eIF3 engages with 3’-UTR termini of highly translated mRNAs
Figures
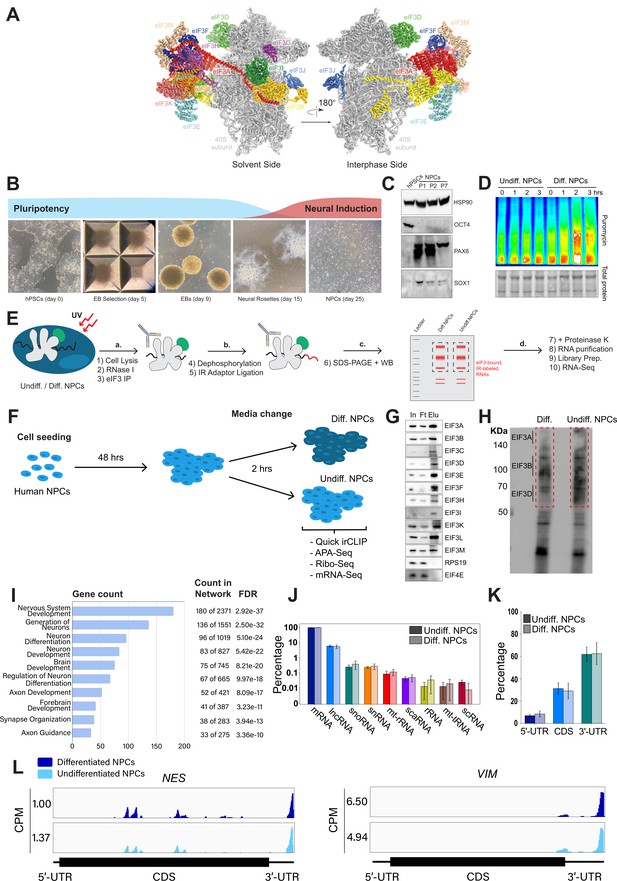
Analysis of Quick irCLIP of eukaryotic initiation factor 3 (eIF3) to RNAs in undifferentiated and differentiated neural progenitor cells.
(A) Structure of the human eIF3 complex as part of the 48 S initiation complex. PDB: 6ZMW (Brito Querido et al., 2020). (B) Generation of neural progenitor cells (NPCs) from human pluripotent stem cells (hPSCs) by embryoid body (EB) and neural rosette selections. (C) Western blots of neural markers Pax6 and Sox1 and pluripotent marker Oct4 from hPSCs and hPSC-derived NPCs (NPCs corresponding to passages 1, 2, and 7 are shown). (D) Western blots of puromycin-treated (15 min) NPCs. Total protein stain is shown as loading control. Differentiated NPCs were treated with forebrain neuron differentiation medium for the indicated time points. Undifferentiated NPCs were treated with NPC medium for the indicated time points. The ratios of puromycin signal to total protein levels can be found in Figure 1—figure supplement 1A. (E) Schematic of Quick-irCLIP. (F) Schematic of the NPC treatment performed in the Quick-irCLIP, polyadenylation sequencing (APA-seq), Ribosome profiling, and mRNA-Seq experiments. Treatment consisted of change of media and incubation at 37 °C for 2 hrs. Differentiation media was used for differentiated (Diff.) NPCs and Basal media for undifferentiated (Undiff.) NPCs. (G) Immunoprecipitation samples of assembled eIF3 complexes from undifferentiated NPCs. 5% inputs (In), flowthroughs (Ft), and eluates (Elu) are shown. Western blot for subunit EIF3G is not shown because we could not identify an effective antibody for its detection. EIF3J, not shown, is usually dissociated from the eIF3 complex upon immunoprecipitation. However, we detected EIF3G by mass spectrometry in the IPs (Supplementary file 1). (H) Infrared (IR) image of IR dye-labeled, eIF3 UV-crosslinked RNA transcripts from Diff. and Undiff. NPCs. Regions marked with red boxes, which correspond to subunits EIF3A through EIF3D, were excised from the blot. (I) Biological Function enrichment determined using the STRING database for the undifferentiated NPC biological replicates of the EIF3A/B/C/D Quick-irCLIP libraries, using the top 500 mRNA hits from the crosslinking analysis. (J) Categories of RNAs crosslinked to eIF3 in undifferentiated and differentiated NPCs, for three replicates plotted in log-scale with standard deviation error bars. (K) mRNA regions that crosslink to eIF3 in undifferentiated and differentiated NPCs. (L) Crosslinking of eIF3 across the transcripts of NES and VIM mRNAs in differentiated and undifferentiated NPCs. Read coverage is provided in Counts Per Million (CPM).
