Nuclear translocation of SIRT4 mediates deacetylation of U2AF2 to modulate renal fibrosis through alternative splicing-mediated upregulation of CCN2
Figures
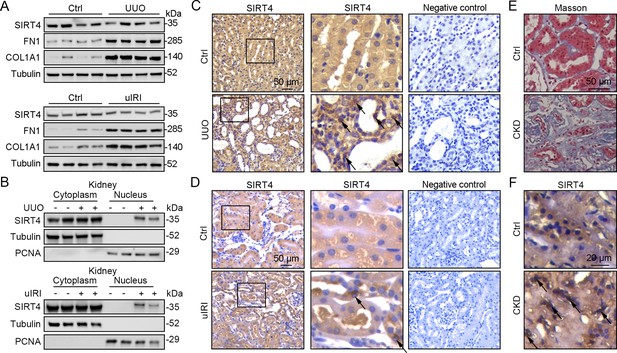
Nuclear accumulation of SIRT4 is increased in renal tubules after injury.
(A) Western blots analysis of SIRT4, FN1, COL1A1, and Tubulin in the kidney of UUO, uIRI, and sham mice (control group). (B) Nuclear fractions were prepared from the kidney of UUO, uIRI, and sham mice. Nuclear PCNA and cytoplasmic tubulin were used as controls. (C, D) Representative images of immunohistochemical staining of SIRT4 (scale bar, 50 μm) in the kidneys from mice that underwent sham surgery, UUO surgery on day 10 post surgery or uIRI surgery on day 28 post surgery. (E, F) Representative images of Masson’s trichrome staining (the upper panel; scale bar = 50 μm) and SIRT4 immumohistochemical staining (the bottom panel; scale bar = 20 μm) in the kidney sections from patients with CKD (n=8) and minimal change disease (control group, n=1).
-
Figure 1—source data 1
Original files for western blot analysis displayed in Figure 1A, B.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig1-data1-v1.zip
-
Figure 1—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 1A and B.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig1-data2-v1.zip
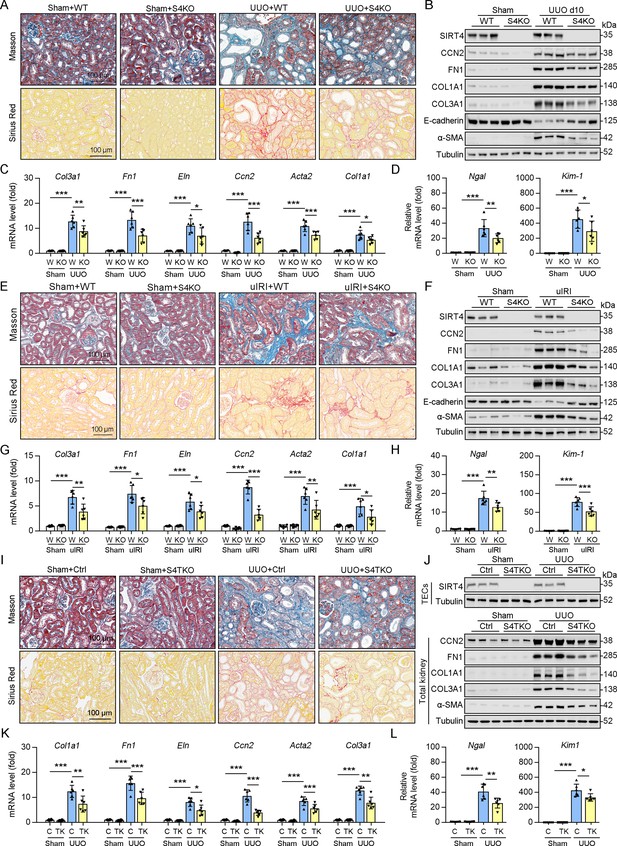
Knockout of Sirt4 or targeted deletion of Sirt4 in renal TECs alleviates renal fibrosis induced by UUO or uIRI.
