A host beetle pheromone regulates development and behavior in the nematode Pristionchus pacificus
Figures
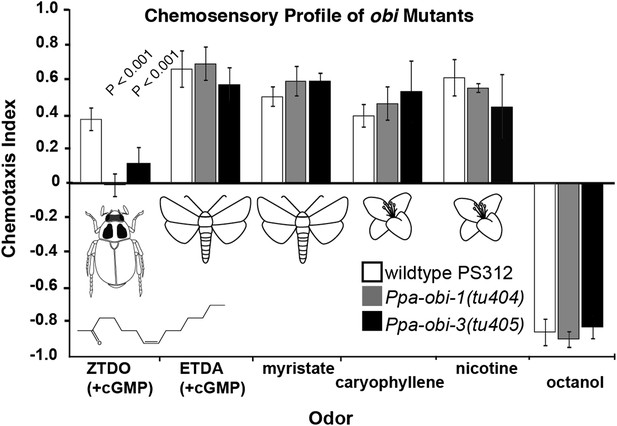
Chemosensory behavior of oriental beetle pheromone insensitive mutants.
Ppa-obi-1 and Ppa-obi-3 mutant animals are specifically defective for sensing the host oriental beetle sex pheromone, Z-7-tetradece-2-one. The ZTDO structure is depicted below the beetle. In contrast, obi mutants' response is wild type towards another cGMP-dependent attractant, E-11-tetradecenyl acetate (ETDA, a lepidopteran pheromone), as well as myristate (methyl tetradecanoate, an insect allomone), plant volatiles beta-caryophyllene and nicotinic acid, and the repellent 1-octanol. N ≥ 15 replicates in ≥3 trials. Significant differences to wild type are indicated by p values (Dunnett's test) and error bars represent SEM.
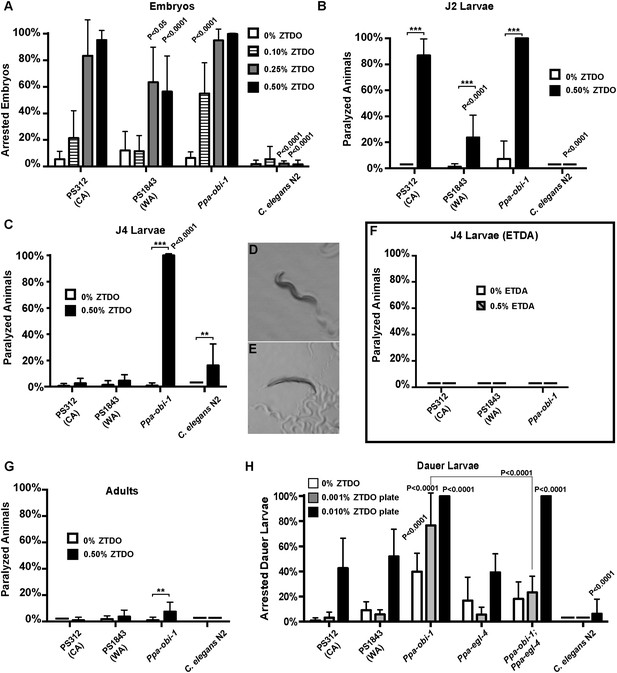
The oriental beetle pheromone ZTDO inhibits P. pacificus development and induces paralysis.
