Opposing roles for Bmp signalling during the development of electrosensory lateral line organs
Figures
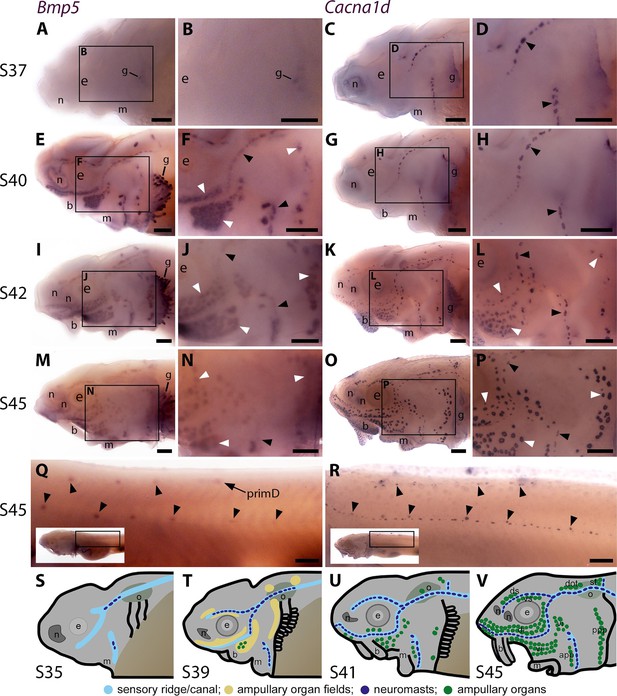
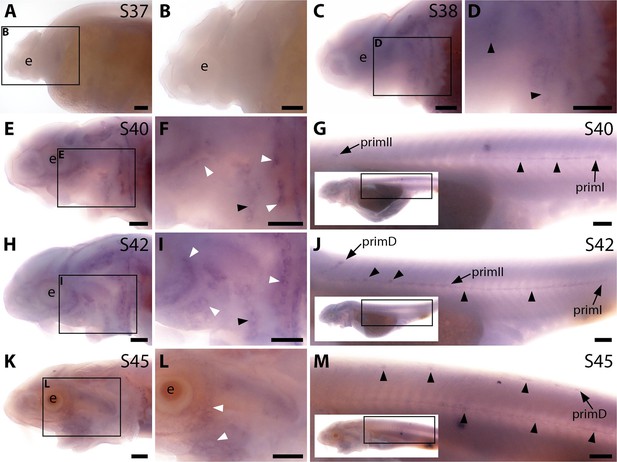
Sterlet Bmp5 is expressed early in developing ampullary organs and transiently in mature neuromasts.
(A–R) In situ hybridisation in sterlet for Bmp5 or the hair cell and electroreceptor marker Cacna1d, which labels mature neuromasts and ampullary organs (also expressed in taste buds on the barbels). Black arrowheads indicate examples of developing neuromasts; white arrowheads indicate examples of developing ampullary organs. (A–D) At stage 37, Bmp5 expression is only detectable in developing gill filaments (A,B) although Cacna1d-positive neuromasts are present (C,D). (E–H) At stage 40, Bmp5 is expressed in neuromasts and ampullary organ primordia (E,F); only a few Cacna1d-positive ampullary organs are present at this stage (G, H). (I–L) At stage 42, Bmp5 is expressed in mature ampullary organs and more weakly in neuromasts (I,J); compare with Cacna1d expression (K,L). (M–P) At stage 45 (onset of independent feeding), Bmp5 expression is weaker in ampullary organs and no longer detectable in most neuromasts (M,N); compare with Cacna1d expression (O,P). (Q,R) At stage 45 on the trunk, Bmp5 expression is visible in primII-deposited secondary neuromasts (more strongly in more rostral neuromasts) as well as in primD and neuromasts of the dorsal line (Q). Compare with Cacna1d expression in all neuromasts (R): arrowheads indicate examples of dorsal-line neuromasts and primII-deposited secondary neuromasts (offset a little dorsal to the line of primI-deposited primary neuromasts). Low-power insets show the location of these trunk regions. (S–V) Schematic depictions of sterlet lateral line organ development at similar stages (stages 35, 39, 41, 45), previously published in Minařík et al., 2024a. Abbreviations: app, anterior preopercular ampullary organ field; b, barbel; di, dorsal infraorbital ampullary organ field; dot, dorsal otic ampullary organ field; ds, dorsal supraorbital ampullary organ field; e, eye; gf, gill filaments; m, mouth; n, naris; o, otic vesicle; ppp, posterior preopercular ampullary organ field; prim, migrating lateral line primordium (primI, primary; primII, secondary; primD, dorsal); S, stage; st, supratemporal ampullary organ field; vi, ventral infraorbital ampullary organ field; vs, ventral supraorbital ampullary organ field. Scale bar: 250 μm.
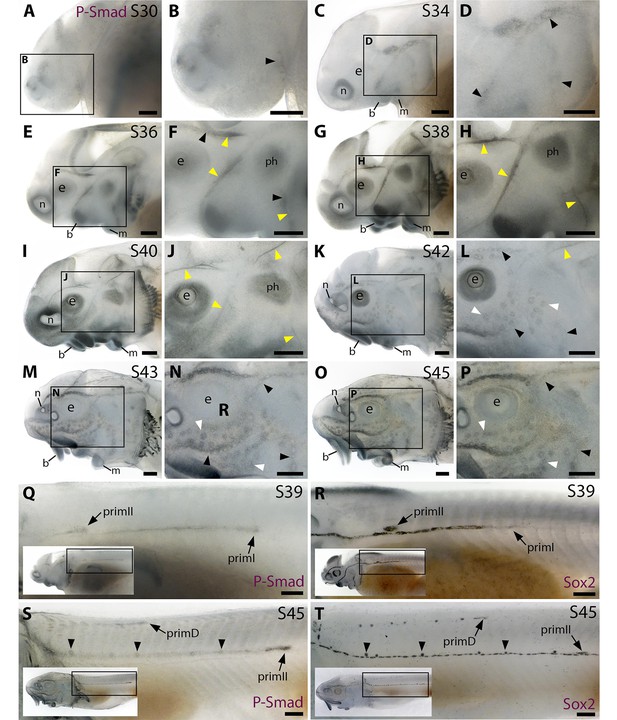
The Bmp signalling pathway is active throughout the developing lateral line system in sterlet.