(A–D) WT or S4KO mice were randomly assigned to sham or UUO surgery according to an established protocol. Kidney samples were obtained from mice on day 10 post UUO or sham surgery (n=6 per group). (A) Representative images of Masson’s trichrome staining and Sirius red in kidneys from mice (scale bar = 100 μm). (B) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL1A1, COL3A1, E-cadherin, α-SMA, and Tubulin in the kidney of mice. (C, D) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, Col3a1, Ngal, and Kim-1 in the kidney of mice. (E–H) WT or S4KO mice received uIRI surgery, and the sham surgery kidneys were used as control. Kidney samples collected from mice after surgery at 28 d (n=6 per group). (E) Representative images of Masson’s trichrome staining and Sirius red in kidneys from mice (scale bar = 100 μm). (F) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL1A1, COL3A1, E-cadherin, α-SMA, and Tubulin in the kidney of mice. (G, H) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, Col3a1, Ngal, and Kim-1 in the kidney of mice. (I–L) WT or S4TKO mice were randomly assigned to sham or UUO surgery according to an established protocol. Kidney samples were obtained from mice on day 10 post UUO or sham surgery (n=6 per group). (I) Representative images of Masson’s trichrome staining and Sirius red in kidneys from mice (scale bar = 100 μm). (J) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL1A1, COL3A1, α-SMA, and Tubulin in the kidney of mice. (K, L) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, Col3a1, Ngal, and Kim-1 in the kidney of mice. For all panels, data are presented as mean ± SD. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 2—source data 1
Original files for western blot analysis displayed in Figure 2B, F and J.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig2-data1-v1.zip
-
Figure 2—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 2B, F and J.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig2-data2-v1.zip
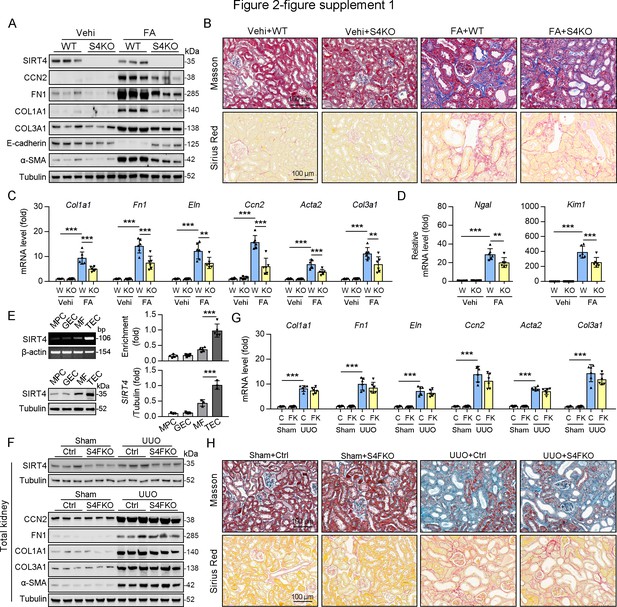
Knockout of SIRT4 aggravated FA-induced kidney fibrosis.
(A–D) WT or S4KO mice were randomly assigned to FA or vehicle according to an established protocol. Kidney samples were obtained from mice on day 14 post FA or vehicle treatment (n=6 per group). (A) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL1A1, COL3A1, E-cadherin, α-SMA, and Tubulin in the kidney of mice. (B) Representative images of Masson’s trichrome staining and Sirius red in kidneys from mice (scale bar, 100 μm). (C, D) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, Col3a1, Ngal, and Kim-1 in the kidney of mice. (E) RT-PCR (n=5) and western blot analysis (n=3) of the expression of SIRT4 in selected mouse renal cells including mouse podocytes (MPC), glomerular endothelial cells (GEC), mouse TECs and mouse fibroblast (MF). (F–H) Control or S4FKO mice were randomly assigned to sham or UUO surgery according to an established protocol. (F) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL1A1, COL3A1, α-SMA, and Tubulin in the kidney of mice. (G) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, Col3a1, Ngal, and Kim-1 in the kidney of mice. (H) Representative images of Masson’s trichrome staining and Sirius red in kidney sections of mice (scale bar, 100 μm). For all panels, data are presented as mean ± SD. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test. For all panels, data are presented as mean ± SD. **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 2—figure supplement 1—source data 1
Original files for western blot analysis displayed in Figure 2—figure supplement 1A, E, F.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
The uncropped gels or blots with 1109 the relevant bands clearly labeled in Figure 2—figure supplement 1A, E, F.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig2-figsupp1-data2-v1.zip
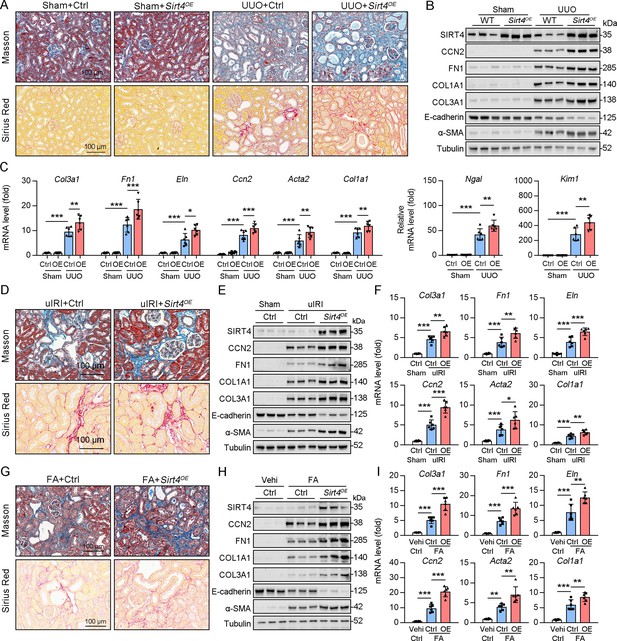
TECs specific SIRT4OE aggravates kidney fibrosis.