(A) Ppa-obi-1 mutant is hypersensitive to 0.10% ZTDO compared to wild type PS312 (California). However, PS312 is more sensitive to 0.5% ZTDO than another strain, PS1843 (Washington). PS312 (n = 372, 82, 90, 320); PS1843 (n = 309, 88, 92, 237); Ppa-obi-1 (n = 332, 69, 71, 389); C. elegans wild-type N2 (n = 316, 63, 84, 174). (B) Both wild-type PS312 and Ppa-obi-1 mutant J2 larvae are more sensitive to ZTDO than PS1843. PS312 (n = 154, 115); PS1843 (n = 110, 102); Ppa-obi-1 (n = 104,100); C. elegans wild-type N2 (n = 23, 73). (C) Ppa-obi-1 J4 larvae is uniquely sensitive to ZTDO compared to PS312 (p < 0.0001). PS312 (n = 209, 290); PS1843 (n = 102, 99); obi-1 (n = 244, 237); C. elegans wild-type N2 (n = 69, 109). (D) 90 min exposure of a Ppa-obi-1(tu404) J4 larvae to ethanol control compared to (E) 0.50% ZTDO resulted in a rigid fully paralyzed posture. (F) Adults are not paralyzed by ZTDO. PS312 (n = 108, 113); PS1843 (n = 114, 107); Ppa-obi-1 (n = 109, 109); C. elegans wild-type N2 (n = 62, 72). (G) 0.01% ZTDO prevents resumption of post-dauer development in approximately 40% of wild-type animals and nearly all Ppa-obi-1(tu404) mutants. This ZTDO inhibition of dauer exit is dependent on the cGMP-dependent PKG, Ppa-egl-4. PS312 (n = 384, 654, 267); PS1843 (n = 195, 139, 101); Ppa-obi-1 (n = 339, 343, 146); Ppa-egl-4(tu374) (n = 123, 181, 77); Ppa-obi-1;Ppa-egl-4 (n = 132, 144, 63); N2 (n = 131, 109, 115). (H) Exposure to 0.50% ETDA moth pheromone from Spodoptera littoralis does not paralyze Ppa-obi-1 mutant and wild-type J4 larvae. PS312 (n = 40, 103); PS1843 (n = 40, 64); Ppa-obi-1 (n = 40, 193). p values indicate Dunnett's multiple comparisons test to wild-type PS312. ***p < 0.001; **p < 0.01 indicate difference between ethanol control and ZTDO exposed groups (pairwise unpaired t test). Error bars are SEM and n is the number of nematodes tested.
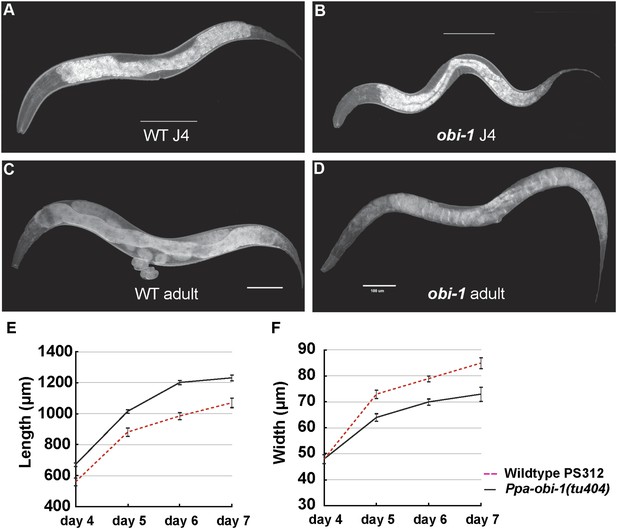
Ppa-obi-1 mutants are long and slim.
(A and B) Wild-type J4 larva and young adult. (C and D) Ppa-obi-1 J4 larva and young adult. (E and F) The body lengths and widths of wild-type (dotted red line) versus Ppa-obi-1 mutants (solid black line) recorded over 4 days post J4 stage (μm); n = 15–30 animals for each time point. Error bars are SEM. Scale bars denote 100 μm.
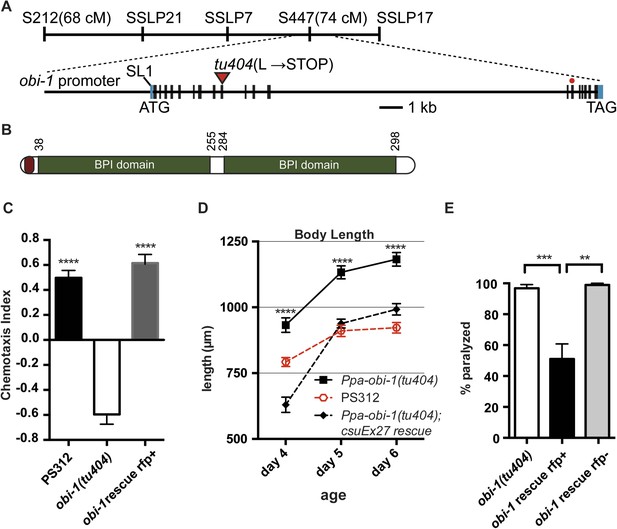
Ppa-obi-1 gene structure and protein comparisons.