Immunostaining in sterlet. Black arrowheads indicate examples of developing neuromasts; white arrowheads indicate examples of developing ampullary organs; yellow arrowheads indicate lateral line nerves. (A–P) Immunoreactivity on the head for phospho-SMAD1/5/9 (P-Smad) as a proxy for Bmp signalling activity. At stage 30 (A,B), weak immunoreactivity is seen in the region of the anteroventral lateral line primordium and by stage 34 (C,D) in lateral line primordia, with a ring pattern around developing neuromast primordia. At stages 36–40 (E–J), immunoreactivity is weak around developing neuromasts and prominent in lateral line nerves (yellow arrowheads). At stage 40 (I,J), diffuse immunoreactivity is also seen in regions flanking the nerves where ampullary organ primordia are forming. Non-lateral line immunoreactivity is present around the mouth and nares, in barbel primordia, gill filaments, and a patch that is most likely the developing muscle m. protractor hyomandibulae. Between stages 42 and 45 (K–P), immunoreactivity disappears in lateral line nerves and is increasingly detected at the periphery of ampullary organs and neuromasts (strongly in supraorbital and infraorbital neuromast lines). (Q–T) Immunostaining on the trunk (boxes on low-power insets indicate the location of the trunk regions shown). At stage 39 (Q,R), pSMAD1/5/9 immunoreactivity is seen in primI and a diffuse trail behind it, and in primII (Q). For comparison, Sox2 is expressed weakly in primI and strongly in primI-deposited neuromasts and interneuromast cells, plus primII (R). At stage 45 (S,T), pSMAD1/5/9 immunoreactivity is seen in primD and primII plus a weak trail behind it, with greater intensity at the periphery of primII-deposited neuromasts (S). For comparison, Sox2 expression is strong in primII, primD and all neuromasts; arrowheads indicate examples of primII-deposited neuromasts (T). Abbreviations: b, barbel; e, eye; f, fin; g, gill filaments; m, mouth; n, naris; ph, m. protractor hyomandibulae; prim, migrating lateral line primordium (primI, primary; primII, secondary; primD, dorsal); S, stage. Scale bar: 250 μm.
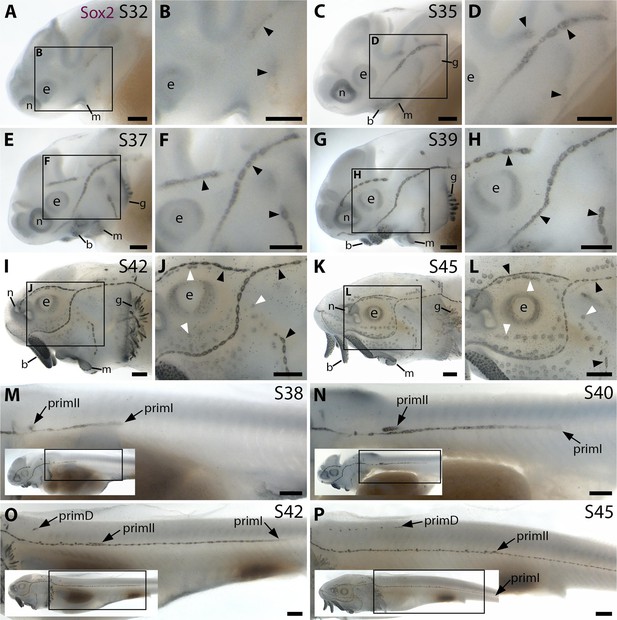
Sox2 expression shows the time-course of sterlet lateral line organ development.
Sterlet embryos and yolk-sac larvae immunostained at selected stages for Sox2, which labels lateral line primordia/sensory ridges and supporting cells in lateral line organs. Black arrowheads indicate examples of developing neuromasts; white arrowheads indicate examples of developing ampullary organs. Non-lateral line expression is seen around the mouth and nares, in the eye, gill filaments and taste buds on the barbels. Scattered individual Sox2-positive skin cells are most likely Merkel cells. (A–D) At stage 32 (A,B) and stage 35 (C,D), Sox2 expression is seen in the otic/anterodorsal and anteroventral lateral line primordia, with a ring pattern around developing neuromast primordia. (E–H) At stage 37 (E,F) and stage 39 (G,H), Sox2 is expressed at the edges of neuromasts and in interneuromast cells. (I–L) At stage 42 (I,J) and stage 45 (K,L), Sox2 is expressed at the periphery of neuromasts and ampullary organs, with stronger expression in neuromasts. The images shown in K,L were previously published in Minařík et al., 2024a. (M–P) Sox2 expression on the trunk at stage 38 (M), stage 39 (N), stage 42 (O) and stage 45 (P) shows the progression of the migrating lateral line primordia (primI, primII and primD). Low-power insets show the location of these trunk regions. Sox2 is expressed only weakly in primI (e.g., M,N), but strongly in primI-deposited neuromasts and interneuromast cells (M–P). PrimII is located a little dorsal to the primI-deposited line (M–P), and primII-deposited neuromasts are slightly offset dorsally from the line of primI-deposited neuromasts (O,P). Sox2 expression only reveals neuromasts deposited by primII and primD, not interneuromast cells (O,P; compare with the primI-deposited line in M-P). Abbreviations: b, barbel; e, eye; g, gill filaments; m, mouth; n, naris; prim, migrating lateral line primordium (primI, primary; primII, secondary; primD, dorsal); S, stage. Scale bar: 250 μm.
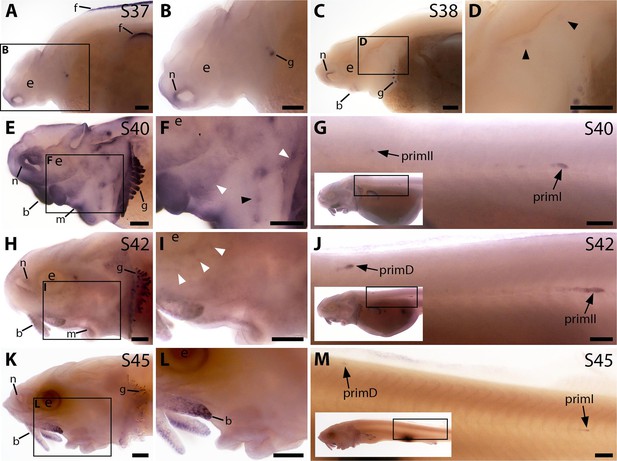
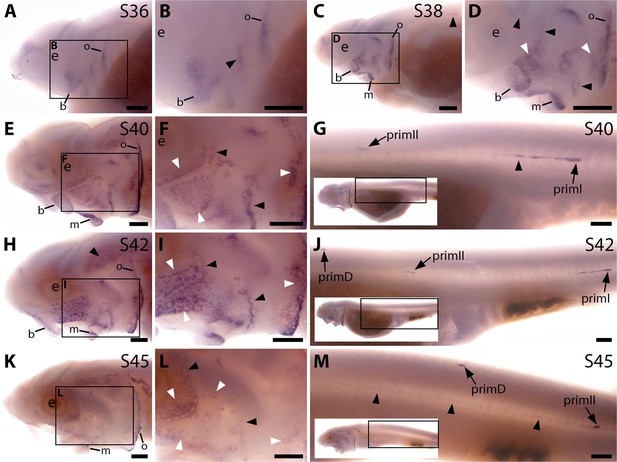
Bmp4 is expressed transiently during sterlet lateral line organ development.