-
Figure 1—source data 1
Files of original western blots for panels C and G, along with gels of the eukaryotic initiation factor 3 (eIF3) crosslinked RNA in the Quick-irCLIP experiment in panel H.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig1-data1-v1.zip
-
Figure 1—source data 2
Files of original blots and gels in panels C, G, and H, with experimental conditions marked.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig1-data2-v1.zip
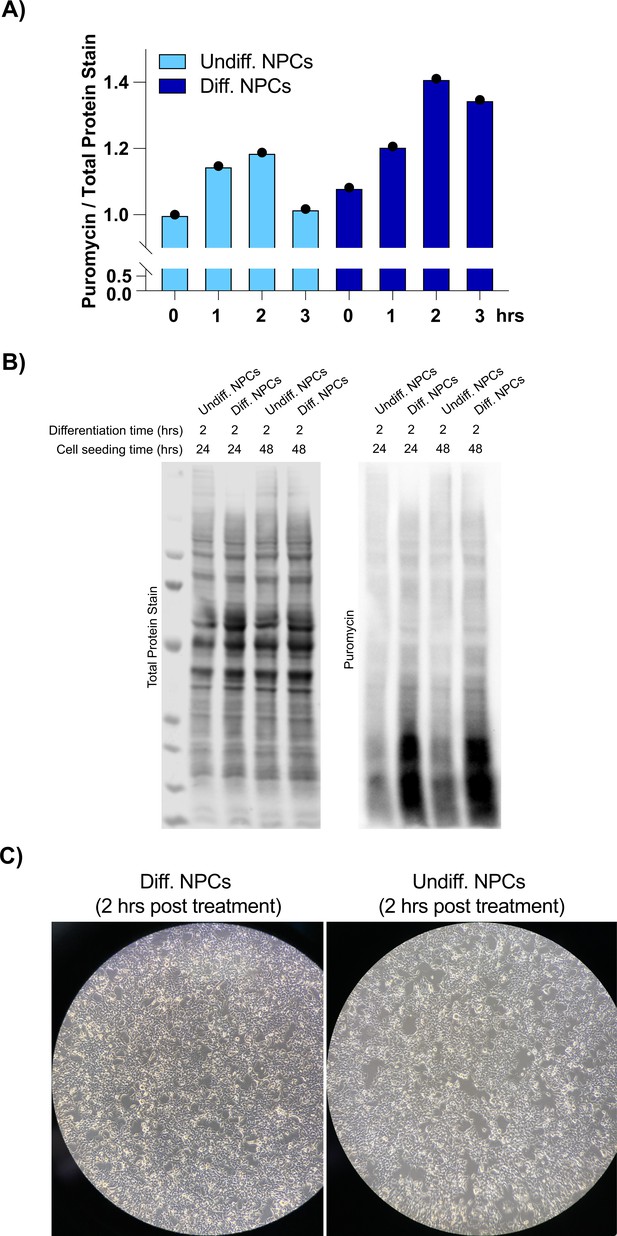
Neural progenitor cells (NPCs) treated with differentiation media or kept in undifferentiated media (2 hr treatment).
(A) Levels of protein synthesis in undifferentiated and differentiated NPCs at different time points, as quantified by puromycin incorporation. Puromycin signal was normalized to total protein levels, which were measured with total protein stain. This is quantification of Figure 1D. (B) A second set of experiments showing changes in levels of protein synthesis in undifferentiated vs. differentiated NPCs. (C) Cellular morphology of the NPCs using dark field microscopy.
-
Figure 1—figure supplement 1—source data 1
Original gel with total protein staining and anti-puromycin western blot in panel B.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Original gel with total protein staining and anti-puromycin western blot in panel B with experimental conditions marked.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig1-figsupp1-data2-v1.zip

Venn Diagram of genes identified by Quick-irCLIP (differentiated neural progenitor cells, NPCs) and PAR-CLIP (HEK293T and activated Jurkat T cells).
The top ~200 mRNA transcripts from the three libraries ranked by total reads mapped to a given gene are compared (See Materials and methods).
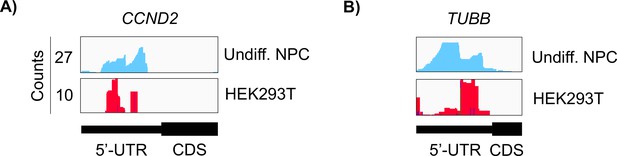
Crosslinking of eukaryotic initiation factor 3 (eIF3) in undifferentiated neural progenitor cells (NPCs) and in HEK293T cells.
Crosslinks in undifferentiated NPCs are in blue, and in HEK294T cells in red. Shown are the 5’-UTR regions of (A) CCND2 and (B) TUBB mRNAs.