(A) The order of mapping markers is depicted on top (not to scale). The Ppa-obi-1 mRNA is SL1 trans-spliced and contains 19 exons spanning ∼19 kb. The 50 bp 5′ UTR and 341 bp 3′ UTR are indicated in light blue. The tu404 allele contains a single base pair deletion in exon 9 that results in a frameshift, converting a codon to a stop codon (inverted triangles). The red dot represents the splice variant. (B) A diagram of the secretion and two BPI lipid-binding domains (PFAM and SMART). (C–E) Transgenic rescue of the Ppa-obi-1(tu404). (C) A 4.3 kb promoter driving Ppa-obi-1 cDNA completely rescued cGMP-dependent ZTDO attraction in Ppa-obi-1(tu404) mutants. PS312 (N = 21); obi-1 (N = 15); obi-1 rescue (N = 12). The Ppa-obi-1 transgenic rescue (D) restored the body length defect in Ppa-obi-1(tu404) mutants, PS312 (n = 15); obi-1 (n = 15); obi-1 rescue (n = 19) as well as (E) significantly reduced ZTDO-induced paralysis in Ppa-obi-1(tu404) J4 larvae. PS312 (N = 11); obi-1 rescue rfp+ (N = 12); obi-1 rescue rfp− (N = 5). **p < 0.01; ***p < 0.001; ****p < 0.0001 (Tukey's multiple comparisons test). Error bars represent SEM. N is the number of assays performed and n is the number of individual worms scored for each time point.
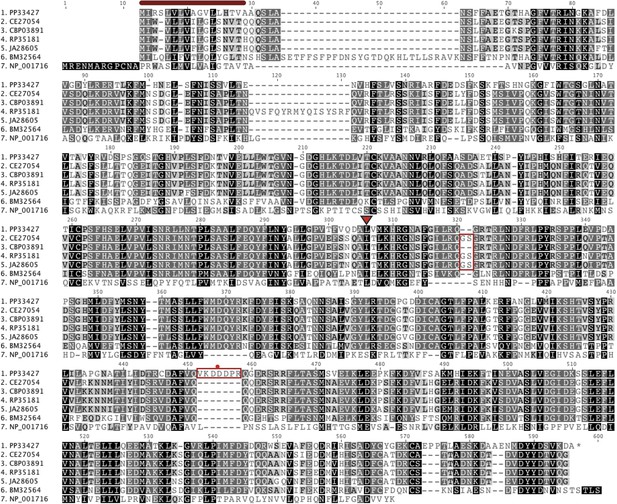
An alignment of putative Ppa-OBI-1 protein orthologs.
Caenorhabditis elegans (C06G1.1), C. briggsae (CBP03891), C. remanei (RP35181), C. japonica (JA26805), Brugia malayi (BM32564), and a human Bactericidal Permeability Increasing protein (NP_001716). The predicted protein secretion signal peptides for Ppa-OBI-1(PP33427) and C06G1.1(CE27054) are 17–18 amino acids long at the N terminus (red bar). The longest predicted protein splice forms are used in the alignment, with orthologs in Caenorhabditis species containing ‘GS’ (red boxed) and Ppa-OBI-1 in P. pacificus containing ‘VKDDDPR’ peptides (red boxed with a red dot). The single base pair deletion in Ppa-obi-1(tu404) converts the leucine codon to a stop codon (inverted red triangle).
