Cytosolic and endoplasmic reticulum chaperones inhibit wt-p53 to increase cancer cells' survival by refluxing ER-proteins to the cytosol
Figures
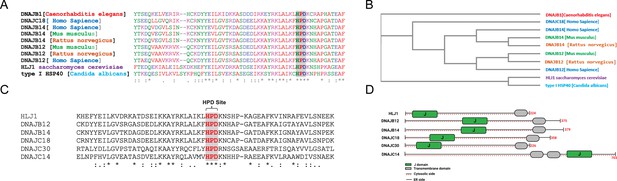
High copy lethal J-protein (HLJ1) is conserved from yeast to humans.
(A) Alignment shows the conservation of yeast-HLJ1 and their HPD domain from yeast to humans. (B) Phylogenic analysis showing the conservation of HLJ1 in different species. (C) The HPD motif within the J-domain is conserved in HLJ1 and its putative human orthologs DNAJB12, DNAJB14, DNAJC14, DNAJC18, and DNAJC30. (D) Schematic showing the different domains of HLJ1 and its orthologs, including a cytosolic J-domain, transmembrane domain, and endoplasmic reticulum (ER) domain.
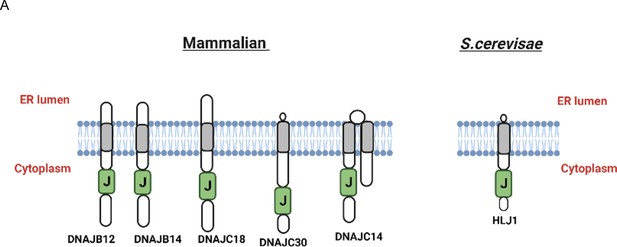
High copy lethal J-protein (HLJ1) putative orthologues share the same topology as HLJ1.
(A) Schematic showing the comparison between HLJ1 topology and the five putative orthologs as described (Piette et al., 2021) with a J-domain facing the cytosol and small endoplasmic reticulum (ER) domain.
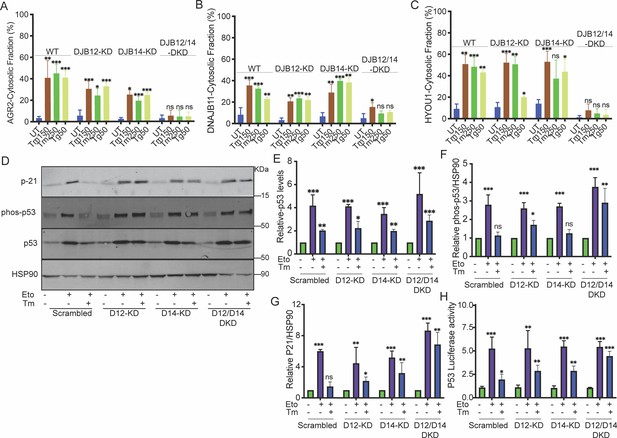
DNAJB12 and DNAJB14 are necessary for AGR2 reflux and wt-p53 inhibition in cancer cells.
(A–C) Quantification of the subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1 in A549 cells as shown in (Figure 2—figure supplement 1) N=3. (D) Representative immunoblot of p-21, phospho-p53, total-p53 (DO-1), and HSP90 in control cells and cells lacking DNAJB12, DNAJB14, or both N=3. (E–G) Quantification of p53, phosph-p53, and P-21 levels as shown in D, respectively. (H) A549 were transfected with scrambled siRNA (scrambled), DNAJB12-targeted siRNA (D12–KD), DNAJB14-targeted siRNA (D14–KD), or both. After 24 hr, cells were transfected with p53-luciferase construct. Cells were treated with etoposide for 2 hr to induce wt-p53, followed by tunicamycin treatment for 16 hr, and luciferase experiments were performed. Biological triplicates, mean ± SD calculated using Prism 9 (GraphPad). (***p<0.001, **p<0.01, *p<0.05).