(A–I) AAV9-Ctrl or AAV9-Ksp-Sirt4 was injected into the kidneys of mice in situ at three independent points. After 2-week transfection, the mice received UUO surgery, uIRI surgery, or FA treatment (vehicle treatment as control) (n=6 per group). (A, D, G) Representative images of Masson’s trichrome staining and Sirius red in kidneys from mice (scale bar = 100 μm). (B, E, H) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL1A1, COL3A1, E-cadherin, α-SMA, and Tubulin in the kidney of mice. (C, F, I) The mRNA level of Col3a1, Fn1, Eln, Ccn2, Acta2, Col1a1, Ngal, and Kim-1 in the kidney of mice. For all panels, data are presented as mean ± SD. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 3—source data 1
Original files for western blot analysis displayed in Figure 3B, E and H.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig3-data1-v1.zip
-
Figure 3—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 3B, E and H.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig3-data2-v1.zip
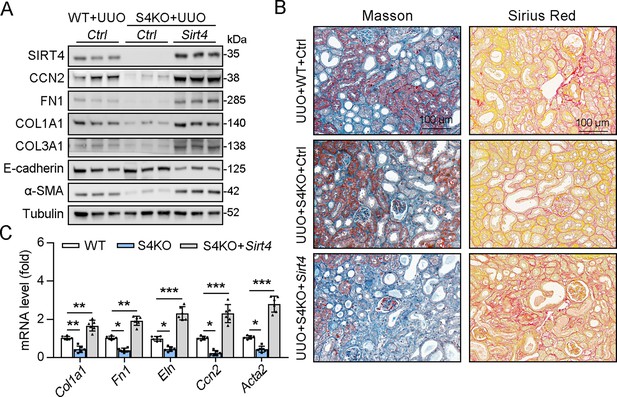
TECs targeted overexpression of SIRT4 reverses S4KO reduced kidney fibrosis in UUO mice.
(A–C) AAV9-Ksp-Sirt4 was injected into kidneys of mice in situ at three independent points in situ (AAV9-Ksp-null used as control). After 2-week transfection, the mice received UUO surgery (WT mice treated with AAV9-Ctrl followed by UUO surgery as control) (n=6 per group). (A) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL1A1, COL3A1, E-cadherin, α-SMA, and Tubulin in the kidney of mice. (B) Representative images of Masson’s trichrome staining and Sirius red in kidney sections of mice (scale bar, 100 μm). (C) The mRNA level of Col1a1, Fn1, Eln, Ccn2, and Acta2. For all panels, data are presented as mean ± SD. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 3—figure supplement 1—source data 1
Original files for western blot analysis displayed in Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig3-figsupp1-data2-v1.zip

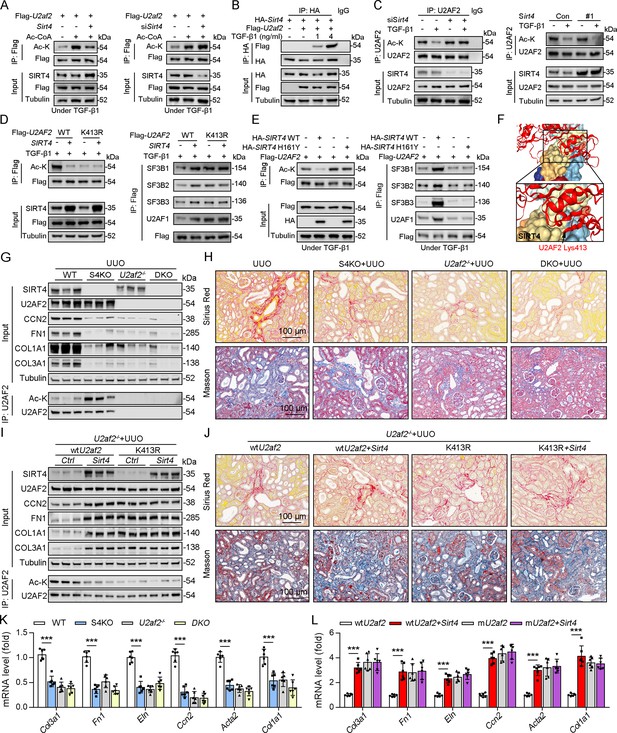
U2AF2 acetylation at K413 is increased under TGF-β1 stimulation.