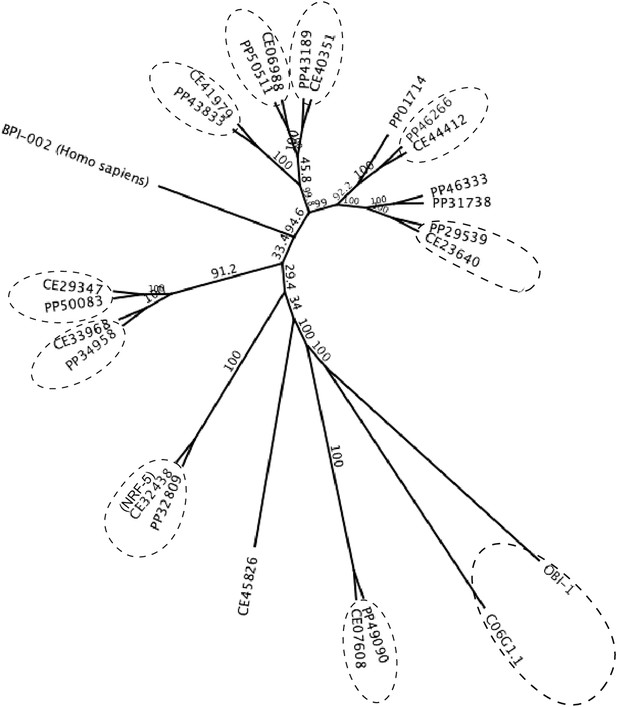
A maximum likelihood relationship of lipid-binding proteins in P. pacificus and C. elegans.
The ‘PP’ and ‘CE’ prefixes denote P. pacificus and C. elegans proteins, respectively (www.wormbase.org). 10 orthologous protein pairs are circled. Ppa-OBI-1 (PP33427) and the 12 other paralogs are also represented in the P. pacificus proteome (Pristionchus.org). A human BPI protein (GenBank NP_001716.2) was used as outgroup. Bootstrap values are indicated along branches.
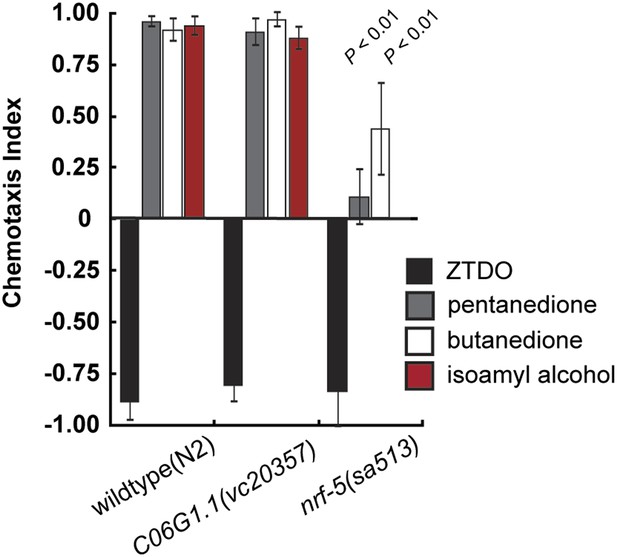
Chemosensory behavior of C. elegans mutants C06G1.1 and nrf-5.
A mutation in C06G1.1(vc20357), the ortholog of Ppa-obi-1, does not alter chemotaxis behavior towards 10% ZTDO, 1% 2,3-pentanedione, 1% 2,3-butanedione (diacetyl), and 1% isoamyl alcohol. However, nrf-5(sa513) mutants have reduced attraction to 1% 2,3-butanedione and 1% isoamyl alcohol. nrf-5(sa513) was not tested for response to 2,3-pentanedione. Significant differences to wild-type are indicated by p values (two sampled t test). N ≥ 10 replicates. Error bars represent SEM.
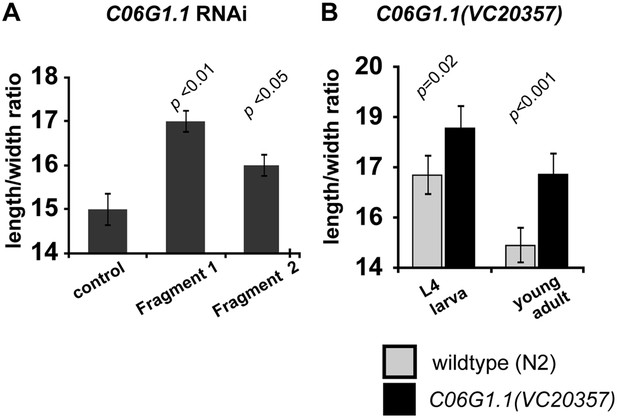
Reduction of C06G1.1 function produce a slimmer body phenotype.