-
Figure 2—source data 1
Original western blots for Figure 2D.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data1-v1.zip
-
Figure 2—source data 2
Original western blots for Figure 2D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data2-v1.zip
-
Figure 2—source data 3
Related to Figure 2A.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data3-v1.xlsx
-
Figure 2—source data 4
Related to Figure 2B.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data4-v1.xlsx
-
Figure 2—source data 5
Related to Figure 2C.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data5-v1.xlsx
-
Figure 2—source data 6
Related to Figure 2E.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data6-v1.xlsx
-
Figure 2—source data 7
Related to Figure 2F.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data7-v1.xlsx
-
Figure 2—source data 8
Related to Figure 2G.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data8-v1.xlsx
-
Figure 2—source data 9
Related to Figure 2H.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-data9-v1.xlsx
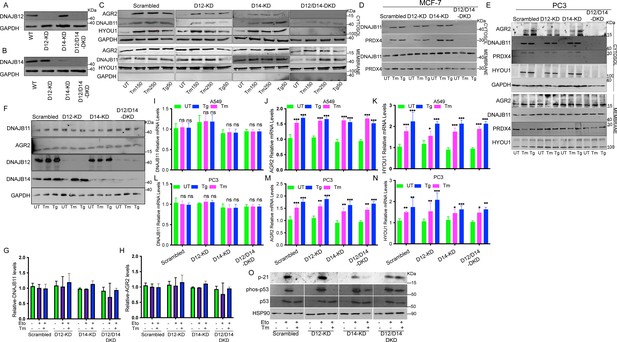
DNAJB12 and DNAJB14 are necessary for ER-protein reflux and wt-p53 inhibition in different cancer cell lines.
(A, B) Immunoblot showing the expression of DNAJB12 and DNAJB14 in A549 cells transfected with siRNA-targeted DNAJB12, DNAJB14, or both. (C–E) Subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1 during endoplasmic reticulum (ER) stress with tunicamycin (Tm) and Thapsigargin in DNAJB12, DNAJB14, or DNAJB12/14 -silenced A549, MCF-7, and PC3 cells, respectively. (F) Total protein levels from whole cell lysates in A549 cells for AGR2, DNAJB11, DNAJB12, DNAJB14. (G, H) quantification of DNAJB11 and AGR2 protein levels as shown in F. (I–K) Relative mRNA levels of AGR2, DNAJB11, and HYOU1 in A549 cells. (L–N) Relative mRNA levels of AGR2, DNAJB11, and HYOU1 in PC3 cells. (O) p21, phosph-p53 and total-p53 levels in DNAJB12, DNAJB14, and double-knock down cells treated with etoposide and tunicamycin.
-
Figure 2—figure supplement 1—source data 1
Original western blots for Figure 2—figure supplement 1A–F and O.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Original western blots for Figure 2—figure supplement 1A–F and O, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data2-v1.zip
-
Figure 2—figure supplement 1—source data 3
Related to Figure 2—figure supplement 1I.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data3-v1.xlsx
-
Figure 2—figure supplement 1—source data 4
Related to Figure 2—figure supplement 1J.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data4-v1.xlsx
-
Figure 2—figure supplement 1—source data 5
Related to Figure 2—figure supplement 1K.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data5-v1.xlsx
-
Figure 2—figure supplement 1—source data 6
Related to Figure 2—figure supplement 1L.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data6-v1.xlsx
-
Figure 2—figure supplement 1—source data 7
Related to Figure 2—figure supplement 1M.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data7-v1.xlsx
-
Figure 2—figure supplement 1—source data 8
Related to Figure 2—figure supplement 1N.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data8-v1.xlsx
-
Figure 2—figure supplement 1—source data 9
Related to Figure 2—figure supplement 1G.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data9-v1.xlsx
-
Figure 2—figure supplement 1—source data 10
Related to Figure 2—figure supplement 1H.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig2-figsupp1-data10-v1.xlsx
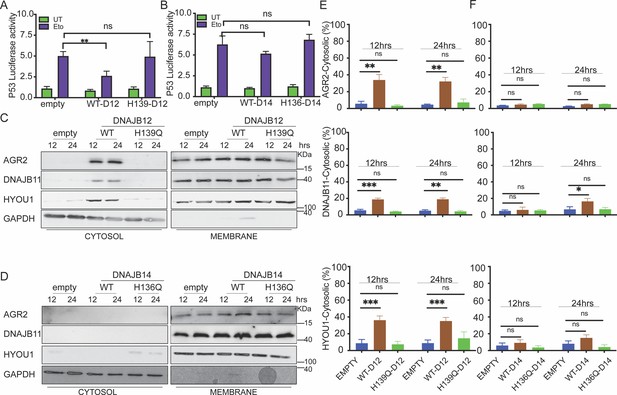
DNAJB12 but not DNAJB14 is sufficient for refluxing AGR2 and other endoplasmic reticulum (ER) resident proteins to inhibit wt-p53 activity.