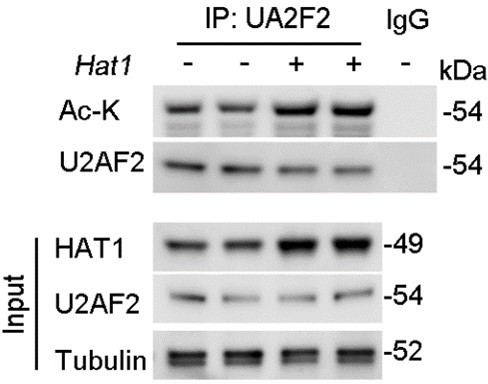
(A) Proteins interacted with SIRT4 in chromatin were identified by RIME. (B) The abundances (Normalized) of PUF60 and U2AF2 were shown. (C) Protein lysates from TECs were exposed to TGF-β1(left panel), kidney from the sham, uIRI, and UUO mice (middle panels), and WT TECs transfected with HA-Sirt4 and/or Flag-U2af2 and then treated with TGF-β1 (right panel), the protein lysates were subjected to Co-IP with anti-SIRT4 antibody, and determined protein expression using western blotting by the indicated antibodies. (D) TECs isolated from WT or U2af2-/- mice and then treated with TGF-β1, cells lysates subjected to Co-IP with anti-SIRT4 antibody, and western blotting using indicated antibodies. (E) WT TECs transfected with Ad-Sirt4 or Ad-null as indicated, and cells lysates subjected to Co-IP with anti-U2AF2 antibody, and western blotting using indicated antibodies. (F) HK2 cells (human renal TECs) transfected with Flag- U2AF2 WT, K70R, K276R, K272R, K292R, K413R, K453R, or K462R under Ac-CoA treatment, cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (G) The conservation of U2AF2 lysine 413 in different species. (H) TECs transfected with Flag-U2af2 WT under TGF-β1 stimulation, cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies (left panel). HK2 cells transfected with Flag-U2AF2 WT or K413R with or without TGF-β1, cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies (right panel). (I) Kidney lysates subjected to Co-IP with anti-U2AF2 antibody in uIRI, and UUO mice, and western blotting using indicated antibodies (middle and right panels). (J) WT TECs transfected with Flag-U2af2 WT and then under serum starvation, Tunicamycin, Poly (I, C), and Cisplatin stimulation. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (K) HK2 cells transfected with Flag-U2AF2 WT or K413R and cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies (left and middle panels). HK2 cells transfected with Flag-U2AF2 WT or K413R with or without RNase stimulation, cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies (right panel). (L) Schematic representation of components of SF3B complex performing pre-mRNA BPS recognition. (M) WT TECs transfected with Flag-U2af2 WT with CBP, p300, HAT1, P/CAF, KAT5, or KAT8, cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (N) Whole-kidney lysates were immunoprecipitated with anti-U2AF2 antibody, and precipitated proteins were detected by indicated antibodies (the upper panel). WT TECs was transfected with HA-Hat1 and/or Flag-U2af2. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (O) WT TECs was transfected with Flag-U2af2 WT with Ad-shHat1 or Ad-Hat1. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (P) HK2 cells were transfected with Flag-U2AF2 WT, Ad-HAT1, or Ad-HAT1 E276Q as indicated in figure. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (Q) WT TECs was transfected with Flag-U2af2, Ad-Hat1 or Ad-shHat1 under TGF-β1 stimulation. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies.
-
Figure 4—source data 1
Original files for western blot analysis displayed in Figure 4C, D, E, F, H, I, J, K, L, M, N, O, P and Q.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig4-data1-v1.zip
-
Figure 4—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 4C, D, E, F, H, I, J, K, L, M, N, O, P and Q.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig4-data2-v1.zip
U2AF2 deacetylation under TGF-β1 is SIRT4 dependent.