In situ hybridisation in sterlet for Bmp4. Black arrowheads indicate examples of neuromast regions; white arrowheads indicate examples of ampullary organ regions. For images of the trunk, boxes on low-power insets delineate the location of the trunk regions shown. (A,B) At stage 37, Bmp4 is not expressed in lateral line regions, although it is present around the nares and in fins and gill-filament primordia. (C,D) At stage 38, two dorsal spots of weak Bmp4 expression may represent sensory patches in the otic vesicle or early-forming neuromast primordia in the otic and supratemporal lines. Expression is also present in the gills, nares and barbel primordia. (E–G) At stage 40, expression is seen on the head in neuromast regions and fields of ampullary organ primordia (E,F; compare with Bmp5 and Cacna1d expression in Figure 1E–H). On the trunk, Bmp4 is expressed in primI and the most recently deposited neuromasts behind it, and in primII (G). (H–J) At stage 42, Bmp4 expression on the head has largely disappeared (H,I), apart from weak expression in the dorsal infraorbital field (arrowheads in I), although expression is still seen in gill filaments and barbels. On the trunk, expression is seen in primD and primII (J). (K–M) At stage 45, no lateral line expression is seen on the head (K,L), although weak expression persists in primD and primI on the trunk (M). Abbreviations: b, barbel; e, eye; f, fin; g, gill filaments; m, mouth; n, naris; prim, migrating lateral line primordium (primI, primary; primII, secondary; primD, dorsal); S, stage. Scale bar: 250 μm.
Sterlet Acvr2a is expressed in developing neuromasts and ampullary organs.
In situ hybridisation in sterlet for the type II receptor gene Acvr2a. Black arrowheads indicate examples of developing neuromasts; white arrowheads indicate examples of developing ampullary organs. For trunk images, boxes on low-power insets delineate the regions shown. (A,B) At stage 37, there is no detectable Acvr2a expression. (C,D) At stage 38, faint expression is seen in developing neuromast regions. (E–J) At stage 40 (E–G) and stage 42 (H–J), Acvr2a is expressed in developing ampullary organ primordia and neuromasts on the head (E,F) as well as in the migrating lateral line primordia and neuromasts on the trunk. (K–M) By stage 45, only faint Acvr2a expression remains around some ampullary organs on the head (K,L), although expression is still seen in the migrating primordia and neuromasts on the trunk (M). Abbreviations: e, eye; prim, migrating lateral line primordium (primI, primary; primII, secondary; primD, dorsal); S, stage. Scale bar: 250 μm.
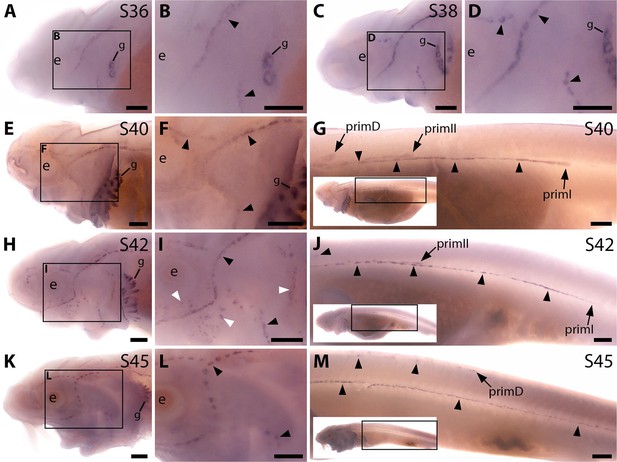
Sterlet Sostdc1 is expressed in neuromasts but only transiently in ampullary organs.
In situ hybridisation in sterlet for Sostdc1, encoding a secreted dual Wnt/Bmp antagonist. Black arrowheads indicate examples of developing neuromasts; white arrowheads indicate examples of developing ampullary organs. For trunk images, boxes on low-power insets delineate the regions shown. Non-lateral line expression is seen in developing gill filaments. (A–D) Sostdc1 expression is seen in neuromasts on the head from stage 36 (A,B), persisting at stage 38 (C,D). (E–G) At stage 40, expression is maintained in neuromasts on the head (E,F) and is also visible in the three migrating lateral line primordia on the trunk and in trunk neuromasts (G). (H–J) At stage 42, Sostdc1 expression persists in cranial neuromasts and is now also seen in ampullary organs (H,I). On the trunk, expression persists in the migrating primordia and neuromasts (J). (K–M) By stage 45, only neuromast expression is observed on the head (K,L). On the trunk, expression continues in the migrating primordia and neuromasts (M). Abbreviations: e, eye; prim, migrating lateral line primordium (primI, primary; primII, secondary; primD, dorsal); S, stage. Scale bar: 250 μm.
Sterlet Apcdd1 is expressed in ampullary organs and neuromasts during development.
In situ hybridisation in sterlet for Apcdd1, encoding a secreted dual Wnt/Bmp antagonist. Black arrowheads indicate examples of developing neuromasts; white arrowheads indicate examples of developing ampullary organs. For trunk images, boxes on low-power insets delineate the regions shown. (A,B) At stage 36, Apcdd1 expression is seen in the region of the preopercular neuromast line, as well as in developing barbel regions, the opercular edge and around the mouth. (C,D) By stage 38, expression in all these locations is more distinct. (E–J) At stage 40 (E–G) and stage 42 (H–J), Apcdd1 is expressed around ampullary organ primordia and neuromasts on the head (E,F,H,I) and is also seen on the trunk in primI and primII and a fairly short line of trailing cells behind primI (G,J). (K–M) At stage 45, this expression pattern largely persists on the head, although appears to be fading in the ventral infraorbital ampullary organ field (K,L). On the trunk, faint expression is seen along the main body line, with strong expression in primD and primII (M). Abbreviations: b, barbels; e, eye; m, mouth; o, operculum edge; prim, migrating lateral line primordium (primI, primary; primII, secondary; primD, dorsal); S, stage. Scale bars: 250 μm.