(A, B) Cells were co-transfected with pCDNA3 plasmid expressing DNAJB12, DNAJB14, or the empty plasmid and p53-luciferase construct after 24 hr. Cells were treated with etoposide for 4 hr to induce wt-p5,3, and luciferase experiments were performed. Biological triplicates (N=3), mean ± SD calculated using Prism 9 (GraphPad). (***p<0.001, **p<0.01, *p<0.05). (C, D) Subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1in cells overexpressing WT and QPD mutant of DNAJB12 and DNAJB14, respectively, at different time points. (E, F) Quantification of the subcellular protein fractionation of AGR2, DNAJB11, and HYOU1 in A549 cells as shown in (C, D) Biological quadruplicate (N=4).
-
Figure 3—source data 1
Original western blots for Figure 3C and D.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-data1-v1.zip
-
Figure 3—source data 2
Original western blots for Figure 3C and D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-data2-v1.zip
-
Figure 3—source data 3
Related to Figure 3A.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Related to Figure 3B.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-data4-v1.xlsx
-
Figure 3—source data 5
Related to Figure 3E.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-data5-v1.xlsx
-
Figure 3—source data 6
Related to Figure 3F.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-data6-v1.xlsx
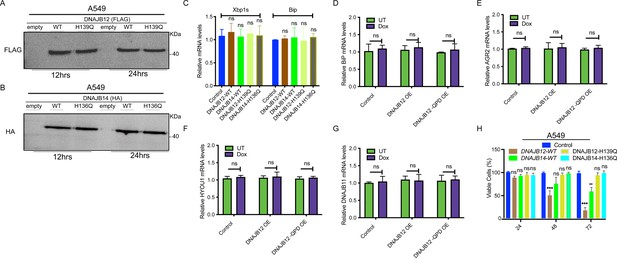
DNAJB12 overexpression decreases cell viability without inducing ER stress.
(A, B) Immunoblot showing the expression of FLAG-tagged DNAJB12 and HA-tagged DNAJB14 in A549 cells overexpressing either WT and QPD mutant of DNAJB12 or DNAJB14, respectively for 12 hr (left) or 24 hr (right). (C) Relative mRNA levels of spliced Xbp1 and Bip (Unfolded Protein Response markers) in A549 cells overexpressing WT and QPD mutant of DNAJB12 or DNAJB14. (D) Relative BiP mRNA levels in -Rex293 cells overexpressing DAJB12 and the QPD mutant with and without doxycycline (Dox). (E–G) Relative AGR2, HYOU1, and DNAJB11 mRNA levels in A549 cells overexpressing DNAJB12 WT or QPD mutant. (H) Cell viability of A549 cells overexpressing either WT and QPD mutant of DNAJB12 or DNAJB14 for the indicated time points.
-
Figure 3—figure supplement 1—source data 1
Original western blots for Figure 3—figure supplement 1A, B.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Original western blots for Figure 3—figure supplement 1A, B, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data2-v1.zip
-
Figure 3—figure supplement 1—source data 3
Related to Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data3-v1.xlsx
-
Figure 3—figure supplement 1—source data 4
Related to Figure 3—figure supplement 1D.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data4-v1.xlsx
-
Figure 3—figure supplement 1—source data 5
Related to Figure 3—figure supplement 1E.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data5-v1.xlsx
-
Figure 3—figure supplement 1—source data 6
Related to Figure 3—figure supplement 1F.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data6-v1.xlsx
-
Figure 3—figure supplement 1—source data 7
Related to Figure 3—figure supplement 1G.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data7-v1.xlsx
-
Figure 3—figure supplement 1—source data 8
Related to Figure 3—figure supplement 1H.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig3-figsupp1-data8-v1.xlsx
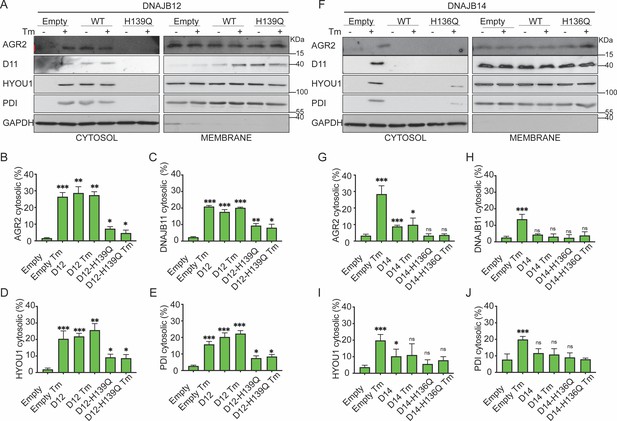
Functional DNAJB12 and 14 are necessary for endoplasmic reticulum (ER) protein reflux during ER stress.