(A) RNAi using two dsRNA fragments of C06G1.1 fed to C. elegans rrf-3(pk1426) resulted in a longer and thinner body proportion. (B) Genetic mutation in C06G1.1(VC20357) resulted in significantly slimmer body proportion (increase length/width) similar to RNAi against C06G1.1 and the Ppa-obi-1(tu404) mutant. n ≥ 13 animals. Note that y-axes do not begin at zero. Significant differences to wild-type are indicated by p values (two sampled t-test). Error bars represent SEM.
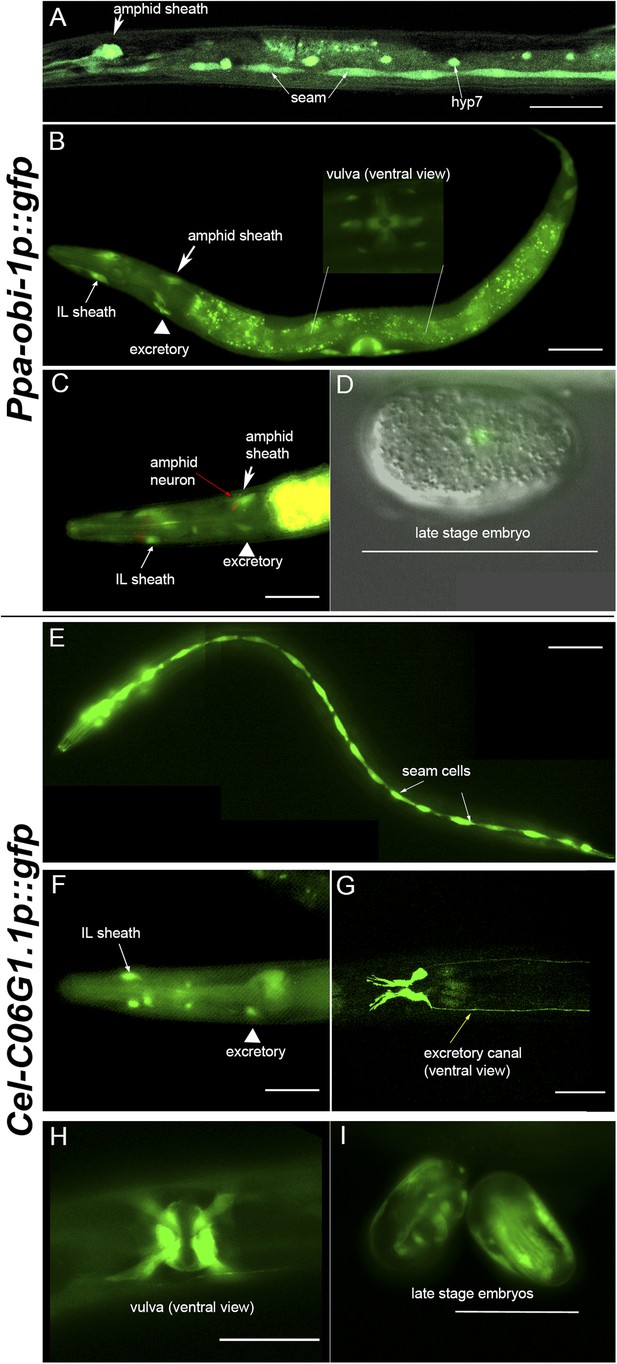
Ppa-obi-1 and C06G1.1 expression in their respective nematode species.