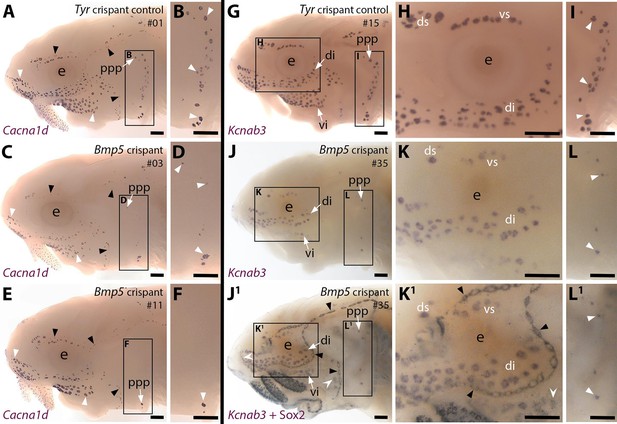
CRISPR/Cas9-mediated targeting of Bmp5 leads to fewer ampullary organs developing.
Sterlet crispants at stage 45 after in situ hybridisation (ISH) for the hair cell and electroreceptor marker Cacna1d (also expressed in taste buds on the barbels) or the electroreceptor-specific marker Kcnab3. All crispants shown are from the same batch of siblings/half-siblings (in vitro fertilisation used a mix of sperm from three different males). Black arrowheads indicate examples of neuromasts; white arrowheads indicate examples of ampullary organs. Crispants are numbered for cross-referencing with data provided for each crispant in Supplementary file 2. (A,B) In a control Tyr crispant, Cacna1d expression shows the normal pattern of neuromast lines flanked by fields of ampullary organs. The higher power view shows the posterior preopercular ampullary organ field. (C–F) In Bmp5 crispants, Cacna1d expression reveals fewer ampullary organs (compare C,E with A); this phenotype is particularly prominent in the posterior preopercular ampullary organ field (compare D,F with B). (G–I) In a control Tyr crispant, electroreceptor-specific Kcnab3 expression shows the normal distribution of ampullary organs. (J–L1) In a Bmp5 crispant, Kcnab3 expression shows fewer ampullary organs (compare J-L with G-I). Post-ISH Sox2 immunostaining for supporting cells (J1,K1,L1) demonstrates that neuromasts have formed normally. Very few "additional" ampullary organs appeared (i.e., Sox2-positive, Kcnab3-negative ampullary organs: compare J1,K1,L1 with I,J,K); examples are indicated with indented white arrowheads. (Non-lateral line Sox2 expression is also seen in gill filaments and in taste buds on the barbels and around the mouth.) Abbreviations: di, dorsal infraorbital ampullary organ field; ds, dorsal supraorbital ampullary organ field; e, eye; ppp, posterior preopercular ampullary organ field; S, stage; vi, ventral infraorbital ampullary organ field; vs, ventral supraorbital ampullary organ field. Scale bar: 250 μm.
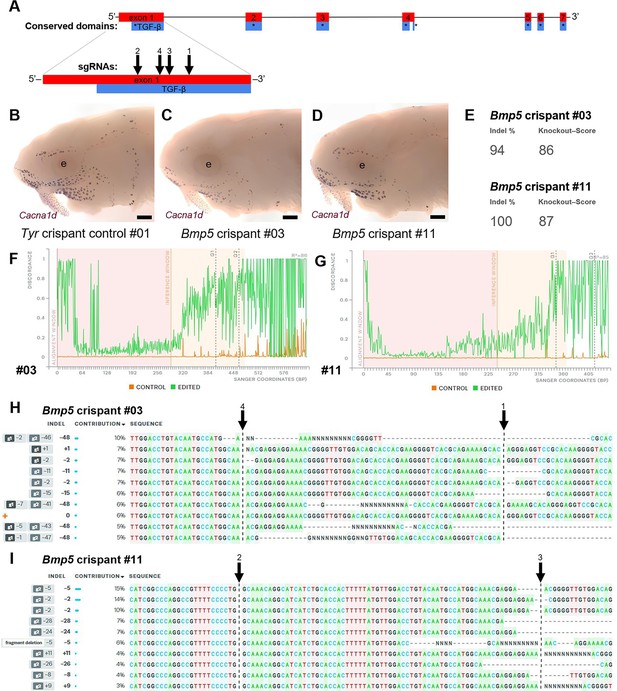
Examples of successful disruption of sterlet Bmp5 by CRISPR/Cas9-mediated mutagenesis in G0-injected embryos.
(A) Schematic showing the exon structure of the sterlet Bmp5 gene relative to conserved domains and the target sites of Bmp5 sgRNAs (Table 1). (B–D) Sterlet crispants at stage 45 after in situ hybridisation for the hair cell and electroreceptor marker Cacna1d (also expressed in taste buds on the barbels). In comparison to the control Tyr crispant (B), the two Bmp5 crispants (C,D) have fewer ampullary organs (this is particularly clear on the operculum). These images are also shown in Figure 4A, C and E. (E–I) Outputs are shown from Synthego’s 'Inference of CRISPR Edits' (ICE) tool (Conant et al., 2022). This tool was used to analyse Sanger sequence data for the targeted region of genomic DNA extracted from the trunk of each of the Bmp5 crispants shown in panels C and D. ‘Indel %’ (E) gives the percentage of insertions and/or deletions among the inferred sequences in the CRISPR-edited population. ‘Knockout-Score’ (E) shows the proportion of indels introducing a frameshift, or that are at least 21 bp in length. Discordance plots (F,G) show the level of discordance between the control sample trace file (orange) and the edited sample Sanger trace file (green). The expected cut sites for the sgRNAs are shown by vertical dotted lines. A successful CRISPR edit is indicated by the increase in discordance near the expected cut site. Panels H and I show nucleotide sequences from the Sanger trace files and their inferred relative contributions to the edited mosaic population. Vertical dotted lines indicate the expected cut sites for the sgRNAs. The wild-type sequence (0) is marked by an orange ‘+’ symbol in H, but is absent in panel I because 100% of the sequence was edited in this crispant. Abbreviation: e, eye. Scale bar: 250 μm.
