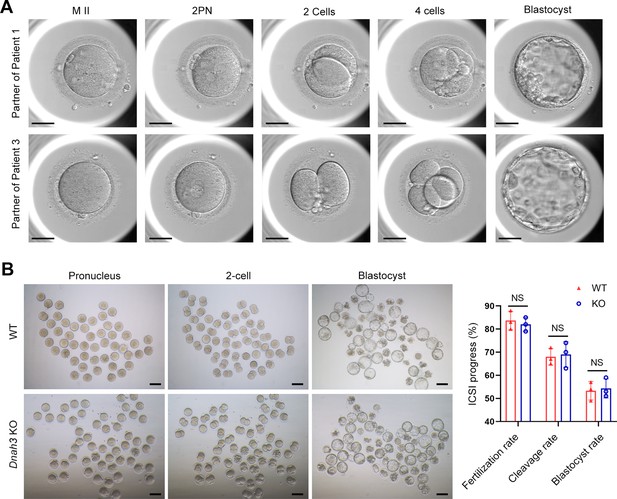
ICSI outcomes of DNAH3-deficient patients and Dnah3 KO mice.
(A) The embryonic development of Patient 1 and Patient 3 after ICSI treatment. MII, metaphase II; PN, pronucleus; scale bars, 40 μm. (B) There was no difference in the fertilization rate or 2 cell and blastocyst embryo formation rates between the Dnah3 KO and WT groups (n=three biologically independent WT mice or KO mice; Student’s t test; NS, no significance; error bars, s.e.m.).
Tables
Semen analysis of the patients in the present study.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Reference | ||
|---|---|---|---|---|---|---|
| Semen parameters | Semen volume (ml) | 4.3 | 3.3 | 0.8 | 4.6 | ≥1.5 |
| Semen concentration (106 /ml) | 2.0 | 0.5 | 11.0 | 21.0 | ≥15.0 | |
| Motility (%) | 6.0 | 3.0 | 0 | 0 | ≥40.0 | |
| Progressive motility (%) | 0 | 2.3 | 0 | 0 | ≥32.0 | |
| Sperm morphology | Normal (%) | 1.3 | 1.1 | 1.8 | 1.2 | ≥4.0 |
| Tail defects (%) | 91.3 | 87.5 | 96.5 | 97.5 | - |
-
-,not applicable.
Variants analysis of the patients in the present study.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Variant | cDNA mutation * | c.3590C>T | c.3590C>G | c.4837G>T | c.5587del | c.10355C>T | c.2314C>T | c.4045G>A | |
| Protein alteration | p.Pro1197Leu | p.Pro1197Arg | p.Ala1613Ser | p.Leu1863* | p.Ser3452Leu | p.Arg772Trp | p.Asp1349Asn | ||
| Mutation type | Missense | Missense | Missense | Nonsense | Missense | Missense | Missense | ||
| Allele frequency | ExAC_EAS | 0.0001 | 0 | 0.004165 | 0 | 0.0006 | 0.0019 | 0.0065 | |
| GnomAD_EAS | 0.00005016 | 0 | 0.00277415 | 0 | 0.0008 | 0.002 | 0.007 | ||
| 1000 Genomes Project_EAS | 0 | 0 | 0.0050 | 0 | 0 | 0.0040 | 0.0069 | ||
| Function prediction | SIFT | Deleterious | Deleterious | Deleterious | / | Tolerated | Deleterious | Deleterious | |
| Polyphen-2 | Probably damaging | Probably damaging | Probably damaging | / | Probably damaging | Probably damaging | Probably damaging | ||
| Mutation Taster | Disease causing | Disease causing | Disease causing | / | Disease causing | Disease causing | Disease causing | ||
| CADD † | 33 | 29.5 | 27.5 | / | 25.4 | 27.9 | 34 | ||
-
/,not applicable.
-
*
NM_017539.2.
-
†
score >4.0 is predicted to be damaging.
Semen analysis using CASA in the Dnah3 KO mice.
| WT | KO | p* value | |
|---|---|---|---|
| Semen parameters | |||
| Sperm concentration (106/ml) † | 112.32±18.26 | 105.17±11.15 | 0.059 |
| Motility (%) * | 71.56±3.97 | 4.37±1.15 | <0.01 |
| Progressive motility (%) * | 60.36±4.32 | 4.37±1.15 | <0.01 |
| Sperm locomotion parameters | |||
| Curvilinear velocity (VCL) (μm/s)* | 67.54±6.79 | 9.07±1.22 | <0.01 |
| Straight-line velocity (VSL) (μm/s)* | 28.91±4.86 | 2.68±0.52 | <0.01 |
| Average path velocity (VAP) (μm/s)* | 39.02±5.31 | 3.85±0.82 | <0.01 |
| Amplitude of lateral head displacement (ALH) (μm)* | 0.71±0.03 | 0.13±0.04 | <0.01 |
| Linearity (LIN)* | 0.43±0.07 | 0.30±0.02 | 0.037 |
| Wobble (WOB,=VAP/VCL)* | 0.58±0.06 | 0.42±0.05 | 0.024 |
| Straightness (STR,=VSL/VAP) | 0.74±0.22 | 0.70±0.13 | 0.80 |
| Beat-cross frequency (BCF) (Hz)* | 4.86±0.12 | 0.73±0.08 | <0.01 |
-
*
A significant difference, two-sided student’s t-test. n=3 biologically independent WT mice or KO mice.
-
†
Epididymides and vas deferens.
