Cryogenic Electron Microscopy: MagIC beads for scarce macromolecules
Cryogenic electron microscopy (or cryo-EM for short) has revolutionized our understanding of biology. The technique – which uses a beam of electrons fired at frozen samples to work out their structure – has revealed the three-dimensional arrangements of many large complexes that drive essential biological processes such as chromatin regulation (Nogales and Mahamid, 2024).
To prepare samples for this type of microscopy, high quality biological materials, such as protein complexes, undergo multiple stages of purification and concentration before being applied to a metal mesh grid in a thin layer. The grid is then rapidly frozen at cryogenic temperatures (Frank, 2017). High concentrations of the target macromolecules are needed to properly analyze the sample, yet most of these macromolecules are lost during the application to the grid, limiting the technique to samples with abundant molecules. To broaden the reach of cryo-EM – especially for complexes that are challenging to purify or concentrate – further advancements in simplifying the handling of a sample and minimizing the amounts of sample needed are essential.
Several strategies have been implemented to expand the scope of samples that can be used for cryo-EM. For example, analyzing samples directly after minimal purification has enabled researchers to characterize larger – albeit more abundant – assemblies, such as the proteasome, an enzyme complex consisting of multiple subunits (Kastritis et al., 2017; Verbeke et al., 2018; Ho et al., 2021). Editing a gene in one component of a macromolecular complex so that it contains the sequence for an affinity tag has further allowed researchers to isolate complexes with minimal processing. These tags can be captured directly from cell extracts using specific molecules or ‘handles’, allowing the tagged protein and the rest of the macromolecular complex to be separated more easily (Herbst et al., 2021).
Similarly, cryo-EM grids containing ‘handles’ such as Ni-NTA, streptavidin or the SpyTag/SpyCatcher system can attract the desired molecules to the grid, thereby lowering the required concentration of a sample (Wang et al., 2008; Kelly et al., 2008; Han et al., 2016; Wang et al., 2020). However, no general solution exists to date that can bypass completely the standard steps of sample purification and concentration.
Now, in eLife, Yasuhiro Arimura, Hide Konishi and Hironori Funabiki of The Rockefeller University report on the development of custom magnetic beads (known as MagIC-cryo-EM beads) to separate low concentration assemblies from a complex mixture and apply them directly to cryo-EM (Arimura et al., 2024). With this technique, the team were able to resolve the structure of a fundamental unit of chromatin (known as a chromatosome) formed inthe egg extract of the African clawed frog, despite this complex only being present at very dilute concentrations.
The chromatosome complex was purified from frog egg extracts using the MagIC-cryo-EM beads. The researchers then used a strong magnet to attract the magnetic beads to the cryo-EM grid membrane, which enabled them to generate sufficient cryo-EM data from a dilute sample. The MagIC-cryo-EM beads further included a protein spacer to distance the sample from the magnetic beads and to avoid interferences from the metallic core during imaging (Figure 1).
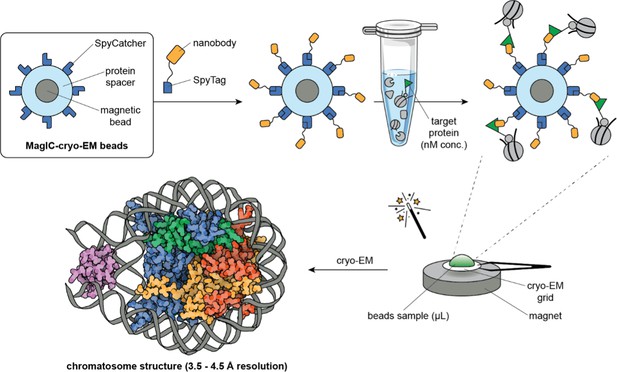
High-resolution structural imaging using customised magnetic beads.
Magnetic Isolation and Concentration-cryo-EM (MagIC-cryo-EM) beads (top left box) consist of a magnetic bead (gray circle) coated with a protein spacer (light blue ring), which has SpyCatcher proteins attached to it (dark blue shapes). To isolate target proteins from a sample, a SpyTag peptide attached to a nanobody (yellow shape) that binds specifically to the target protein tag (green triangle) is added to the MagIC-cryo-EM beads using SpyCatcher/SpyTag technology. This allows the beads to capture the target proteins – and any stable complexes they are a part of – directly from cell extracts. A magnet helps to pull the beads and their cargo onto cryogenic electron microscopy (cryo-EM) grids (bottom right), which are then imaged directly, bypassing the standard intermediate purification and concentration steps. Thanks to the protein spacer, the enriched complexes can be characterized accurately without interference from the magnetic beads. Arimura et al. used this technique to reveal the chromatosome structure of the African clawed frog (bottom left, H1: purple, H2A: yellow, H2B: red, H3: blue, H4: green, DNA: grey). Chromatosome model represents PDB 4QLC (Zhou et al., 2015). Microtube adapted from Bioicons.com (CC BY 4.0).