(A) WT TECs were transfected with Flag-U2af2 WT, Ad-Sirt4 or Ad-shSirt4 under Ac-CoA treatment. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (B) WT TECs were transfected with Flag-U2af2 WT and HA-Sirt4 under TGF-β1 stimulation. Cells lysates subjected to Co-IP with anti-HA antibody and Western blotting using indicated antibodies. (C) WT TECs were transfected with Ad-shSirt4 or Ad-Sirt4 with or without TGF-β1 stimulation. Cells lysates subjected to Co-IP with anti-U2AF2 antibody, and western blotting using indicated antibodies. (D) HK2 cells were transfected with Flag-U2AF2 WT or K413R, Ad-shSIRT4 under TGF-β1 stimulation. Cells lysates subjected to Co-IP with anti-Flag antibody, and Western blotting using indicated antibodies. (E) HK2 cells were transfected with HA-SIRT4 WT or H161Y and Flag-U2AF2 under TGF-β1 treatment. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (F) SIRT4-U2AF2 docking with the HDOCK server. High magnification of boxed areas is presented on the left. Arrow indicates K413 of U2AF2 protein. (G, H, K) WT, S4KO, U2AF2-/- or DKO mice were randomly assigned to UUO surgery according to an established protocol. Kidney samples were obtained from mice on day 10 post surgery or sham surgery (n=6 per group). (G) Western blot analysis of the expression of SIRT4, U2AF2, CCN2, FN1, COL1A1, COL3A1, and Tubulin in the kidney from mice. Kidney lysates subjected to Co-IP with anti-U2AF2 antibody, and western blotting using indicated antibodies. (H) Representative images of Masson’s trichrome staining and Sirius red in kidneys from mice (scale bar, 100 μm). (K) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2 and Col3a1 in the kidney of mice. (I, J, L) U2af2-/- mice received in situ renal injection of AAV9-Ksp-U2af2 WT (wtU2af2), wtU2af2 plus Sirt4, AAV9-Ksp-U2af2 K413R (mutU2af2), K413R plus Sirt4 at 6 weeks of age. After 2 weeks, the mice were randomly assigned to sham surgery or UUO surgery according to an established protocol. Kidney samples were obtained from mice on day 10 post surgery or sham surgery (n=6 per group). (I) Western blot analysis of the expression of SIRT4, U2AF2, CCN2, FN1, COL1A1, COL3A1, and Tubulin in the kidney from mice. Kidney lysates subjected to Co-IP with anti-U2AF2 antibody, and western blotting using indicated antibodies. (J) Representative images of Masson’s trichrome staining and Sirius red in kidneys from mice (scale bar, 100 μm). (L) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, and Col3a1 in the kidney of mice. For all panels, data are presented as mean ± SD. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 5—source data 1
Original files for western blot analysis displayed in Figure 5A, B, C, D, E, G, I.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig5-data1-v1.zip
-
Figure 5—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 5A, B, C, D, E, G I.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig5-data2-v1.zip
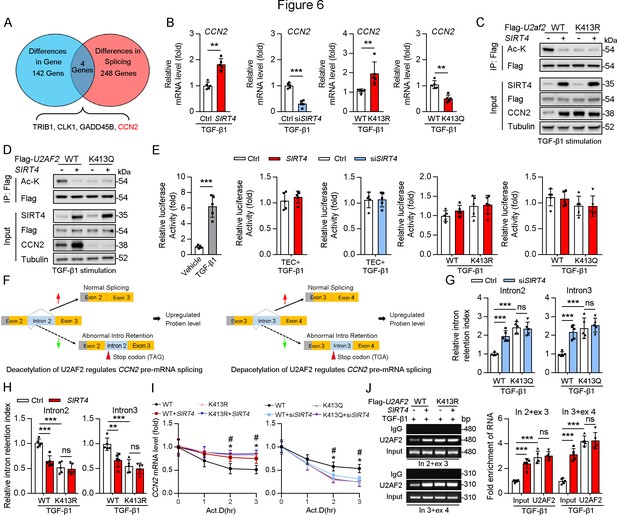
Acetylation of U2AF2 at K413 regulates CCN2 gene expression and pre-mRNA splicing.
(A) The crosstalk of differences of the gene expression profiling analysis and splicing analysis. (B) The Ccn2 mRNA level in HK2 cells (n=5). (C, D) HK2 cells were transfected with Flag-U2AF2 WT, K413R, or K413Q, and Ad-SIRT4 or SIRT4 siRNA (siSIRT4) as indicated in figure. Cells lysates subjected to Co-IP with anti-Flag antibody, and western blotting using indicated antibodies. (E) Luciferase activity of Ccn2 promoter in HK2 cells transfected with Ad-SIRT4, Ad-shSIRT4, WT U2AF2, U2AF2 K413R, or U2AF2 K413Q under TGF-β1 stimulation was measured (n=5). (F) Schematics of Ccn2 alternative splicing pattern regulated by hyperacetylated U2AF2. Full line and dotted line represent normal splicing and abnormal intron retention, respectively. Grey and yellow boxes represent untranslated region (UTR) and protein coding region (CDS), respectively. The first stop codon in intron 2 or 3 was indicated with red arrow. (G, H) Quantitative real-time PCR analysis of normal or abnormal splicing isoforms of CCN2 (n=5). (I) Actinomycin D (Act D, 2 mg/mL) treatment and quantitative real-time PCR were performed to measure CCN2 mRNA levels (n=5). (J) HK2 cells were transfected with Flag-U2AF2 WT, Flag-U2AF2 WT, or Flag-U2AF2 K413R with or without Ad-SIRT4 under TGF-β1 stimulation. Cells lysates subjected to RNA Binding Protein Immunoprecipitation (RIP) with anti-U2AF2 antibody (left panel). Quantitative real-time PCR results of RIP assays (right panel). For all panels, data are presented as mean ± SD. **p< 0.01, ***p< 0.001, by Student’s t-test for B. **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test for E, G, H, I, and J.