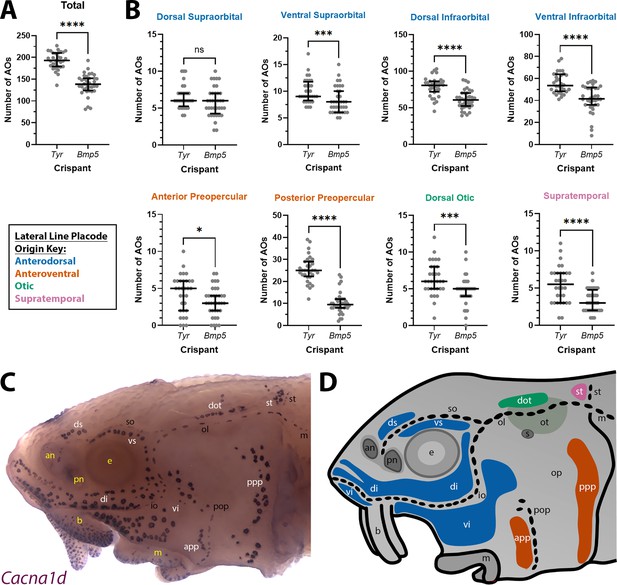
Bmp5 crispants have significantly fewer ampullary organs than control Tyr crispants.
(A) Scatter plot showing median and interquartile range for the total number of ampullary organs on one side of the head at stage 45 in Bmp5 sterlet crispants (counted after in situ hybridisation [ISH] for Cacna1d or Kcnab3; n=36) versus control Tyr crispants (counted after ISH for Cacna1d or Kcnab3; n=32). Bmp5 crispants have significantly fewer ampullary organs overall than control Tyr crispants (p<0.0001; two-tailed Mann-Whitney test). Supplementary file 2 provides the sgRNA combination, injection batch and raw counts for each crispant. All the Bmp5 crispants and 20 of the Tyr crispants used for statistical analysis were from the same batch. (B) Scatter plots showing median and interquartile range for the number of ampullary organs in each individual ampullary organ field on one side of the head at stage 45 in Bmp5 crispants (n=36) versus control Tyr crispants (n=32). The raw counts are provided in Supplementary file 2. For the location of each field, see panel C (Cacna1d expression) and panel D (schematic). Scatter plots are grouped with differently coloured titles according to lateral line placode (LLp) origin, following Gibbs and Northcutt, 2004: blue, anterodorsal LLp (supraorbital and infraorbital fields); orange, anteroventral LLp (preopercular fields); green, otic LLp (dorsal otic field); pink, supratemporal LLp (supratemporal field). All fields except the dorsal supraorbital field have significantly fewer ampullary organs in Bmp5 crispants versus control Tyr crispants (two-tailed Mann-Whitney tests). Symbols on plots represent p values: ns, not significant, p>0.05; *, p≤0.05; ***, p≤0.001; ****, p≤0.0001. Dorsal supraorbital: not significant, p=0.1207. Ventral supraorbital: p=0.0008. Dorsal infraorbital: p<0.0001. Ventral infraorbital: p<0.0001. Anterior preopercular: p=0.0466. Posterior preopercular: p<0.0001. Dorsal otic: p=0.0008. Supratemporal: p<0.0001. (C) Stage 45 sterlet head after ISH for the hair cell and electroreceptor marker Cacna1d (also expressed in taste buds on the barbels). Labels are white for ampullary organ fields; black for neuromast lines; yellow for anatomical landmarks. (D) Schematic of a stage 45 sterlet larval head. Ampullary organ fields are represented by coloured patches flanking the neuromast lines, which are represented as dotted lines. The different field colours indicate their lateral line placode origin (consistent with scatter plot titles in B). Abbreviations for ampullary organ fields: app, anterior preopercular; di, dorsal infraorbital; dot, dorsal otic; ds, dorsal supraorbital; ppp, posterior preopercular; st, supratemporal; vi, ventral infraorbital; vs, ventral supraorbital. Abbreviations for neuromast lines: io, infraorbital; m, middle; ol, otic; pop, preopercular; so, supraorbital; st, supratemporal. Abbreviations for anatomical landmarks: an, anterior naris; b, barbel; e, eye; m, mouth; op, operculum; ot, otic vesicle; pn, posterior naris; s, spiracle (first gill cleft).
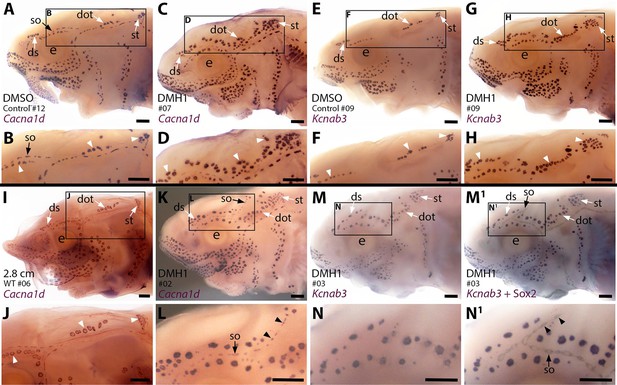
Sterlet larvae in which Bmp signalling was blocked prior to ampullary organ formation have supernumerary ampullary organs and ectopic supraorbital neuromasts.
Sterlet larvae after in situ hybridisation (ISH) for the hair cell and electroreceptor marker Cacna1d (also expressed in taste buds on barbels) or the electroreceptor-specific marker Kcnab3. Black arrowheads indicate examples of neuromasts; white arrowheads indicate examples of ampullary organs. (A–H) Stage 45 larvae that had been treated for 20 hr from stage 36 (i.e., from hatching to approximately stage 38, just prior to the onset of ampullary organ development) with either DMH1 or DMSO as controls. Larvae are numbered for cross-referencing with ampullary organ counts in Supplementary file 3. ISH for Cacna1d (A–D) or Kcnab3 (E–H) shows that, relative to DMSO-treated controls (A,B,E,F), DMH1-treated larvae have many more ampullary organs (C,D,G,H). This phenotype is particularly prominent in the three dorsal-most ampullary organ fields, where the dorsal supraorbital, dorsal otic and supratemporal fields - clearly separate in DMSO-treated larvae (A,B,E,F) - almost fuse together in DMH1-treated larvae (C,D,G,H). (I,J) A much older wild-type larva (2.8 cm in length, ~65 dpf) after ISH for Cacna1d. The dorsal supraorbital, dorsal otic and supratemporal ampullary organ fields are clearly separated, suggesting the supernumerary ampullary organs in this region in DMH1-treated larvae (C,D,G,H) are ectopic, not precocious. (K–N1) Most DMH1-treated larvae also develop an ectopic offshoot from the supraorbital neuromast line. This is visible after ISH for Cacna1d (K,L; compare with DMSO control in A,B) and confirmed to represent neuromasts in DMH1-treated larvae via ISH for electroreceptor-specific Kcnab3 (M,N) followed by immunostaining for the supporting cell marker Sox2 to reveal neuromasts (M1,N1). Abbreviations: dot, dorsal otic ampullary organ field; ds, dorsal supraorbital ampullary organ field; e, eye; S, stage; so, supraorbital neuromast line; st, supratemporal ampullary organ field; WT, wild type. Scale bar: 250 μm.