The MagIC-cryo-EM beads have a protein on their surface known as 'SpyCatcher', which irreversibly binds to its corresponding 'SpyTag' peptide. In the system designed by Arimura et al., the SpyTag peptide is bound to a nanobody that specifically binds to a tag in the complex of interest. This allowed the chromatosome complex within frog egg extracts to bind to the MagIC-cryo-EM beads.
Cryo-EM showed that the chromatosome contained fragments of DNA wrapped around a core of histone proteins (H2A, H2B, H3 and H4) that were further bound by the linker histone H1.8. The resulting high-resolution structures also highlighted that, contrary to a previous hypothesis, there were no positional differences of H1.8 on the chromatosome between the interphase and metaphase of a cell cycle.
Arimura et al. also devised a new protocol, called Duplicated Selection to Exclude Rubbish particles (or DuSTER for short), which helps identify useful cryo-EM signals and eliminate cell debris. This provided, for the first time, structural evidence that a group of proteins that support protein folding and are known as chaperones (in particular NPM2) were involved in binding the H1.8 dissociated from the DNA. The distinct conformational states during the different phases of the cell cycle could lead to potential changes in the binding affinities of H1.8 for NPM2.
The new cryo-EM protocol designed by Arimura et al. makes it possible to study macromolecular complexes extracted directly from cell extracts at low concentrations, eliminating the need for intermediate fractionation, purification or concentration steps. In addition, the modular design of the MagIC-cryo-EM beads allows the tool to be used with a range of nanobodies to target different tags or even native proteins. Together, these methods hold promise towards a broad application and constitute a valuable resource that will help to illuminate the nanoscopic organization of life.
References
-
Long shelf-life streptavidin support-films suitable for electron microscopy of biological macromoleculesJournal of Structural Biology 195:238–244.https://doi.org/10.1016/j.jsb.2016.06.009
-
Structure of the human SAGA coactivator complexNature Structural & Molecular Biology 28:989–996.https://doi.org/10.1038/s41594-021-00682-7
-
Capturing protein communities by structural proteomics in a thermophilic eukaryoteMolecular Systems Biology 13:936.https://doi.org/10.15252/msb.20167412
-
The Affinity Grid: a pre-fabricated EM grid for monolayer purificationJournal of Molecular Biology 382:423–433.https://doi.org/10.1016/j.jmb.2008.07.023
-
Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collectionJournal of Structural Biology 164:190–198.https://doi.org/10.1016/j.jsb.2008.07.008
Article and author information
Author details
Publication history
Copyright
© 2025, Moreno-Yruela and Fierz
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 622
- views
-
- 41
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Chromosomes and Gene Expression
- Evolutionary Biology
Repression of retrotransposition is crucial for the successful fitness of a mammalian organism. The domesticated transposon protein L1TD1, derived from LINE-1 (L1) ORF1p, is an RNA-binding protein that is expressed only in some cancers and early embryogenesis. In human embryonic stem cells, it is found to be essential for maintaining pluripotency. In cancer, L1TD1 expression is highly correlative with malignancy progression and as such considered a potential prognostic factor for tumors. However, its molecular role in cancer remains largely unknown. Our findings reveal that DNA hypomethylation induces the expression of L1TD1 in HAP1 human tumor cells. L1TD1 depletion significantly modulates both the proteome and transcriptome and thereby reduces cell viability. Notably, L1TD1 associates with L1 transcripts and interacts with L1 ORF1p protein, thereby facilitating L1 retrotransposition. Our data suggest that L1TD1 collaborates with its ancestral L1 ORF1p as an RNA chaperone, ensuring the efficient retrotransposition of L1 retrotransposons, rather than directly impacting the abundance of L1TD1 targets. In this way, L1TD1 might have an important role not only during early development but also in tumorigenesis.
-
- Chromosomes and Gene Expression
RNA interference (RNAi) is a conserved pathway that utilizes Argonaute proteins and their associated small RNAs to exert gene regulatory function on complementary transcripts. While the majority of germline-expressed RNAi proteins reside in perinuclear germ granules, it is unknown whether and how RNAi pathways are spatially organized in other cell types. Here, we find that the small RNA biogenesis machinery is spatially and temporally organized during Caenorhabditis elegans embryogenesis. Specifically, the RNAi factor, SIMR-1, forms visible concentrates during mid-embryogenesis that contain an RNA-dependent RNA polymerase, a poly-UG polymerase, and the unloaded nuclear Argonaute protein, NRDE-3. Curiously, coincident with the appearance of the SIMR granules, the small RNAs bound to NRDE-3 switch from predominantly CSR-class 22G-RNAs to ERGO-dependent 22G-RNAs. NRDE-3 binds ERGO-dependent 22G-RNAs in the somatic cells of larvae and adults to silence ERGO-target genes; here we further demonstrate that NRDE-3-bound, CSR-class 22G-RNAs repress transcription in oocytes. Thus, our study defines two separable roles for NRDE-3, targeting germline-expressed genes during oogenesis to promote global transcriptional repression, and switching during embryogenesis to repress recently duplicated genes and retrotransposons in somatic cells, highlighting the plasticity of Argonaute proteins and the need for more precise temporal characterization of Argonaute-small RNA interactions.