-
Figure 6—source data 1
Original files for western blot and Chip analysis displayed in Figure 6C, D and J.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig6-data1-v1.zip
-
Figure 6—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 6C, D and J.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig6-data2-v1.zip
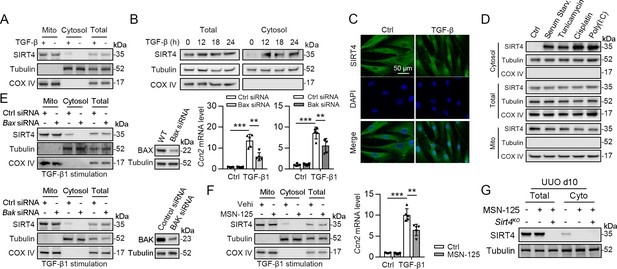
SIRT4 is released through BAX/BAK pore in a TGF-β1-dependent manner.
(A) Organelle separation experiment and immunoblot analysis detected the expression/localization of SIRT4, Tubulin, and COX IV. (B) TECs were treated with TGF-β1 for indicated time. Organelle separation experiment and immunoblot analysis detected the localization of SIRT4, Tubulin, and COX IV. (C) Representative image of immunofluorescent staining of SIRT4 in TECs treated with TGF-β1 or not (scale bar, 50 μm). (D) TECs were treated with Tunicamycin, Cisplatin, Poly(I:C), or under serum starvation. Organelle separation experiment and immunoblot analysis detected the localization of SIRT4, Tubulin, and COX IV. (E) WT TECs incubated with Bax siRNA or Bak siRNA under TGF-β1 stimulation. Organelle separation experiment and immunoblot analysis detected the localization of SIRT4, Tubulin, and COX IV (left panels). Western blot analysis of the expression of BAX, BAK and Tubulin in TECs (middle panels). The mRNA level of Ccn2 was detected by qPCR (n=5) (right panels). (F) WT TECs were treated with MSN-125 (10 μM) or vehicle. Organelle separation experiment and immunoblot analysis detected the localization of SIRT4, Tubulin, and COX IV (left panel). The mRNA level of Ccn2 was detected by qPCR (n=5) (right panel). (G) Kidney tissues from WT or S4KO mice were treated with MSN-125 or vehicle. The mRNA level of Ccn2 was detected by qPCR in the kidney of mice (n=5). For all panels, data are presented as mean ± SD. **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 7—source data 1
Original files for western blot analysis displayed in Figure 7A, B, D, E, F and G.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig7-data1-v1.zip
-
Figure 7—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 7A, B, D, E, F and G.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig7-data2-v1.zip
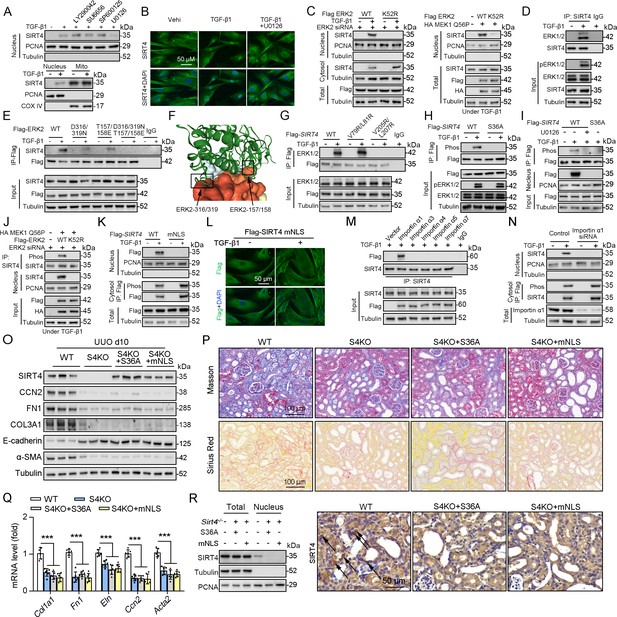
ERK2 phosphorylates SIRT4 at S36 and promotes it nucleus translocation.