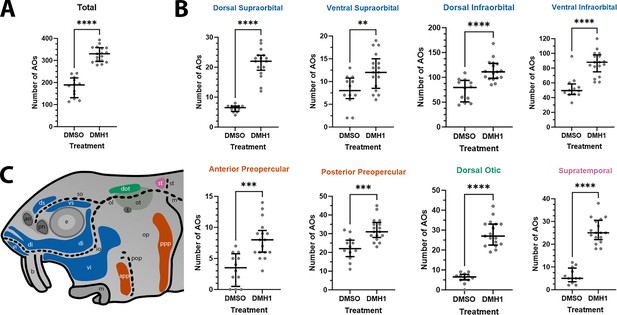
DMH1-treated larvae have significantly more ampullary organs than DMSO controls.
(A) Scatter plot showing median and interquartile range for the total number of ampullary organs on one side of the head in stage 45 sterlet larvae that had been treated for 20 hours from stage 36 (i.e., from hatching to approximately stage 38, just prior to the onset of ampullary organ development) with DMH1 (n=17) or DMSO as controls (n=12). DMH1-treated larvae have significantly more ampullary organs (p<0.0001; two-tailed Mann-Whitney test). Ampullary organs were counted after in situ hybridisation [ISH] for Cacna1d or Kcnab3; raw counts are provided in Supplementary file 3. (B) Scatter plots showing median and interquartile range for the number of ampullary organs in each individual ampullary organ field on one side of the head in stage 45 sterlet larvae that had been treated for 20 hr from stage 36 with DMH1 (n=17), versus with DMSO as controls (n=12). Raw counts are provided in Supplementary file 3. For the location of each field, see schematic in panel C (reproduced from Figure 5D). Scatter plots are grouped with differently coloured titles according to lateral line placode (LLp) origin, following Gibbs and Northcutt, 2004: blue, anterodorsal LLp origin (supraorbital and infraorbital fields); orange, anteroventral LLp origin (preopercular fields); green, otic LLp origin (dorsal otic field); pink, supratemporal LLp origin (supratemporal field). All fields have significantly more ampullary organs in DMH1-treated larvae (n=17) than in DMSO controls (n=12; two-tailed Mann-Whitney tests). Asterisks on plots represent p values: **, p≤0.01; ***, p≤0.001; ****, p≤0.0001. p values for all fields are <0.0001 except for the ventral supraorbital field (p=0.0074), anterior preopercular field (p=0.0002) and posterior preopercular field (p=0.0003). (C) Schematic of a stage 45 sterlet larval head. Ampullary organ fields are represented by coloured patches flanking the neuromast lines, which are represented as dotted lines. The different field colours indicate their lateral line placode origin (consistent with scatter plot titles in B). Abbreviations for ampullary organ fields: app, anterior preopercular; di, dorsal infraorbital; dot, dorsal otic; ds, dorsal supraorbital; ppp, posterior preopercular; st, supratemporal; vi, ventral infraorbital; vs, ventral supraorbital. Abbreviations for neuromast lines: io, infraorbital; m, middle; ol, otic; pop, preopercular; so, supraorbital; st, supratemporal. Abbreviations for anatomical landmarks: an, anterior naris; b, barbel; e, eye; m, mouth; op, operculum; ot, otic vesicle; pn, posterior naris; s, spiracle (first gill cleft).
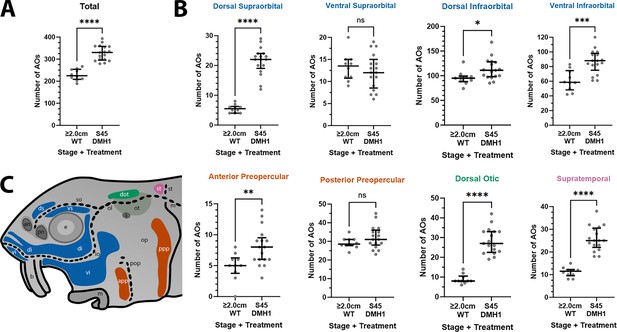
DMH1-treated larvae have significantly more ampullary organs at stage 45 than older wild-type larvae.
(A,B) Scatter plots showing median and interquartile range for the number of ampullary organs on one side of the head in stage 45 sterlet larvae that had been treated for 20 hr from stage 36 with DMH1 (n=17) vs 2.0/2.8 cm wild-type larvae (~50/65 dpf; n=10). Raw counts are provided in Supplementary file 3. Two-tailed Mann-Whitney tests were used for statistical analysis. DMH1-treated larvae have significantly more ampullary organs overall at stage 45 than wild-type older larvae (p<0.0001; panel A). In panel B, scatter plots are grouped with differently coloured titles according to lateral line placode (LLp) origin, following Gibbs and Northcutt, 2004: blue, anterodorsal LLp (supraorbital and infraorbital fields); orange, anteroventral LLp (preopercular fields); green, otic LLp (dorsal otic field); pink, supratemporal LLp (supratemporal field). DMH1-treated larvae have significantly more ampullary organs at stage 45 than older wild-type larvae in all fields except the ventral supraorbital and posterior preopercular fields. Symbols on plots represent p values: ns, not significant, (p>0.05; *, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001). Dorsal supraorbital: p<0.0001. Ventral supraorbital: not significant (p=0.5109). Dorsal infraorbital: p=0.0123. Ventral infraorbital: p=0.0002. Anterior preopercular: p=0083. Posterior preopercular: not significant (p=0.1789). Dorsal otic: p<0.0001. Supratemporal: p<0.0001. (C) Schematic of a stage 45 sterlet larval head. Ampullary organ fields are represented by coloured patches flanking the neuromast lines, which are represented as dotted lines. The different field colours indicate their lateral line placode origin (consistent with scatter plot titles in B). Abbreviations for ampullary organ fields: app, anterior preopercular; di, dorsal infraorbital; dot, dorsal otic; ds, dorsal supraorbital; ppp, posterior preopercular; st, supratemporal; vi, ventral infraorbital; vs, ventral supraorbital. Abbreviations for neuromast lines: io, infraorbital; m, middle; ol, otic; pop, preopercular; so, supraorbital; st, supratemporal. Other abbreviations: an, anterior naris; b, barbel; e, eye; m, mouth; op, operculum; ot, otic vesicle; pn, posterior naris; s, spiracle (first gill cleft); WT, wild type.