(A) Subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1in cells overexpressing either WT and QPD mutant of DNAJB12 during ER stress with tunicamycin (Tm). (B–E) Quantification of the subcellular protein fractionation of AGR2, DNAJB11, and HYOU1 in A549 cells as shown in A and Figure 4—figure supplement 1A. Biological triplicates (N=3) (F) Subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1 in cells overexpressing WT and QPD mutant of DNAJB14 during ER stress with tunicamycin (Tm). (G–J) Quantification of the subcellular protein fractionation of AGR2, DNAJB11, and HYOU1 in A549 cells as shown in F and Figure 4—figure supplement 1B, C. Biological quadruplicates (N=4) (***p<0.001, **p<0.01, *p<0.05).
-
Figure 4—source data 1
Original western blots for Figure 4A and F.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data1-v1.zip
-
Figure 4—source data 2
Original western blots for Figure 4A and F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data2-v1.zip
-
Figure 4—source data 3
Related to Figure 4B.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data3-v1.xlsx
-
Figure 4—source data 4
Related to Figure 4C.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data4-v1.xlsx
-
Figure 4—source data 5
Related to Figure 4D.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data5-v1.xlsx
-
Figure 4—source data 6
Related to Figure 4E.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data6-v1.xlsx
-
Figure 4—source data 7
Related to Figure 4G.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data7-v1.xlsx
-
Figure 4—source data 8
Related to Figure 4H.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data8-v1.xlsx
-
Figure 4—source data 9
Related to Figure 4I.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data9-v1.xlsx
-
Figure 4—source data 10
Related to Figure 4J.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-data10-v1.xlsx
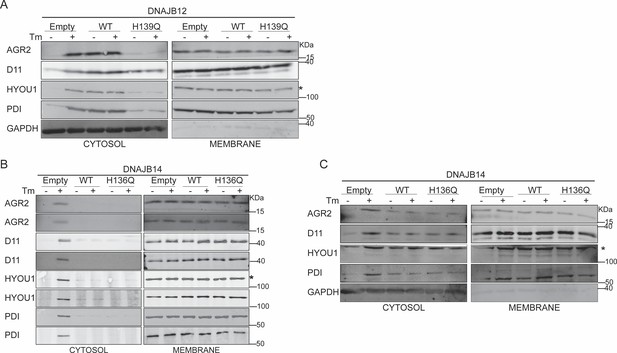
Functional DNAJB12 and 14 are necessary for ER protein reflux during ER stress.
(A) Subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1 in A549 cells overexpressing WT and QPD mutant of DNAJB12. (B) Subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1in cells overexpressing either WT and QPD mutant of DNAJB14 during endoplasmic reticulum (ER) stress with tunicamycin (Tm). (C) Subcellular protein fractionation (Digitonin fraction) of AGR2, DNAJB11, and HYOU1 in cells overexpressing WT and QPD mutant of DNAJB14 during ER stress with Tm.
-
Figure 4—figure supplement 1—source data 1
Original western blots for Figure 4—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original western blots for Figure 4—figure supplement 1A–C, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig4-figsupp1-data2-v1.zip
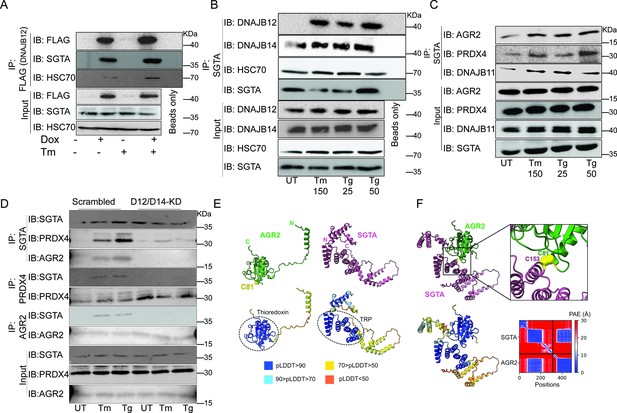
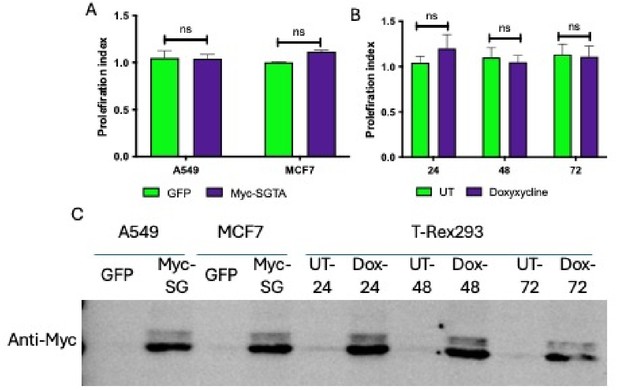
The cytosolic cochaperone SGTA is recruited by DAJB12 and interacts with the refluxed endoplasmic reticulum (ER) protein in the cytosol.