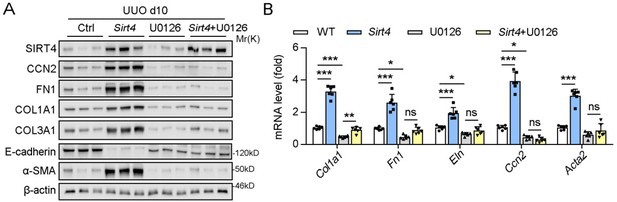
(A) (The upper panel) Nuclear fractions were prepared from TECs pretreated with LY290042 (30 μM), SU6656 (4 μM), SP600125 (25 μM), and U0126 (20 μM) for 30 min before TGF-β1 (2 ng/ml) for 12 hr. Nuclear PCNA and cytoplasmic Tubulin were used as controls. (The bottom panel) TECs were treated with TGF-β1 (2 ng/ml) for 12 hr. Organelle separation experiment and immunoblot analysis detected the localization of SIRT4, PCNA, and Tubulin. (B) TECs were pretreated with or without U0126 (20 μM) for 30 min, then treated with TGF-β1 (2 ng/ml) for 12 hr. Immunofluorescence analyses were performed with the indicated antibodies. (C) HK2 cells were stably transfected with Flag-ERK2, Flag-ERK2 K52R, or siRNA ERK2 (left panel) or transiently transfected with HA-MEK1 Q56P and indicated Flag-tagged ERK2 proteins (right panel). The cells were treated with or without TGF-β1 (2 ng/ml) for 12 hr, and the total cell lysates, cytosol and nuclear fractions were prepared for determination of indicated proteins by western blot. (D) TECs were treated with TGF-β1, and cells lysates subjected to Co-IP with anti-SIRT4 antibody, and western blotting using indicated antibodies. (E) HK2 cells transfected with vectors expressing the indicated Flag-tagged ERK proteins were treated with or without TGF-β1 (2 ng/ml) for 12 hr. (F) SIRT4-ERK2 docking with the HDOCK server. (G) HK2 cells expressing the indicated Flag-SIRT4 proteins were treated with or without TGF-β1 (2 ng/ml) for 12 hr. (H, I) HK2 cells transfected with Flag-Sirt4 WT or S36A and then incubated with TGF-β1 and/or U0126 as indicated in figure, the total cell lysates and nuclear fractions were prepared. Total cells lysates subjected to Co-IP with anti-Flag antibody, and Western blotting using indicated antibodies. (J) HK2 cells were transfected with MEK1 Q56P, Flag-ERK2 WT or K52R, and ERK2 siRNA as indicated in figure, and the total cell lysates and nuclear fractions were prepared. Total cells lysates subjected to Co-IP with SIRT4 antibody and western blotting using indicated antibodies. (K, L) HK2 cells transfected with Flag-Sirt4 WT or mNLS with or without TGF-β1 treatment, and the total cell lysates, cytosol, and nuclear fractions were prepared. Western blotting using indicated antibodies (K). Immunofluorescence analyses were performed with the indicated antibodies (L). (M) The indicated Flag-importin α proteins were transfected in HK2 cells, then treated with TGF-β1 (2 ng/ml) for 12 h. Flag-importin α proteins were immunoprecipitated with an anti-Flag antibody. (N) HK2 cells transfected with importin α1 were treated with or without TGF-β1 (2 ng/ml) for 12 h. The total cell lysates, cytosolic and nuclear fractions were prepared and western blotting using indicated antibodies. (O–R) S4KO mice received in situ renal injection of AAV9-Ksp-Sirt4 S36A (S36A) or AAV9-Ksp-Sirt4 mNLS (mNLS) at 6 weeks of age. After 2 weeks, the mice were randomly assigned to sham surgery or UUO surgery according to an established protocol. O: Western blot analysis of the expression of SIRT4, CCN2, FN1, COL3A1, E-cadherin, α-SMA and Tubulin in mouse kidney. P: Representative images of Masson’s trichrome staining and Sirius red staining in kidney sections from mice (scale bar, 100 μm). Q: The mRNA level of Col1a1, Fn1, Eln, Ccn2 and Acta2 in the kidney of mice. R: The total cell lysates and nuclear fractions were prepared from kidney and western blotting using indicated antibodies (left panel). (Right panel) Representative images of immunohistochemical staining of SIRT4 in kidneys from mice (scale bar, 100 μm). For all panels, data are presented as mean ± SD. ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 8—source data 1
Original files for western blot analysis displayed in Figure 8A, C, D, E, G, H, I, J, K, M, N, O and R.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-data1-v1.zip
-
Figure 8—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 8A, C, D, E, G, H, I, J, K, M, N, O and R.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-data2-v1.zip
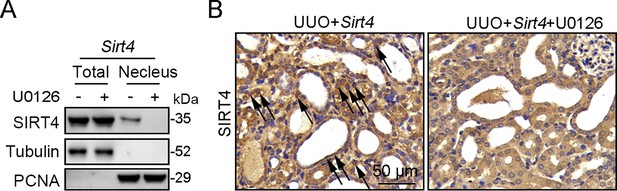
U0126 prevents SIRT4 overexpression induced kidney fibrosis in UUO mice.
(A, B) AAV9-Ksp-Sirt4 was injected into kidneys of mice in situ at three independent points in situ. After 2-week transfection, the mice received UUO surgery, and then mice were treated with U0126 (10 mg/kg body weight) or vehicle for 10 days (n=6 per group). (A) Organelle separation experiment and immunoblot analysis detected the localization of SIRT4 and Tubulin in the kidneys of mice. (B) Representative images of immunohistochemical staining of SIRT4 (scale bar, 50 μm) in the kidney sections of mice.