(A–D) An integrated Ppa-obi-1 promoter::gfp transgene (csuIs01) in P. pacificus. (A) Lateral view of a single animal showing prominent expression in an amphid sheath cell, seam cells, and the syncytial hypodermal cell (hyp7) in a J4 hermaphrodite. (B) Medial view of a transgenic animal with expression in the putative dorsal and ventral inner labial (IL) or outer labial (OL) neuronal support cells (socket or sheath), the duct and excretory cells on the ventral side, the amphid sheath cell on the dorsal side (arrow head), and the vulval muscles. The inset shows a ventral view of the young adult vulva in another animal. Autofluorescence is visible in the intestine. (C) A DiI-labeled neuron (red arrow) anterior to a Ppa-obi-1p::gfp expressing amphid sheath cell (white arrow) is dorsal to an excretory cell (arrow head). A putative IL or OL socket or sheath cell (arrow) is also visible. (D) DIC and fluorescent overlay image of expression in early P. pacificus embryo. (E–I) C06G1.1 promoter::gfp transgenes (csuEx02, csuEx03) in C. elegans. (E) Lateral view of a seam cell expression in a L3 hermaphrodite. (F) Medial view of putative inner or outer labial neuron support cells (arrow) and excretory cells (arrowhead). No amphid sheath shows gfp expression. (G) Ventral view of head region shows prominent expression in excretory cells and canal (arrow). (H) Ventral view of young adult vulva. (I) Embryonic expression in C. elegans embryos appears later, stronger, and in more cells than in Ppa-obi-1. Anterior is left and the scale bars represent 50 μm. Each panel is representative of ≥50 animals.
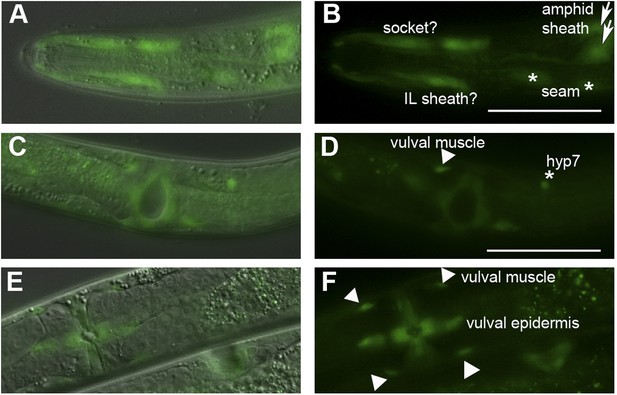
Ppa-obi-1 is expressed in putative amphid socket cells, amphid sheath cells, and vulval cells.
Dorsal and ventral views of Ppa-obi-1p::gfp expression in roller prl-1(tu92) background with merged DIC and fluorescence (A, C, E) and corresponding fluorescence images (B, D, F). (A and B) Dorso-lateral view of head region shows possible Ppa-obi-1 expression in a socket cell (top), an IL sheath cell (bottom), two seam cells (asterisks), and two amphid sheath cells (arrows). (C and D) Ventral view of a mid-J4 larvae vulva shows Ppa-obi-1p::gfp expression in P(5–7).p vulval epidermal cell descendants, a putative vulval muscle (triangle), and a hyp7 cell (asterisk). (E and F) Ventral view of a young adult vulva (top left) shows Ppa-obi-1p::gfp expression in P(5–7).p vulval epidermal cell descendents and four vulval muscles (triangles) and a lateral view of a mid-J4 vulva (bottom right) in another animal. Anterior is left and the scale bars represent 50 μm.
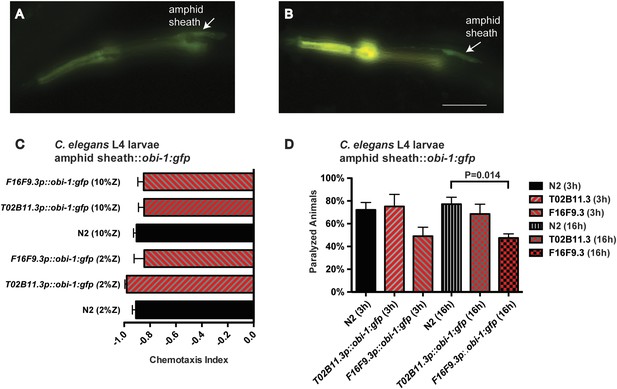
Targeted misexpression of Ppa-obi-1 in C. elegans.
L2 larvae show amphid sheath cell expression using (A) Cel-T02B11.3 promoter::Ppa-obi-1:gfp (csuEx29) and (B) Cel-F16F9.3 promoter::Ppa-obi-1:gfp (csuEx30). The co-injection marker myo-2p::DsRED is visible in the anterior pharyngeal bulb due to bleeding over into the GFP spectrum. Arrows indicate amphid sheath cell bodies. (C) Chemotaxis response to the ZTDO beetle pheromone remains unchanged towards 2% or 10% ZTDO. (D) ZTDO-induced paralysis in L4 larvae shows that transgenic Cel-F16F9.3p::Ppa-obi-1::gfp are better protected than wild-type animals in a 16 hr assay. N2 (N = 8), T02B11.3p::Ppa-obi-1 (N = 6), F16F9.3p::Ppa-obi-1 (N = 6). Anterior is left and the scale bar represents 25 μm. p value indicates Dunnett's multiple comparisons test to wild-type N2. Error bars are SEM and N is the number of assays performed.