Tables
sgRNAs used in this study.
The target sequences and sgRNA combinations used in this study are shown. The Tyr sgRNAs were previously published (preprint, Minařík et al., 2024b); the asterisk against Tyr sgRNAs 7 and 8 indicates that these sgRNAs were originally designed and published by Stundl et al., 2022 as their tyr sgRNAs 3 and 4, respectively.
| Target Gene | sgRNA | Target Sequence | PAM | Combinations Used |
|---|---|---|---|---|
| Bmp5 | 1 | TCACGCAGAAAAGCACAGGG | AGG | 1+2 + 3, 1+4 |
| 2 | AGATGATGCCTGTTTGCCAG | GGG | 1+2 + 3, 2+3 | |
| 3 | GGCAAACGAGGAGGAAAACG | GGG | 1+2 + 3, 2+3 | |
| 4 | GTACAATGCCATGGCAAACG | AGG | 1+4 | |
| Tyr | 1 | GGTGCCAAGGCAAAAACGCT | GGG | 1+2, 1+2 + 3+4 |
| 2 | GATATCCCTCCATACATTAT | TGG | 1+2, 1+2 + 3+4 | |
| 3 | GATGTTTCTAAACATTGGGG | TGG | 1+2 + 3+4 | |
| 4 | GCTATGAATTTATTTTTTTC | AGG | 1+2 + 3+4 | |
| 5 | GCAAGGTATACGAAAGTTGA | CGG | 5+6 | |
| 6 | GATTGCAAGTTCGGCTTCTT | AGG | 5+6 | |
| 7* | GGTTAGAGACTTTATGTAAC | GGG | 7+8 | |
| 8* | GGCTCCATGTCTCAAGTCCA | AGG | 7+8 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Acipenser ruthenus) | Acvr2a | NCBI_Gene (RRID:SCR_002473) | GeneID: 117427134 | |
| Gene (A. ruthenus) | Apcdd1 | NCBI_Gene (RRID:SCR_002473) | GeneID: 117394164 | |
| Gene (A. ruthenus) | Bmp4 | NCBI_Gene (RRID:SCR_002473) | GeneID: 117395489 | |
| Gene (A. ruthenus) | Bmp5 | NCBI_Gene (RRID:SCR_002473) | GeneID: 117402601 | |
| Gene (A. ruthenus) | Cacna1d | NCBI_Gene (RRID:SCR_002473) | GeneID:117413950 | |
| Gene (A. ruthenus) | Kcnab3 | NCBI_Gene (RRID:SCR_002473) | GeneID:117404443 | |
| Gene (A. ruthenus) | Sostdc1 | NCBI_Gene (RRID:SCR_002473) | GeneID:117400407 | |
| Biological sample (A. ruthenus) | Fertilised sterlet sturgeon eggs and embryos/larvae (A. ruthenus) | Research Institute of Fish Culture and Hydrobiology, Faculty of Fisheries and Protection of Waters, University of South Bohemia in České Budějovice Vodňany, Czech Republic | ||
| Antibody | anti-Sox2 (rabbit monoclonal) | Abcam | Cat.#:ab92494; RRID:AB_10585428 | (1:200) |
| Antibody | anti-human Phospho-SMAD1/5/9 (rabbit monoclonal) | Cell Signalling Technology | Cat.#:13820; RRID:AB_2493181 | (1:100) |
| Antibody | horseradish peroxidase-conjugated goat anti-rabbit IgG | Jackson ImmunoResearch | Cat.#:111-035-003; RRID:AB_2313567 | (1:300) |
| Recombinant DNA reagent | pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) (plasmid) | Addgene (Cong et al., 2013; DOI: 10.1126/science.1231143) | RRID:Addgene_42335 | Used to synthesize DNA templates containing the single guide (sg)RNA scaffold |
| Sequence-based reagent | Bmp4 riboprobe forward primer (F) | This paper | PCR primers | GGGGCCGCAAGAAAAACCGGAA |
| Sequence-based reagent | Bmp4 riboprobe reverse primer (R) | This paper | PCR primers | TCGCAGTGAGCCTTGCCCATTT |
| Sequence-based reagent | Bmp5 riboprobe F | This paper | PCR primers | ACCCAGTGGTTGCTTGTAGC |
| Sequence-based reagent | Bmp5 riboprobe R | This paper | PCR primers | ATTCTGGGCTTACCACATCG |
| Sequence-based reagent | Cacna1d riboprobe F | This paper | PCR primers | GCCACGAACATCACTCCGCCAA |
| Sequence-based reagent | Cacna1d riboprobe R | This paper | PCR primers | TCATGTCGCAAGCGTCCGCAAT |
| Sequence-based reagent | Kcnab3 riboprobe F | Minařík et al., 2024a (DOI: https://doi.org/10.3389/fcell.2024.1327924) | PCR primers | GGTAAATTCAGCGTGGAGGA |
| Sequence-based reagent | Kcnab3 riboprobe R | Minařík et al., 2024a (DOI: https://doi.org/10.3389/fcell.2024.1327924) | PCR primers | ACCTTCGATGATGTGCTTCC |
| Sequence-based reagent | Sostdc1 riboprobe F | This paper | PCR primers | CCACGCCTGGTTAATCCTGTGGA |
| Sequence-based reagent | Sostdc1 riboprobe R | This paper | PCR primers | GTGCTTGCCCGTCTTGCCTGAT |
| Sequence-based reagent | sgRNA scaffold R | Pers. comm., Dr Ahmed Elewa, Karolinska Institutet, Stockholm, Sweden | PCR primers | AAAAAAGCACCGACTCGGTGCC |
| Sequence-based reagent | Bmp5 sgRNA F1 | This paper | PCR primers | GATCACTAATACGACTCACTATAG TCACGCAGAAAAGCACAGGGGTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Bmp5 sgRNA F2 | This paper | PCR primers | GATCACTAATACGACTCACTAT AGAGATGATGCCTGTTTGCCAGGTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Bmp5 sgRNA F3 | This paper | PCR primers | GATCACTAATACGACTCACTAT AGGCAAACGAGGAGGAAAACGGTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Bmp5 sgRNA F4 | This paper | PCR primers | GATCACTAATACGACTCACTAT AGTACAATGCCATGGCAAACGGTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F1 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | GATCACTAATACGACTCACTAT AGGTGCCAAGGCAAAAACGCTGTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F2 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | GATCACTAATACGACTCACTAT AGATATCCCTCCATACATTATG TTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F3 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | GATCACTAATACGACTCACTAT AGATGTTTCTAAACATTGGGG GTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F4 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | GATCACTAATACGACTCACTA TAGCTATGAATTTATTTTTTTC GTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F5 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | GATCACTAATACGACTCACTAT AGCAAGGTATACGAAAGTTGA GTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F6 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | GATCACTAATACGACTCACTAT AGATTGCAAGTTCGGCTTCTT GTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F7 | Stundl et al., 2022 (DOI: https://doi.org/10.3389/fcell.2022.750833) | PCR primers | GATCACTAATACGACTCACTAT AGGTTAGAGACTTTATGTAAC GTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Tyr sgRNA F8 | Stundl et al., 2022 (DOI: https://doi.org/10.3389/fcell.2022.750833) | PCR primers | GATCACTAATACGACTCACTAT AGGCTCCATGTCTCAAGTCC AGTTTTAGAGCTAGAAAT |