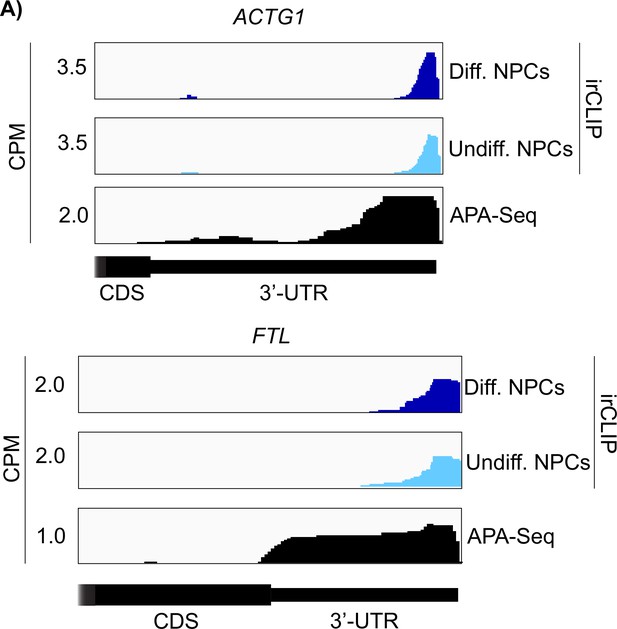
Crosslinking of eukaryotic initiation factor 3 (eIF3) in differentiated and undifferentiated NPCs across the 3’-UTR regions of ACTG1 and FTL mRNAs.
polyadenylation sequencing (APA-Seq) data is also shown for both transcripts.
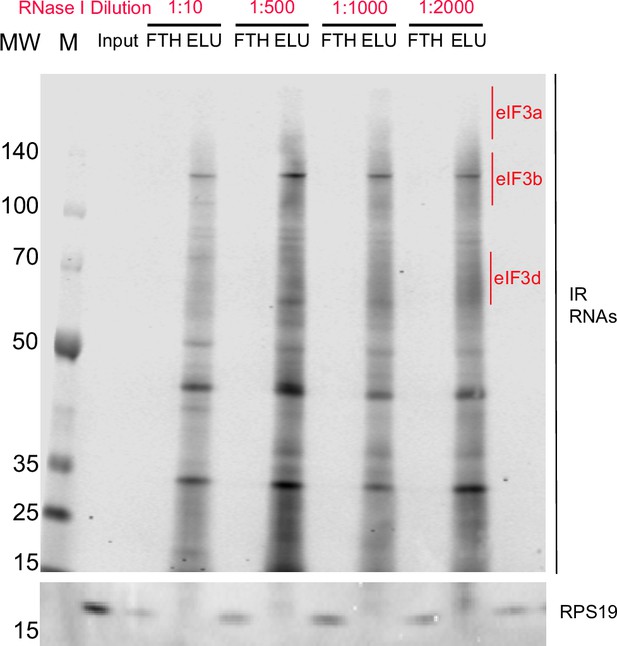
Titration of RNAse I for the Quick-irCLIP experiment.
Crosslinked samples were treated with RNAse I for 3 min and subject to anti-EIF3B IP. Shown are 5% input of the lysate, flowthrough (FTH) and elution (ELU) for each concentration of RNAse I. Molecular weight markers (MW, M) are shown to the left.
-
Figure 1—figure supplement 5—source data 1
Original gels of labeled RNAs from the eukaryotic initiation factor 3 (eIF3) pulldowns.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig1-figsupp5-data1-v1.zip
-
Figure 1—figure supplement 5—source data 2
Original gels of labeled RNAs from the eukaryotic initiation factor 3 (eIF3) pulldowns, with conditions indicated.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig1-figsupp5-data2-v1.zip

Correlation of Quick-irCLIP replicates.
Spearman correlation coefficients (Rho) of irCLIP bioreplicates (n=3) for differentiated (average Rho: 0.7855, standard deviation: 0.0092) and undifferentiated neural progenitor cells (NPCs) (average Rho: 0.834, standard deviation: 0.0058) demonstrate high reproducibility.