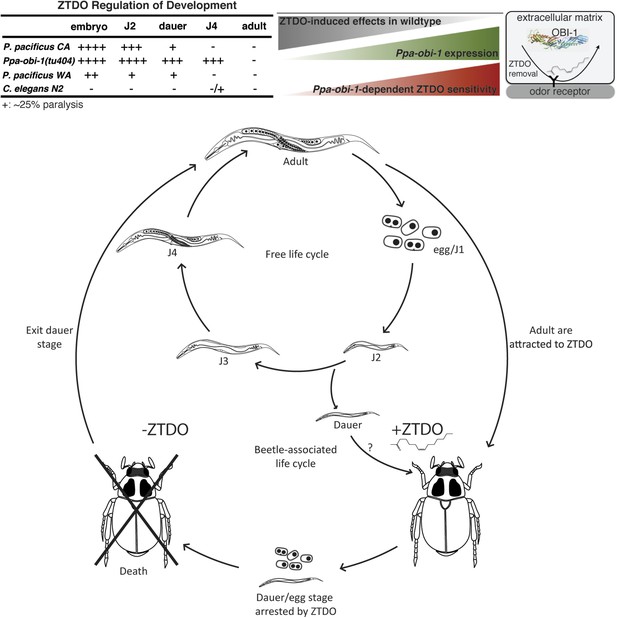
Summary and model of the oriental beetle pheromone ZTDO on P. pacificus development and lifecycle.
(Top) Peak Ppa-obi-1 expression in the wild-type J4 larvae coincides with the least sensitive developmental stage to the paralytic effects of the beetle pheromone ZTDO (at 0.5%). Ppa-obi-1 mutants are most sensitive to ZTDO-induced paralysis in the J4 stage. Our working model on Ppa-OBI-1 functions in the J4 larvae hypothesizes that Ppa-OBI-1 proteins in the extracellular space, such as the lumen of the amphid sheath cell, may be required to remove ZTDO bound to odor receptors on neurons. (Bottom) This working model shows both P. pacificus adults and dauer larvae from non-beetle populations associate with the host oriental beetle through attraction to the beetle's sex pheromone ZTDO, whereby the progeny born on the beetle are arrested as eggs or dauer larvae. Death of the host allows microorganism to grow, which then encourages dauer larvae to exit and resume reproductive development. Several iterations of asynchronous nematode populations likely complete each dispersal cycle.
Tables
Ppa-obi-1 affects hatching rate and brood size
| Hatching rate (%) | Brood size (3 days) | n | |
|---|---|---|---|
| Wild-type(PS312) | 95 ± 3 | 156 ± 16 | 18 |
| Ppa-obi-1(tu404) | 85 ± 3 | 103 ± 17 | 15 |
Primers
| Primer | Primer sequence (5′→3′) | Template target |
|---|---|---|
| RHL101 | TGAACGGGCTTTTAATCTGG | Ppa-obi-1 gDNA |
| RHL102 | GTGTCGAGATAGTCGGGCAT | Ppa-obi-1 gDNA |
| RHL104 | ATGACTGCTCCAAAGAAG | Ppa-obi-1 cDNA |
| RHL107 | ATTTGCCCTTCGTTCCAC | Ppa-obi-1 for RNAi |
| RHL108 | CTGTCTCAGTATGCCGAATG | Ppa-obi-1 for RNAi |
| RHL109 | ATTCAGCCGTGTTACAAATC | Ppa-obi-1 cDNA |
| RHL110 | CTGCATGTGTTCCGGTCTCG | Ppa-obi-1 cDNA |
| RHL113 | AAACCTCTGACACATGCAGC | Ppa-obi-1 cDNA |
| RHL121 | AGTTTCAAAGTTGCATGG | Ppa-obi-1 promoter |
| RHL124 | CTTCTTTGGAGCAGTCATGTAGCGATAATCAGGAGT | gfp; Ppa-obi-1 promoter |
| RHL132 | ATGACTGCTCCAAAGAAG | gfp |
| RHL136 | CAACCGTGTGCAGTAGAACC | Ppa-obi-1 5′ RACE |
