Gut Microbes: Regulating uric acid
Improvements in the quality of life have led to an increase in the incidence of hyperuricemia, a medical condition that can lead to kidney stones and gout, with cases increasingly affecting younger individuals (Johnson et al., 2018; Zhang et al., 2019). Hyperuricemia – the presence of abnormally high levels of uric acid in the blood – arises from interactions between the liver, the kidneys and the gut, which has a role in removing uric acid from the body (Dalbeth et al., 2021; Niu et al., 2018; Yun et al., 2017). Studies indicate that gut microbes are crucial to uric acid metabolism, and interventions such as probiotics, prebiotics and fecal microbiota transplants can help reduce hyperuricemia by altering the gut microbiota (Cao et al., 2022a; Wang et al., 2022; Zhao et al., 2022).
It has been shown that various strains of bacteria can alleviate hyperuricemia through two mechanisms: the direct hydrolysis of uric acid, and the hydrolase-mediated degradation of nucleosides that are the precursors of uric acid in the intestine. Limosilactobacillus fermentum JL-3 – a strain isolated from Chinese mud water – is capable of the hydrolysis of uric acid (Wu et al., 2021), whereas various strains of Lactobacillus, a well-known genus of bacteria, reduce uric acid levels through the hydrolysis of nucleosides in the intestine: these strains include L. paracasei (X11; Cao et al., 2022b) and strains of L. plantarum derived from Chinese sauerkraut (DM9218-A; Li et al., 2014) and Chinese mustard (GKM3; Hsu et al., 2019).
Recent studies have revealed that gene cloning can be used to identify specific hydrolases involved in the degradation of nucleosides for L. plantarum and L. aviarius (Li et al., 2023b; Li et al., 2023a). However, the precise mechanisms underlying the hydrolysis of the nucleoside precursors of uric acid have remained unclear. Now, in eLife, Wence Wang (South China Agricultural University), Qiang Tu (Shandong University) and colleagues – including Yang Fu as first author – report the results of in vitro studies and experiments on geese and mice that shed new light on the hydrolysis of these precursors (Fu et al., 2024).
The team isolated a strain called L. plantarum SQ001 from geese with hyperuricemia, and a genome-wide analysis revealed the presence of four genes that code for nucleoside hydrolysis-related enzymes (iunH, yxjA, rihA, rihC). In vitro experiments revealed that one of these enzymes, iunH, effectively catalyzes the hydrolysis of nucleosides, such as inosine and guanosine, converting them to nucleobases, as evidenced by metabolomics analysis. The hydrolysis mechanism was further validated through experiments that involved knocking out the gene for iunH in L. plantarum SQ001, and expressing it in E. coli. Although nucleosides are hydrolyzed to produce nucleobases, the direct link between this process and the reduction of uric acid remains unclear, possibly due to the transport of nucleosides and nucleobases in the gut. It may be that the lower uptake of these substances reduces the synthesis and accumulation of uric acid.
The team validated the functionality of L. plantarum SQ001 by establishing models of hyperuricemia in both geese and mice (Figure 1), and showed that this particular strain significantly enhanced the abundance of Lactobacillus in the gut of the host, which alleviated the symptoms of hyperuricemia by reducing the synthesis of uric acid and increasing its excretion. The fact that hyperuricemia was alleviated in mice may help with efforts to develop new ways to treat hyperuricemia and gout in humans.
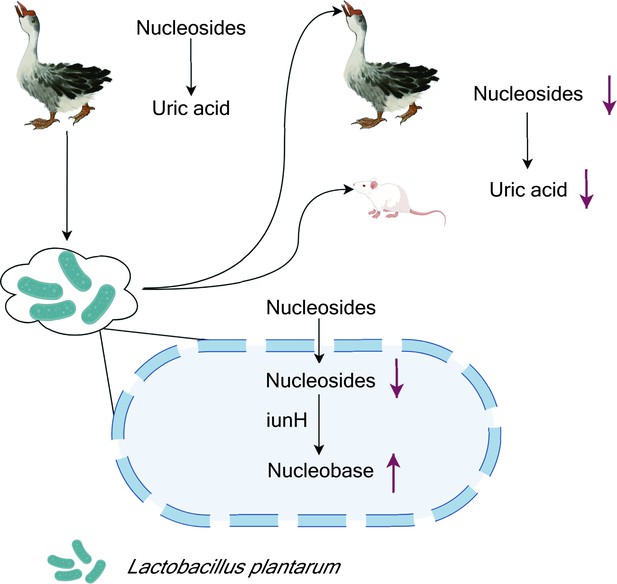
Lactobacillus plantarum reduces uric acid synthesis through the hydrolysis of nucleosides.