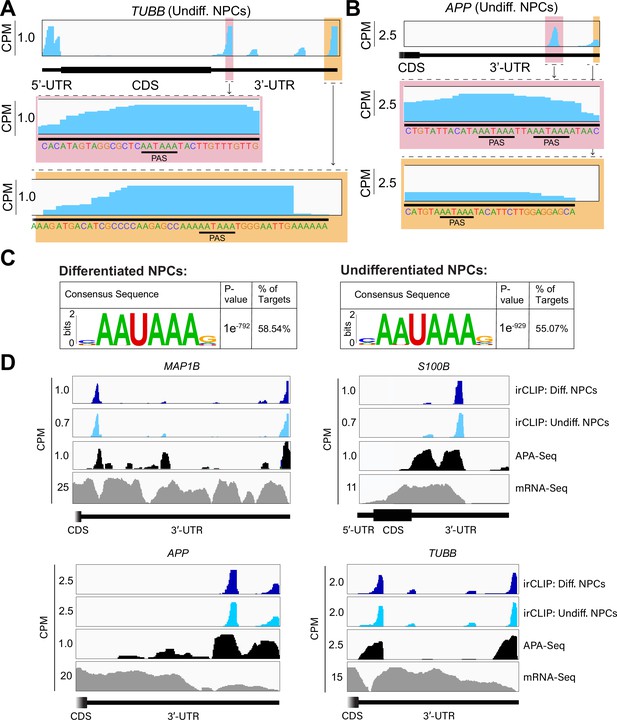
Crosslinking of eukaryotic initiation factor 3 (eIF3) to mRNA 3’-UTRs adjacent to the poly(A) tail.
(A) Crosslinking of eIF3 across the TUBB mRNA in undifferentiated neural progenitor cells (NPCs). A zoomed-in view of TUBB mRNA 3’-UTR terminus with the eIF3 crosslinks is shown. The polyadenylation signal (PAS) is marked. (B) Crosslinking of eIF3 across the APP mRNA 3’-UTR in undifferentiated NPCs. A zoomed-in view of the two 3’-UTR regions presenting eIF3 crosslinks are shown as well as their PAS sequences. (C) Sequence logo of the eIF3 crosslinks located at 3’-UTRs in differentiated and undifferentiated NPCs. (D) Crosslinking of eIF3, polyadenylation sequencing (APA-seq) peaks, and mRNA-seq across the MAP1B, S100B, TUBB, and APP mRNAs in differentiated and undifferentiated NPCs.
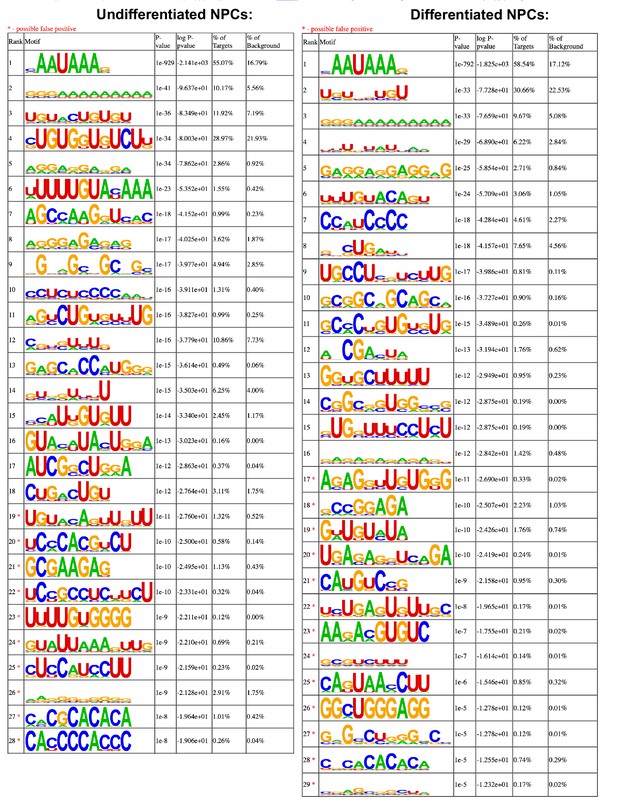
Motif enrichment of eukaryotic initiation factor 3 (eIF3) 3’-UTR crosslinks for undifferentiated and differentiated neural progenitor cells (NPCs).
Motifs were identified with HOMER (v4.11).
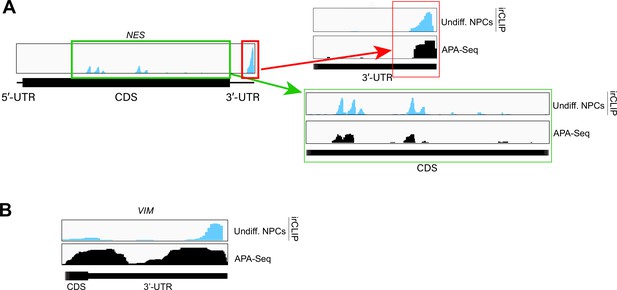
Examples of eukaryotic initiation factor 3 (eIF3) crosslinking to 3’-UTR regions of mRNAs.