(A) T-Rex293 cell lines overexpressing FLAG-tagged DNAJB12 using the Tet-On system for Doxycycline-inducible gene expression were treated with doxycycline to induce DNAJB12 expression. FLAG-DNAJB12 interaction with SGTA was analyzed using coimmunoprecipitation assay N=3. (B) Representative immunoblot showing the interaction between SGTA in one hand and DNAJB12, DNAJB14, and HSC70 in A549 cells treated with tunicamycin and thapsigargin as indicated. Quantified in Figure 5—figure supplement 1I–N. N=3 (C) A representative immunoblot showing the interaction between SGTA and the ER-resident proteins AGR2, PRDX4, and DNAJB11 in A549 cells treated with tunicamycin and thapsigargin as indicated. N=3. (D) A representative immunoblot showing the interaction between SGTA and the ER-resident proteins AGR2, PRDX4, and DNAJB11 in A549 cells depleted of DNAJB12/14 treated with tunicamycin and thapsigargin as indicated. (E) AlphaFold2 modeling of AGR2 and SGTA. N and C termini are shown, as well as cysteine 81 in AGR2. Bottom-coloring DNAJB12-silenced cells according to confidence in prediction according to per-residue confidence (pLDDT) scale (see legend). Note the high confidence in the predictions of the thioredoxin domain in AGR2 and the TRP domain in SGTA (circled in dashed lines). (F) ColabFold modeling of the SGTA-AGR2 interaction interface. The unstructured N-terminal region of AGR2 was removed for clarity. Top – cys81 in AGR2 and cys153 in SGTA are found within bonding distance, shown in stick presentation and colored yellow. Right–zoom into the boxed region, showing the sulfur atoms of the cysteine residues as spheres—bottom - coloring according to confidence in prediction according to pLDDT. Inter predicted aligned error (PAE) plot is shown with regions of low PAE (blue), which indicates a confident prediction.
-
Figure 5—source data 1
Original western blots for Figure 5A–D.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-data1-v1.zip
-
Figure 5—source data 2
Original western blots for Figure 5A–D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-data2-v1.zip
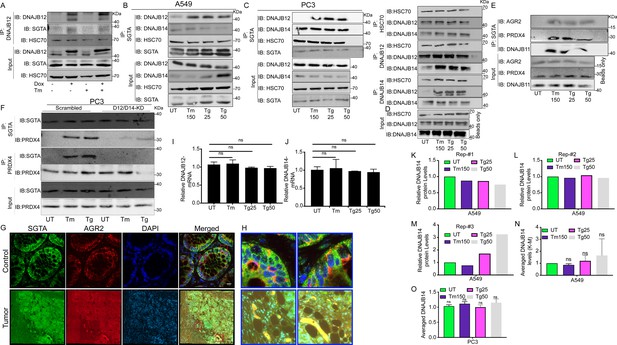
The cytosolic cochaperone SGTA interacts with the refluxed ER protein in different cancer cell lines and in tumors isolated from colorectal cancer patients.