| Sequence-based reagent | Bmp5 genotyping F | This paper | PCR primers | GGAACACAGTCGCTGAAGTG |
| Sequence-based reagent | Bmp5 genotyping R | This paper | PCR primers | GTTGCATACATGCCCAGATG |
| Sequence-based reagent | Tyr genotyping F1 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | GCGTCTCTCCAGTCCCAATA |
| Sequence-based reagent | Tyr genotyping R1 | Minařík et al., 2024b (DOI: https://doi.org/10.1101/2023.04.15.537030) | PCR primers | AGAGAGAAGTGGCCCTTGGT |
| Peptide, recombinant protein | Cas9 protein with NLS | PNA Bio | Cat.#:CP01-200 | |
| Peptide, recombinant protein | Q5 High-Fidelity DNA Polymerase | New England Biolabs | Cat.#:M0491S | |
| Commercial assay or kit | Ambion Turbo DNA-free kit | Thermo Fisher Scientific | Cat.#:AM1907 | |
| Commercial assay or kit | High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat.#:4368814 | |
| Commercial assay or kit | QIAGEN PCR Cloning Kit | QIAGEN | Cat.#:231124 | |
| Commercial assay or kit | MinElute Gel Extraction Kit | QIAGEN | Cat.#:28604 | |
| Commercial assay or kit | EnzMet kit | Nanoprobes | Cat.#:6010 | |
| Commercial assay or kit | PCRBIO Rapid Extract PCR Kit | PCR Biosystems | Cat.#:PB10.24–08 | |
| Commercial assay or kit | HS Taq Mix Red | PCR Biosystems | Cat.#:PB10.23–02 | |
| Commercial assay or kit | Monarch PCR & DNA Cleanup Kit | New England Biolabs | Cat.#:T1030 | |
| Commercial assay or kit | Monarch RNA Cleanup Kit | New England Biolabs | Cat.#:T2040 | |
| Commercial assay or kit | HiScribe T7 High Yield RNA Synthesis Kit | New England Biolabs | Cat.#:E2040S | |
| Chemical compound, drug | DMH1 (dorsomorphin homolog 1) | Cayman Chemical | Cat.#:CAY16679 | |
| Software, algorithm | Benchling | Benchling | RID:SCR_013955 | |
| Software, algorithm | Primer3Plus | Primer3Plus | RRID:SCR_003081 | |
| Software, algorithm | Inference of CRISPR Edits (ICE) | Synthego | RRID:SCR_024508 | |
| Software, algorithm | NCBI BLAST | National Institutes of Health (NIH) | RRID:SCR_004870 | |
| Software, algorithm | Helicon Focus | Helicon Soft | RRID:SCR_014462 | |
| Software, algorithm | Adobe Photoshop | Adobe | RRID:SCR_014199 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | QCapture Pro 7.0 | QImaging | RRID:SCR_014432 | |
| Software, algorithm | Ocular | Teledyne Photometrics | RRID:SCR_024490 |
Additional files
-
Supplementary file 1
Breakdown by sgRNA mix of CRISPR/Cas9 experiments targeting Bmp5 or tyrosinase (control).
For each sgRNA mix, the table shows the number of independent batches, markers used for analysis, percentage with a phenotype, and genotyping information including genotyping primer sequences. Note: Tyr crispant data are shared between this study and Minařík et al., 2024b. Tyr sgRNAs 7 and 8 were designed and published by Stundl et al., 2022 as their tyr sgRNA 3 and tyr sgRNA 4, respectively.
- https://cdn.elifesciences.org/articles/99798/elife-99798-supp1-v3.xlsx
-
Supplementary file 2
Breakdown by individual crispant of CRISPR/Cas9 experiments targeting Bmp5 or tyrosinase (control).
For each crispant, the table lists the sgRNA mix used, the batch number, the ICE genotyping results and the number of ampullary organs within each field. Descriptive statistics are also shown for each ampullary organ field and across all ampullary organ fields for both Bmp5 crispants and Tyr crispants.
- https://cdn.elifesciences.org/articles/99798/elife-99798-supp2-v3.xlsx
-
Supplementary file 3
Breakdown of ampullary organ counting data for DMH1 experiments and older wild-type larvae.
The table shows the number of ampullary organs within each field for each DMH1-treated embryo, DMSO-treated embryo (control) or older wild-type larva. Descriptive statistics are also given for each ampullary organ field and across all ampullary organ fields for DMH1-treated, DMSO-treated (control) and older wild-type larvae.
- https://cdn.elifesciences.org/articles/99798/elife-99798-supp3-v3.xlsx
-
Supplementary file 4
Riboprobe information.
For each riboprobe used during in situ hybridisation, the table lists the primer sequences used to clone the cDNA template, the GenBank accession number of the top-matched ohnolog, and the nucleotide region targeted by the riboprobe. It also shows the chromosomal location, the percentage identity of the riboprobe and the genome annotation of both the top-matched ohnolog and the second ohnolog (if present).
- https://cdn.elifesciences.org/articles/99798/elife-99798-supp4-v3.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/99798/elife-99798-mdarchecklist1-v3.docx