(A) Crosslinking of eIF3 in undifferentiated neural progenitor cells (NPCs) as indicated across the 3’-UTR region of NES mRNA. APA-seq data is also shown. (B) Crosslinking of eIF3 in undifferentiated NPCs as indicated across the 3’-UTR region of VIM mRNA. APA-seq data is also shown.
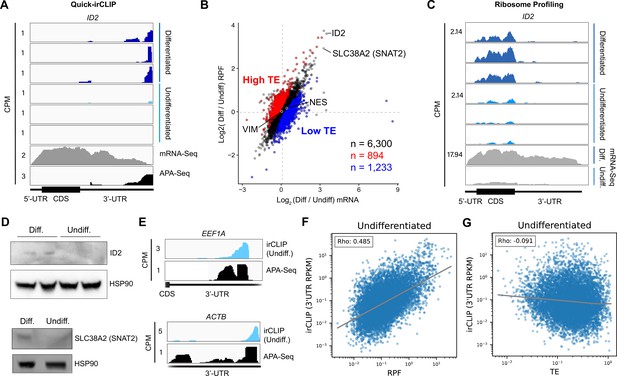
Ribosome profiling of undifferentiated and differentiated neural progenitor cells (NPCs), and comparisons to eukaryotic initiation factor 3 (eIF3) Quick irCLIP crosslinking.
(A) Crosslinking of eIF3 across the ID2 mRNA in undifferentiated and differentiated NPCs. Polyadenylation sequencing (APA-seq) and mRNA-seq data are also shown. (B) log2 fold change of ribosomal footprints and log2 fold change of mRNA transcript levels upon NPC differentiation. Transcripts with a higher TE in differentiated NPCs compared to undifferentiated NPCs (high TE) are indicated in blue. Transcripts with a lower TE in differentiated NPCs compared to undifferentiated NPCs (low TE) are shown in red. The number of samples within each condition is shown. (C) Ribosomal footprints across the ID2 mRNA in undifferentiated and differentiated NPCs. mRNA-seq data are also shown. (D) Western blots of ID2, SLC38A2 (SNAT2), and Hsp90 in undifferentiated and differentiated NPCs. (E) Crosslinking of eIF3 to the 3’-UTR of the EEF1A and ACTB mRNAs in undifferentiated NPCs. APA-seq data is also shown. (F) Comparison of irCLIP reads in mRNA 3’-UTRs to ribosome protected fragments (RPF) in undifferentiated NPCs. (G) Comparison of irCLIP reads in mRNA 3’-UTRs to translation efficiency (TE) in undifferentiated NPCs. Outliers exceeding the TE’s 99th percentile were removed, to make the log-log plot more readable. In panels (F) and (G), the Spearman correlation of average irCLIP 3’-UTR RPKM with mean RPF or TE across replicates (n=3) is shown.
-
Figure 3—source data 1
Original western blots for Figure 3D.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig3-data1-v1.zip
-
Figure 3—source data 2
Original western blots for Figure 3D with neural progenitor cell (NPC) treatment conditions indicated.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig3-data2-v1.zip
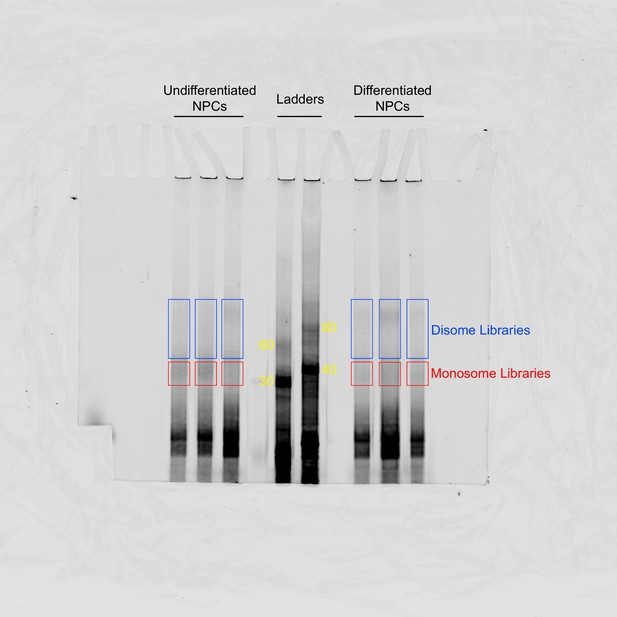
TBE 8% Urea-PAGE of the Ordered Two-Template Relay (OTTR) reaction products of cDNA libraries constructed from undifferentiated and differentiated neural progenitor cells (NPCs).
OTTR was also performed with control samples that yield marker products of 30, 40, 60, 80 nts in size (marked in yellow) so they could be used as references for gel excision for monosome and disome libraries. Gel excisions performed are shown in red (monosome) and blue (disome) rectangles.