(A) T-Rex293 cell line overexpressing DNAJB12 using the Tet-On system for Doxycycline-inducible gene expression were treated with doxycycline to induce DNAJB12 expression. DNAJB12 interaction with SGTA was analyzed by coimmunoprecipitation assay. (B) Representative immunoblot showing the interaction between SGTA in one hand and DNAJB12, DNAJB14, and HSC70 in A549 cells treated with tunicamycin and thapsigargin as indicated- quantified in M. (C) Representative immunoblot showing the interaction between SGTA in one hand and DNAJB12, DNAJB14, and HSC70 in PC3 cells treated with tunicamycin and thapsigargin as indicated. (D) Coimmunoprecipitation assay for HSC70, DNAJB12, and DNAJB14 during endoplasmic reticulum (ER) stress with tunicamycin and thapsigargin as indicated. (E) A representative immunoblot showing the interaction between SGTA on one hand and the ER-resident proteins AGR2, PRDX4, and DNAJB11 in A549 cells treated with tunicamycin and thapsigargin as indicated. (F) A representative immunoblot showing the interaction between SGTA and PRDX4 in DNAJB12/14 double knockout PC3 cells treated with tunicamycin and thapsigargin as indicated. (G) Colocalization of SGTA and AGR2 in tumor from Colorectal cancer (CRC) patients. Green SGTA Alexa-Flour 680, Red AGR2 Alexa-Flour 555, scale bar=17µM. (H) Zoom-in on the rectangular zones in (G). (I, J) Relative mRNA levels of DNJB12 and DNAJB14 in A549 cells, respectively. (K–M) Relative DNAJB14 protein levels in A549 cells treated with Tm and Tg in A549 cells as shown in B and Figure 5B (N=3). (N) Average of the DNAJB14 levels calculated in K-M and shown. (O) Averaged DNAJB14 protein levels in PC3 cells (represented in C) treated with tunicamycin (Tm) and Tg (N=3).
-
Figure 5—figure supplement 1—source data 1
Original western blots for Figure 5—figure supplement 1A–F.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Original western blots for Figure 5—figure supplement 1A–F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-figsupp1-data2-v1.zip
-
Figure 5—figure supplement 1—source data 3
Related to Figure 5—figure supplement 1I.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-figsupp1-data3-v1.xlsx
-
Figure 5—figure supplement 1—source data 4
Related to Figure 5—figure supplement 1J.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-figsupp1-data4-v1.xlsx
-
Figure 5—figure supplement 1—source data 5
Related to Figure 5—figure supplement 1K–N.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-figsupp1-data5-v1.xlsx
-
Figure 5—figure supplement 1—source data 6
Related to Figure 5—figure supplement 1O.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig5-figsupp1-data6-v1.xlsx
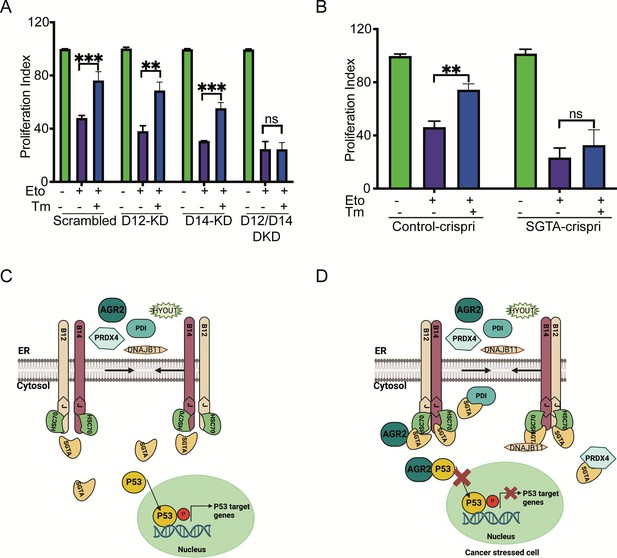
Silencing DNAJB12 and DNAJB14 or SGTA inhibits the rescue cell proliferation under subtoxic endoplasmic reticulum (ER) stress conditions.
(A) XTT assay in A549 transfected with scrambled siRNA (scrambled), DNAJB12-targeted siRNA (D12–KD), DNAJB14-targeted siRNA (D14–KD), or both. After 24 hr, cells were treated with etoposide in the presence or absence of Tm for 48 hr. Biological triplicates. (B) XTT assay in SGTA-silenced A549 cells using CRISPRi and treated with etoposide in the presence or absence of tunicamycin for 48 hr. Biological triplicates, mean ± SD calculated using Prism 9 (GraphPad). (***p<0.001, **p<0.01, *p<0.05). Biological triplicates (C, D) Cartoon showing our working model in cancer cells.in normal conditions, there is almost full compartmentation between the ER and the cytosol (C). Under ER stress conditions, DNAJB12 and DNAJB14 recruit cytosolic chaperones and cochaperones (SGTA) to reflux AGR2 and other ER resident proteins to the cytosol and inhibit wt-p53 activity (D). Created with BioRender.com.