A strain of the bacterium L. plantarum was isolated from the large intestine of geese with hyperuricemia, a condition caused by the presence of abnormally high levels of uric acid in the blood (top left). In vitro experiments showed that the presence of the bacteria led to an increase in the degradation of nucleosides that are precursors of uric acid. Administering the bacteria to healthy geese and mice (top right) also led to a reduction in the levels of uric acid in the blood. Other experiments showed that L. plantarum absorbed the nucleosides, and that an enzyme called iunH broke down the nucleosides to produce nucleobases (bottom).
Figure created with figdraw.com.
References
-
Lactobacillus paracasei X11 ameliorates hyperuricemia and modulates gut microbiota in miceFrontiers in Immunology 13:940228.https://doi.org/10.3389/fimmu.2022.940228
-
Antiobesity and uric acid-lowering effect of Lactobacillus plantarum GKM3 in high-fat-diet-induced obese ratsJournal of the American College of Nutrition 38:623–632.https://doi.org/10.1080/07315724.2019.1571454
-
Inhibition of 3,5,2’,4’-tetrahydroxychalcone on production of uric acid in hypoxanthine-induced hyperuricemic miceBiological & Pharmaceutical Bulletin 41:99–105.https://doi.org/10.1248/bpb.b17-00655
-
The gut microbiota as a target to control hyperuricemia pathogenesis: Potential mechanisms and therapeutic strategiesCritical Reviews in Food Science and Nutrition 62:3979–3989.https://doi.org/10.1080/10408398.2021.1874287
-
Hyperuricemia and cardiovascular diseaseCurrent Pharmaceutical Design 25:700–709.https://doi.org/10.2174/1381612825666190408122557
-
The potential of probiotics in the amelioration of hyperuricemiaFood & Function 13:2394–2414.https://doi.org/10.1039/D1FO03206B
Article and author information
Author details
Publication history
Copyright
© 2024, Hu
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 643
- views
-
- 54
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Microbiology and Infectious Disease
Saprolegnia parasitica is one of the most virulent oomycete species in freshwater aquatic environments, causing severe saprolegniasis and leading to significant economic losses in the aquaculture industry. Thus far, the prevention and control of saprolegniasis face a shortage of medications. Linalool, a natural antibiotic alternative found in various essential oils, exhibits promising antimicrobial activity against a wide range of pathogens. In this study, the specific role of linalool in protecting S. parasitica infection at both in vitro and in vivo levels was investigated. Linalool showed multifaceted anti-oomycetes potential by both of antimicrobial efficacy and immunomodulatory efficacy. For in vitro test, linalool exhibited strong anti-oomycetes activity and mode of action included: (1) Linalool disrupted the cell membrane of the mycelium, causing the intracellular components leak out; (2) Linalool prohibited ribosome function, thereby inhibiting protein synthesis and ultimately affecting mycelium growth. Surprisingly, meanwhile we found the potential immune protective mechanism of linalool in the in vivo test: (1) Linalool enhanced the complement and coagulation system which in turn activated host immune defense and lysate S. parasitica cells; (2) Linalool promoted wound healing, tissue repair, and phagocytosis to cope with S. parasitica infection; (3) Linalool positively modulated the immune response by increasing the abundance of beneficial Actinobacteriota; (4) Linalool stimulated the production of inflammatory cytokines and chemokines to lyse S. parasitica cells. In all, our findings showed that linalool possessed multifaceted anti-oomycetes potential which would be a promising natural antibiotic alternative to cope with S. parasitica infection in the aquaculture industry.
-
- Genetics and Genomics
- Microbiology and Infectious Disease
Polyamines are biologically ubiquitous cations that bind to nucleic acids, ribosomes, and phospholipids and, thereby, modulate numerous processes, including surface motility in Escherichia coli. We characterized the metabolic pathways that contribute to polyamine-dependent control of surface motility in the commonly used strain W3110 and the transcriptome of a mutant lacking a putrescine synthetic pathway that was required for surface motility. Genetic analysis showed that surface motility required type 1 pili, the simultaneous presence of two independent putrescine anabolic pathways, and modulation by putrescine transport and catabolism. An immunological assay for FimA—the major pili subunit, reverse transcription quantitative PCR of fimA, and transmission electron microscopy confirmed that pili synthesis required putrescine. Comparative RNAseq analysis of a wild type and ΔspeB mutant which exhibits impaired pili synthesis showed that the latter had fewer transcripts for pili structural genes and for fimB which codes for the phase variation recombinase that orients the fim operon promoter in the ON phase, although loss of speB did not affect the promoter orientation. Results from the RNAseq analysis also suggested (a) changes in transcripts for several transcription factor genes that affect fim operon expression, (b) compensatory mechanisms for low putrescine which implies a putrescine homeostatic network, and (c) decreased transcripts of genes for oxidative energy metabolism and iron transport which a previous genetic analysis suggests may be sufficient to account for the pili defect in putrescine synthesis mutants. We conclude that pili synthesis requires putrescine and putrescine concentration is controlled by a complex homeostatic network that includes the genes of oxidative energy metabolism.