-
Figure 3—figure supplement 1—source data 1
Original gels of Ordered Two-Template Relay (OTTR) reaction products used for ribosome profiling cDNA library construction.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Original gels of Ordered Two-Template Relay (OTTR) reaction products used for ribosome profiling cDNA library construction, with locations of regions cut out of gel indicated.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig3-figsupp1-data2-v1.zip
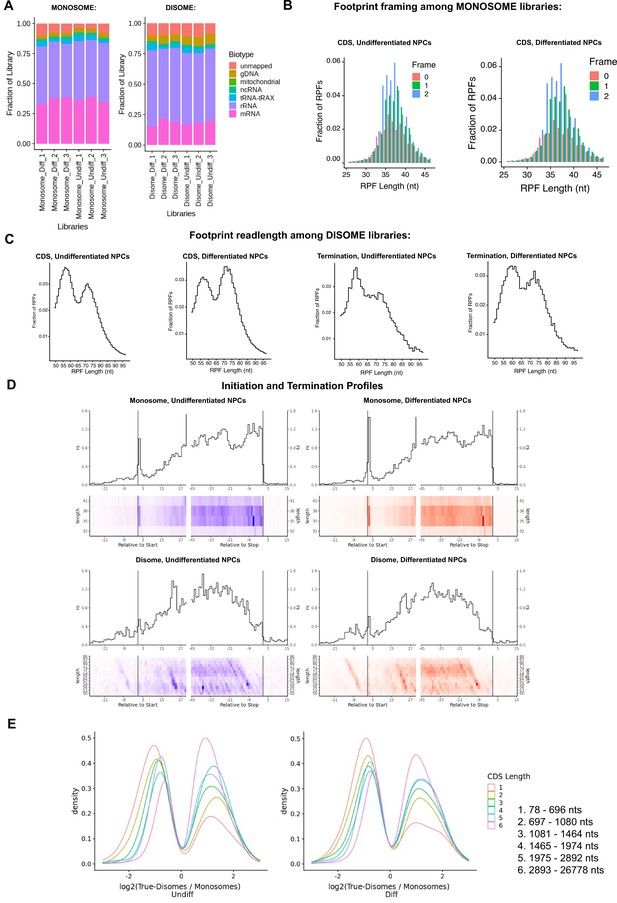
Ribosome profiling of undifferentiated and differentiated neural progenitor cells (NPCs).
(A) Fraction of sequencing reads mapped to each transcript type from monosome and disome profiling from P1 nuclease digested NPCs (undifferentiated and differentiated) and Ordered Two-Template Relay (OTTR) library cDNA synthesis. (B) Framing among monosome libraries in undifferentiated and differentiated NPCs. (C) Readlength of footprints aligned to CDS or terminating codons among disome libraries for undifferentiated and differentiated NPCs. (D) A-site corrected initiation and termination profiles for genes with sufficient coverage (≥1 read per codon) for monosome and disome libraries. (E) DESeq2 log2 fold change in CDS occupancy for disome libraries with respect to monosome libraries, stratified by CDS length in nucleotides.
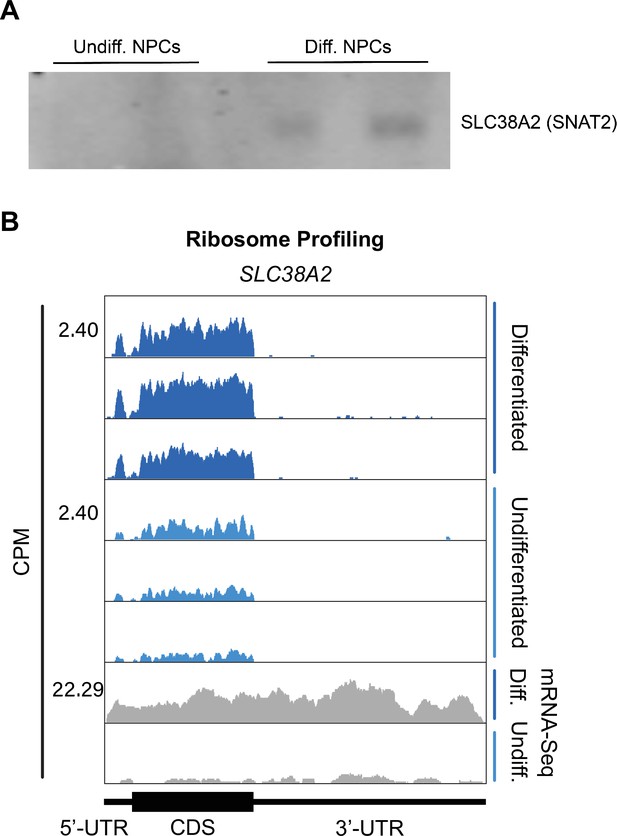
SLC38A2 expression in differentiated and undifferentiated neural progenitor cells (NPCs).