| RHL149 | ACTATCCTATCATCGGAAGC | Ppa-obi-1 promoter |
| RHL151 | AGGGCATTGACAAATCATCG | Ppa-obi-1 cDNA |
| RHL154 | CACGACGTTGTAAAACGACG | Ppa-obi-1 cDNA |
| RHL169 | CGCGCTAAACCAATTCCGCC | C06G1.1 promoter |
| RHL181 | GTTGAGAGAAGAGGTGGAGTCC | C06G1.1 promoter |
| RHL187 | CTTCTTTGGAGCAGTCATCAGATCTGAAAATTTGACACATTG | gfp; C06G1.1 promoter |
| RHL267 | GGAACTAACATTAATGTCAC | C06G1.1 RNAi |
| RHL268 | GAGAAGTGCATAGTTGATG | C06G1.1 RNAi |
| RHL477 | GCGCTACAGGGGATTTTGTC | T02B11.3 promoter |
| RHL480 | TGAAAGATGCAAGTAAAGGAGAAGAACTTTTC | p95.75 GFP |
| RHL481 | AAGGGCCCGTACGGCCGACTA | p95.75 GFP |
| RHL482 | GTAGGAAACAGTTATGTTTGG | p95.75 GFP |
| RHL484 | CTTCAGAAAATGATCAGGAGTCTGGTTC | obi-1 cDNA |
| RHL487 | AGTGAAAAGTTCTTCTCCTTTACTTGCATCTTTCACGGAGTC | obi-1 cDNA; gfp |
| RHL490 | AACGAGAACCAGACTCCTGATCATTTTCTGAAGAAAGTTGAAAAAC | T02B11.3 promoter; obi-1 cDNA |
| RHL498 | GCGATAAGATCGGTCAATCTGAG | F16F9.3 promoter |
| RHL499 | GCCAGTAAGGGCTAGTAAGTG | F16F9.3 promoter |
| RHL500 | AACGAGAACCAGACTCCTGATCATATTTTGTTTCTTACTGTCTTG | F16F9.3 promoter |
| RHL501 | ATGGGTACGTTTCTGGGTATAG | T02B11.3 promoter; obi-1 cDNA |
| VR86 | GACTCCTGATTATCGCTACG | Ppa-obi-1 cDNA |
| VR87 | AGATAGTCAAACTATGCATC | Ppa-obi-1 cDNA |
| VR88 | CTAATAATCTACACTAAATG | Ppa-obi-1 cDNA |
| AG11112 | CTCGGAGGAGGAACTGGATC | Ppa-beta-tubulin RT-qPCR |
| AG11113 | GACCGTGTCAGAGACCTTAG | Ppa-beta-tubulin RT-qPCR |
| RH12548 | GCAGGAACATATATTCGCAG | Ppa-egl-4 RT-qPCR |
| RH12549 | TGTCACGGAACGTTTTGTAC | Ppa-egl-4 RT-qPCR |
| SL1 | GGTTTAATTACCCAAGTTTGAG | SL1 for 5′ RACE |
| SSLP21F | ACTGGTGCTCATCGCAAGA | Mapping marker |
| SSLP21R | CCTTCTGTTTTCCTACCCCC | Mapping marker |
| SSLP7F | TCTTGCATAACACCGAACAAA | Mapping marker |
| SSLP7R | AGGGCGTTACGTGAATAAGC | Mapping marker |
| SSLP17F | CAATGCAGTAGCCGAATCAT | Mapping marker |
| SSLP17R | CAATATTGGCCTTCACCCTG | Mapping marker |
| S447F | TGGGGATAGCGAAAATCATC | Mapping marker |
| S447R | CGCATCGTTATTCACGAAATTC | Mapping marker |
-
Letters in bold are based on different templates for fusion PCR.