-
Figure 6—source data 1
Related to Figure 6A.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Related to Figure 6B.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig6-data2-v1.xlsx
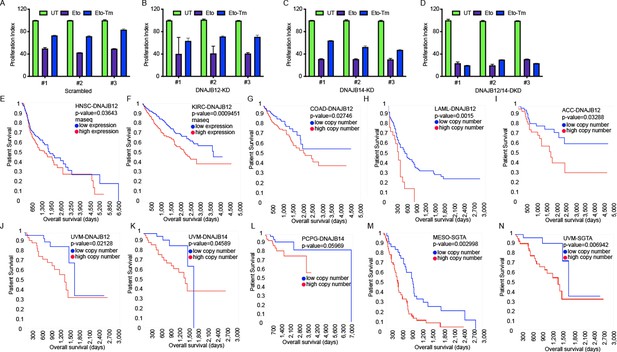
DNAJB12, DNAJB14, and SGTA are unfavorable prognostic markers in different cancer types.
(A–D) XTT assay in A549 transfected with scramble siRNA (scramble), DNAJB12-targeted siRNA (D12–KD), DNAJB14-targeted siRNA (D14–KD), or both. After 24 hr, cells were treated with etoposide in the presence or absence of Tm for 48 hr. (E, F) DNAJB12 is unfavorable prognostic marker in different cancer types including Head and neck squamous cell carcinoma (HNSC), Kidney renal clear cell carcinoma (KIRC) TCGA survival analysis- rnaseq database, red line represents low expression and blue lines represent high expression. (G–J) High copy number of DNAJB12 is unfavorable prognostic marker in different cancer types including colon adenocarcinoma (COAD), acute myeloid leukemia (LAML), adrenocortical carcinoma (ACC), and uveal melanoma (UVM), red line represents high copy number and blue lines represent low copy number (K, L) High copy number of DNAJB14 is unfavorable prognostic marker in different cancer types including Pheochromocytoma and paraganglioma (PCPG) and UVM, red line represents high copy number and blue lines represent low copy number. (M, N) High copy number of SGTA is unfavorable prognostic marker in different cancer types including mesothelioma (MESO) and UVM, red line represents high copy number and blue lines represent low copy number. For all survival analyses: x-axis shows the patients’ survival duration in months; y-axis shows the patients’ survival rate.The p-values have been calculated using the log-rank test.
-
Figure 6—figure supplement 1—source data 1
Related to Figure 6—figure supplement 1A.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig6-figsupp1-data1-v1.xlsx
-
Figure 6—figure supplement 1—source data 2
Related to Figure 6—figure supplement 1B.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig6-figsupp1-data2-v1.xlsx
-
Figure 6—figure supplement 1—source data 3
Related to Figure 6—figure supplement 1C.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig6-figsupp1-data3-v1.xlsx
-
Figure 6—figure supplement 1—source data 4
Related to Figure 6—figure supplement 1D.
- https://cdn.elifesciences.org/articles/102658/elife-102658-fig6-figsupp1-data4-v1.xlsx

(A) eroGFP with transmembrane domain (TM-eroGFP) is highly oxidized in the ER in all the indicated conditions. (B) XTT assay in WT, D12KD, D14KD and the DKD cells after treatment with Etoposide and tunicamycin (Tm). (C) Caspase-3 activity in scrambled cells and in the D12/D14 DKD cells.
Additional files
-
Supplementary file 1
Alignment of the yeast high copy lethal J-protein (HLJ1) protein sequence against many different databases, including human, mouse, rat, zebrafish, fly, mosquito, worm, and fission yeast using the DRSC/TRiP Functional Genomics Resources & DRSC-BTRR, DIOPT version 9, Harvard Medical School.
- https://cdn.elifesciences.org/articles/102658/elife-102658-supp1-v1.xlsx
-
Supplementary file 2
List of the different antibodies, siRNA, andoligos used in this study.
(a) List of the different antibodies used in this study. (b) List of the commercially available siRNA used in this study. (c) List of oligos used for SGTA-CRISPRi from Gilbert et al., 2014. (d) List of oligos used for DNAJB12 and DNAJB14 cloning.
- https://cdn.elifesciences.org/articles/102658/elife-102658-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/102658/elife-102658-mdarchecklist1-v1.docx