(A) Western blots of SLC38A2 for additional replicates of differentiated and undifferentiated NPCs. Gel loading was normalized by total protein amounts determined by Bradford assay. (B) Ribosomal footprints across the SLC38A2 mRNA in undifferentiated and differentiated NPCs. mRNA-seq data are also shown.
-
Figure 3—figure supplement 3—source data 1
Original western blot for Figure 3—figure supplement 3A.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig3-figsupp3-data1-v1.zip
-
Figure 3—figure supplement 3—source data 2
Original western blot for Figure 3—figure supplement 3A with conditions indicated.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig3-figsupp3-data2-v1.zip
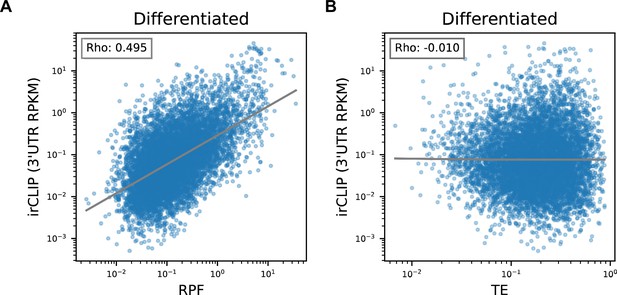
Correlation of Quick-irCLIP with Ribosome Profiling and Translational Efficiency in Differentiated neural progenitor cells (NPCs).
(A) Comparison of irCLIP reads in mRNA 3’-UTRs to ribosome protected fragments (RPF) in differentiated NPCs. (B) Comparison of irCLIP reads in mRNA 3’-UTRs to translation efficiency (TE) in differentiated NPCs. Outliers exceeding the TE’s 99th percentile were removed, to make the log-log plot more readable. The Spearman correlation of average irCLIP 3’-UTR RPKM with mean RPF or TE across replicates (n=3) is shown.
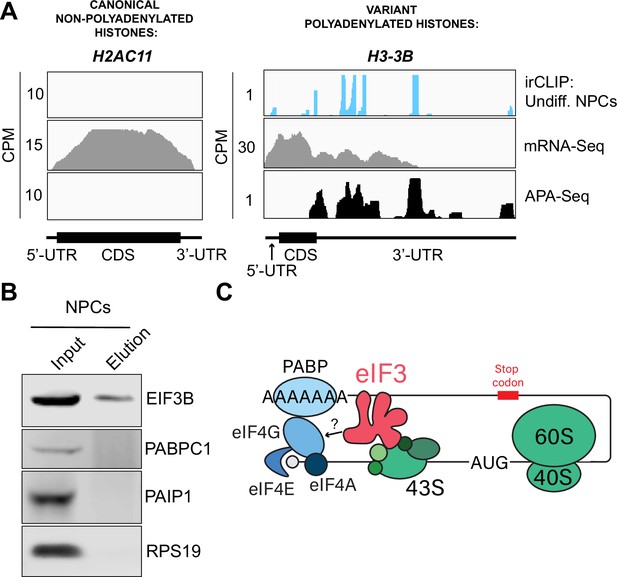
Crosslinking of eukaryotic initiation factor 3 (eIF3) to polyadenylated mRNAs and model for mRNA circularization.
(A) Crosslinking of eIF3 across the canonical non-polyadenylated histone H2AC11 mRNA and the variant polyadenylated histone H3-3B mRNA in undifferentiated neural progenitor cells (NPCs). Polyadenylation sequencing (APA-seq) and mRNA-seq data for these regions are also shown. (B) Western blots of EIF3B immunoprecipitations from undifferentiated NPCs. (C) Model for eIF3 contribution to mRNA circularization in highly translated transcripts.
-
Figure 4—source data 1
Original western blots for Figure 4B and Figure 4–figure supplement 1B.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig4-data1-v1.zip
-
Figure 4—source data 2
Original western blots for Figure 4B and for Figure 4–figure supplement 1B, with experimental conditions indicated.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig4-data2-v1.zip
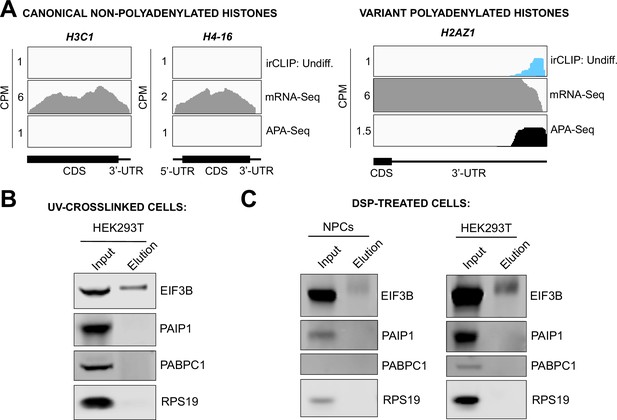
Mechanism of eukaryotic initiation factor 3 (eIF3) binding to mRNA 3’-UTR elements.