-
Figure 8—figure supplement 1—source data 1
Original files for western blot analysis displayed in Figure 8—figure supplement 1A.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-figsupp1-data1-v1.zip
-
Figure 8—figure supplement 1—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 8—figure supplement 1A.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-figsupp1-data2-v1.zip
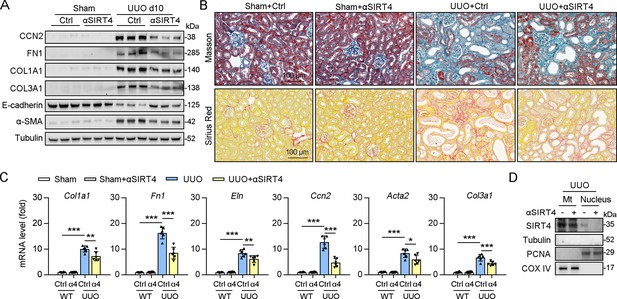
Exosomes contain anti-SIRT4 antibody alleviated UUO-induced kidney fibrosis.
(A–D) After sham or UUO surgery, WT mice were treated with exosomes null or contain anti-SIRT4 for 10 days (n=6 per group). (A) Western blot analysis of the expression of CCN2, FN1, COL1A1, COL3A1, E-cadherin. α-SMA and Tubulin in the kidney from mice. (B) Representative images of Masson’s trichrome staining and Sirius red staining in kidneys sections of mice (scale bar, 100 μm). (C) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, and Col3a1 in the kidney of mice. (D) Organelle separation experiment and immunoblot analysis detected the localization of SIRT4 in the kidneys of mice. For all panels, data are presented as mean ± SD. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 8—figure supplement 2—source data 1
Original files for western blot analysis displayed in Figure 8—figure supplement 2A, D.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-figsupp2-data1-v1.zip
-
Figure 8—figure supplement 2—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 8—figure supplement 2A, D.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-figsupp2-data2-v1.zip
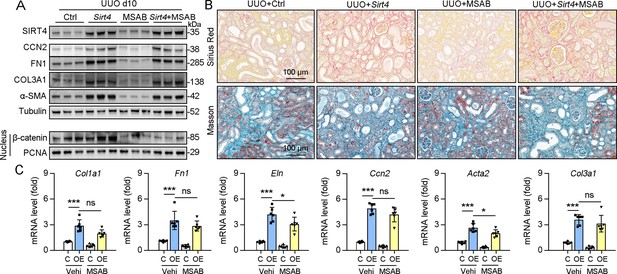
Inhibition of nucleus accumulation of β-catenin failed to suppresses kidney fibrosis induced by SIRT4 overexpression in UUO mice.
(A–C) AAV9-Ksp-Sirt4 or AAV-Ctrl was injected into kidneys of mice in situ at three independent points in situ. After 2-week transfection, the mice received UUO surgery. After UUO surgery, mice were treated with vehicle or MSAB for 10 days (n=6 per group). (A) Western blot analysis of the expression of SIRT4, CCN2, FN1, COL3A1, α-SMA, and Tubulin in the kidneys from mice. (B) Representative images of Masson’s trichrome staining and Sirius red staining in kidney sections of mice (scale bar, 100 μm). (C) The mRNA level of Col1a1, Fn1, Eln, Ccn2, Acta2, and Col3a1 in the kidney of mice. For all panels, data are presented as mean ± SD. ns: not significant difference, *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni correction test.
-
Figure 8—figure supplement 3—source data 1
Original files for western blot analysis displayed in Figure 8—figure supplement 3A.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-figsupp3-data1-v1.zip
-
Figure 8—figure supplement 3—source data 2
The uncropped gels or blots with the relevant bands clearly labeled in Figure 8—figure supplement 3A.
- https://cdn.elifesciences.org/articles/98524/elife-98524-fig8-figsupp3-data2-v1.zip
Additional files
-
Supplementary file 1
Supplementary tables 1a-e.
(a) Differential expressed genes in SIRT4 OE TECs vs Ctrl TECs. Differential expression was performed with DESeq, P<0.01. (b) Genes with significantly different intron retention events in SIRT4 OE TECs vs Ctrl TECs (FDR <0.05). (c) Clinical characteristics of CKD in biopsy samples Data are calculated by One-way ANOVA and presented as mean ± SEM. The number of patients in each group was as indicated. NS: not significant difference, y: year. (d) Primers used in qPCR. (e) Primers used in IR Index.
- https://cdn.elifesciences.org/articles/98524/elife-98524-supp1-v1.xls
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98524/elife-98524-mdarchecklist1-v1.docx