Outcomes of ICSI treatment in the patients with DNAH3 mutations.
| Subjects | Patient | Patient 1 | Patient 2 | Patient 3 | Patient 4 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Female age (y) | 24 | 30 | 30 | 36 | |||||
| Length of primary infertility history (y) | 6 | 1 | 3 | 8 | |||||
| Basal hormones | FSH (IU/L) | 7.1 | 8.5 | 3.49 | 4.3 | ||||
| LH (IU/L) | 4.44 | 2.5 | 4.2 | 2.6 | |||||
| E2 (pg/mL) | 83 | 50 | 43.49 | 68 | |||||
| Prog (ng/mL) | 0.2 | 0.6 | 0.3 | 0.3 | |||||
| ICSI Cycles | Cycle 1 | Cycle 1 | Cycle 2 | Cycle 1 | Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | |
| E2 level on the trigger day (pg/ml) | 3366 | 1519.6 | 1582.4 | >5,000 | 4440 | 5000 | 3152 | 2206 | |
| No. of follicles ≥14 mm on the trigger day | 15 | 6 | 5 | 20 | 13 | 12 | 14 | 11 | |
| No. of follicles ≥18 mm on the trigger day | 10 | 3 | 4 | 8 | 11 | 9 | 7 | 5 | |
| No. of oocytes retrieved | 24 | 6 | 5 | 25 | 16 | 20 | 21 | 14 | |
| ICSI progress | No. of MII oocytes | 21 | 6 | 5 | 20 | 12 | 13 | 8 | 7 |
| Fertilization rate (%) | 80.95 (17/21) | 50 (3/6) | 100 (5/5) | 95 (19/20) | 41.67 (5/12) | 46.15 (6/13) | 50 (4/8) | 42.86 (3/7) | |
| Cleavage Rate (%) | 100 (17/17) | 100 (2/2) | 100 (5/5) | 100 (19/19) | 100 (5/5) | 100 (6/6) | 100 (4/4) | 66.7 (2/3) | |
| Available D3 embryos | 13 | 2 | 5 | 15 | 2 | 2 | 0 | 0 | |
| Blastocyst formation rate (%) | 61.5 (8/13) | 0 | 66.7 (2/3) | 66.7 (10/15) | - | - | - | - | |
| Clinical outcomes | No. of embryos transferred | 2 blastocysts | 0 | 2 D3 | 1 blastocyst | 2 D3 | 2 D3 | - | - |
| Implantation rate (%) | 50 (1/2) | 0 | 0 | 100 (1/1) | 0 | 0 | - | - | |
| Clinical pregnancy | Yes | No | No | Yes | - | - | - | - | |
| No. of live birth | ngoing | - | - | ngoing | - | - | - | - | |
-
-,not applicable.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-DNAH1 (Rabbit polyclonal) | Cusabio | CSB-PA878961LA01HU | IF (1:100) |
| Antibody | Anti-DNAH3 (Rabbit polyclonal) | Cusabio | CSB-PA823461LA01HU | IF (1:100) |
| Antibody | Anti-DNAH3 (Rabbit polyclonal) | Gift from Prof. Yueqiu Tan, Central South University, China. | WB (1:200) | |
| Antibody | Anti-DNAH6 (Rabbit polyclonal) | Proteintech | 18080–1-AP, RRID: AB_2878493 | IF (1:50), WB (1:150) |
| Antibody | Anti-DNAH8 (Rabbit polyclonal) | Atlas | HPA028447, RRID: AB_10599600 | IF (1:200) |
| Antibody | Anti-DNAH17 (Rabbit polyclonal) | Proteintech | 24488–1-AP, RRID: AB_2879568 | IF (1:50) |
| Antibody | Anti-DNAI1 (Rabbit polyclonal) | Proteintech | 12756–1-AP, RRID: AB_10643244 | IF (1:50) |
| Antibody | Anti-DNALI1 (Rabbit polyclonal) | Proteintech | 17601–1-AP, RRID: AB_2095372 | IF (1:50), WB (1:150) |
| Antibody | Anti-TOM20 (Rabbit polyclonal) | Proteintech | 11802–1-AP, RRID: AB_2207530 | IF (1:50) |
| Antibody | Anti-SLC25A4 (Rabbit polyclonal) | Signalway | 32484, RRID: AB_2941094 | IF (1:100) |
| Antibody | Anti-alpha tubulin (Mouse monoclonal) | Abcam | ab7291, RRID: AB_2241126 | IF (1:500) |
| Antibody | Anti-alpha tubulin (Rabbit polyclonal) | Proteintech | 11224–1-AP, RRID: AB_2210206 | WB (1:1000) |
| Antibody | Anti-Rabbit IgG, Alexa Fluor 488 (Goat polyclonal) | Invitrogen | A11008, RRID: AB_143165 | IF (1:1000) |
| Antibody | Anti-Mouse IgG, Alexa Fluor 594 (Goat polyclonal) | Invitrogen | A11005, RRID: AB_141372 | IF (1:1000) |
| Antibody | Anti-acetylated alpha tubulin (Mouse monoclonal) | Abcam | ab24610, RRID: AB_448182 | IF (1:500) |
| Antibody | Anti-Mouse IgG, HRP-conjugated (Goat polyclonal) | Proteintech | SA00001-1, RRID: AB_2722565 | WB (1:5000) |
| Antibody | Anti-Rabbit IgG, HRP-conjugated (Goat polyclonal) | Proteintech | SA00001-2, RRID: AB_2722564 | WB (1:5000) |
| Other | Lectin PNA | Invitrogen | L-32460 | IF (1:50) |