(A) Crosslinking of eIF3 to histone mRNAs in undifferentiated neural progenitor cells (NPCs). Shown are canonical non-polyadenylated histone H3C1 and H4-16 mRNAs and the variant polyadenylated histone H2AZ1 mRNA. (B) Western blots for EIF3B immunoprecipitation samples from UV-crosslinked HEK293T cells. (C) Western blots for EIF3B immunoprecipitation samples from dithiobis[succinimidylpropionate] (DSP)-treated undifferentiated NPCs and HEK293T.
-
Figure 4—figure supplement 1—source data 1
Original western blots in Figure 4—figure supplement 1C.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original western blots in Figure 4—figure supplement 1C, with cells and experimental conditions indicated.
- https://cdn.elifesciences.org/articles/102977/elife-102977-fig4-figsupp1-data2-v1.zip
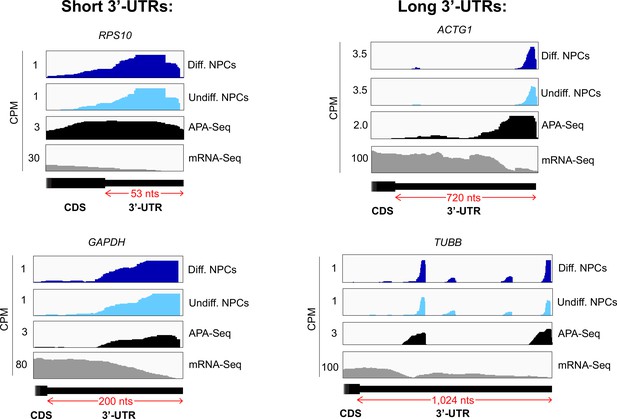
Crosslinking of eukaryotic initiation factor 3 (eIF3) to mRNA 3’-UTRs of various lengths in neural progenitor cells (NPCs).
Crosslinking patterns of eIF3 in differentiated and undifferentiated NPCs across RPS10, GAPDH, ACTG1, and TUBB mRNAs are shown. The length of each 3’-UTR is indicated. Polyadenylation sequencing (APA-seq) and mRNA-seq data are also shown.
Additional files
-
Supplementary file 1
Subunits of eukaryotic initiation factor 3 (eIF3) and proteins pulled down by anti-EIF3B IP.
The IP was performed using lysates from undifferentiated neural progenitor cells (NPCs). Given are protein names, sequence coverage, and number of peptides detected.
- https://cdn.elifesciences.org/articles/102977/elife-102977-supp1-v1.xlsx
-
Supplementary file 2
Annotation of clusters from Quick-irCLIP of eukaryotic initiation factor 3 (eIF3) to RNAs in undifferentiated neural progenitor cells (NPCs).
- https://cdn.elifesciences.org/articles/102977/elife-102977-supp2-v1.xlsx
-
Supplementary file 3
Annotation of clusters from Quick-irCLIP of eukaryotic initiation factor 3 (eIF3) to RNAs in differentiated neural progenitor cells (NPCs).
- https://cdn.elifesciences.org/articles/102977/elife-102977-supp3-v1.xlsx
-
Supplementary file 4
Ribosome profiling and translation efficiency in undifferentiated and differentiated neural progenitor cells (NPCs).
DESeq2 comparisons of ribosome protected fragments (rpf), mRNAs (rna), translation efficiency (te) are given for MANE transcripts.
- https://cdn.elifesciences.org/articles/102977/elife-102977-supp4-v1.xlsx
-
Supplementary file 5
Normalized mRNA counts from DESeq2.
Counts are given for all NPC replicates, using MANE transcripts.
- https://cdn.elifesciences.org/articles/102977/elife-102977-supp5-v1.xlsx
-
Supplementary file 6
Normalized monosome and disome ribosome protected fragments from DESeq2.
Counts are given for all neural progenitor cells (NPC) replicates, using MANE transcripts.
- https://cdn.elifesciences.org/articles/102977/elife-102977-supp6-v1.xlsx
-
Supplementary file 7
List of antibodies used in this study.
- https://cdn.elifesciences.org/articles/102977/elife-102977-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/102977/elife-102977-mdarchecklist1-v1.pdf





