The structure of a LAIR1-containing human antibody reveals a novel mechanism of antigen recognition
Figures
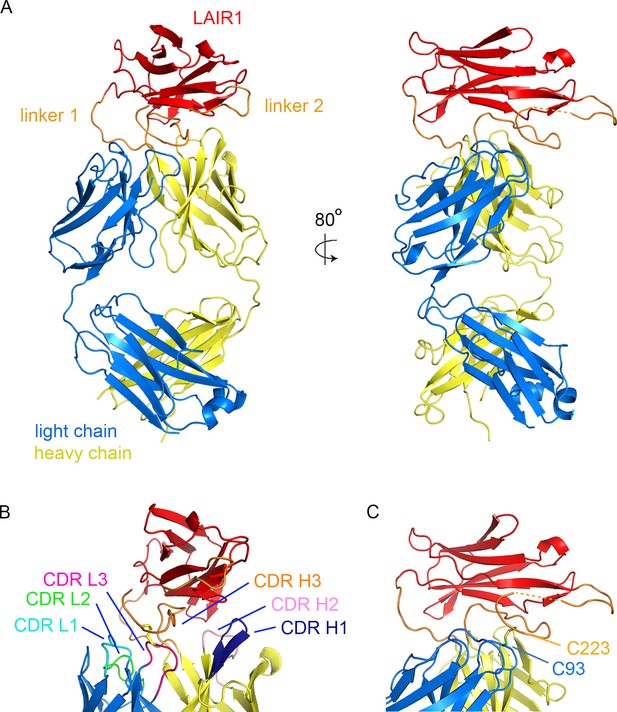
Structure of a LAIR1-containing antibody Fab fragment.
(A) The structure of the Fab fragment. LAIR1 (red) is inserted into the third CDR loop of the heavy chain (yellow) through two extended linkers (orange). The light chain is blue. The dashed orange link represents protein disordered in the structure. (B) The organization of the CDRs. The three CDR loops of the light chain and remaining two CDR loops of the heavy chain directly contact the LAIR1 insert or the linkers. Each of the CDR loops and its corresponding label is a shown in a different colour. (C) A disulphide bond between C93 of the light chain and C223 of the heavy chain stabilizes the interface (cysteine residues are shown as sticks).
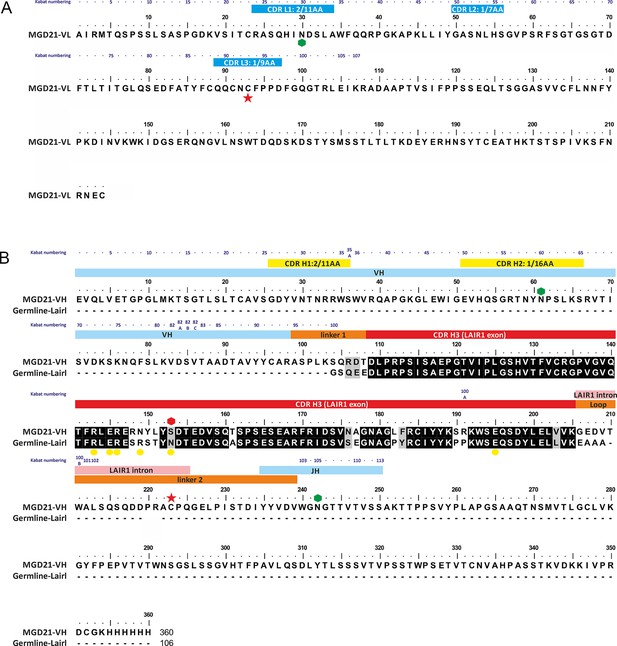
Annotated sequence of antibody MGD21 and its alignment to germ line LAIR1.
(A) The sequence of the light chain of MGD21 with the CDR loops indicated by blue bars. (B) The sequence of the heavy chain of MGD21 aligned to that of germ line LAIR1. Hexagons represent putative glycosyation sites with the red hexagon representing a site lost in MGD21. Yellow circles mark sites in germ line LAIR1 known to affect collagen binding. Red stars represent cysteine residues that make a cross-chain disulphide bond. Residues in the heavy chain are labeled according to whether they derive from the V, J or LAIR1 genes. In both cases, Kabat numbers (Dunbar and Deane, 2016) of residues are given above the sequence. In addition, all the CDRs, with the exception of the third CDR of the heavy chain, are labeled with their canonical class (Martin and Thornton, 1996).
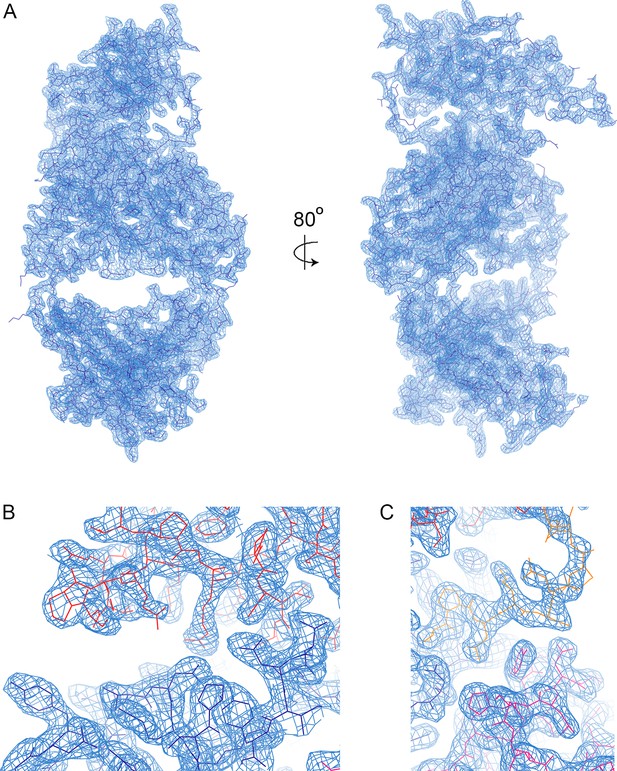
Electron density.
(A) A view of the electron density, showing the 2Fo-Fc map in blue, contoured at 1.0σ. (B) A view of the electron density in the region in which the CDR loops of the heavy chain (dark blue) contact the LAIR1 insert (red). (C) A view of the electron density where the CDR loops of the light chain (magenta) contact linker 2.
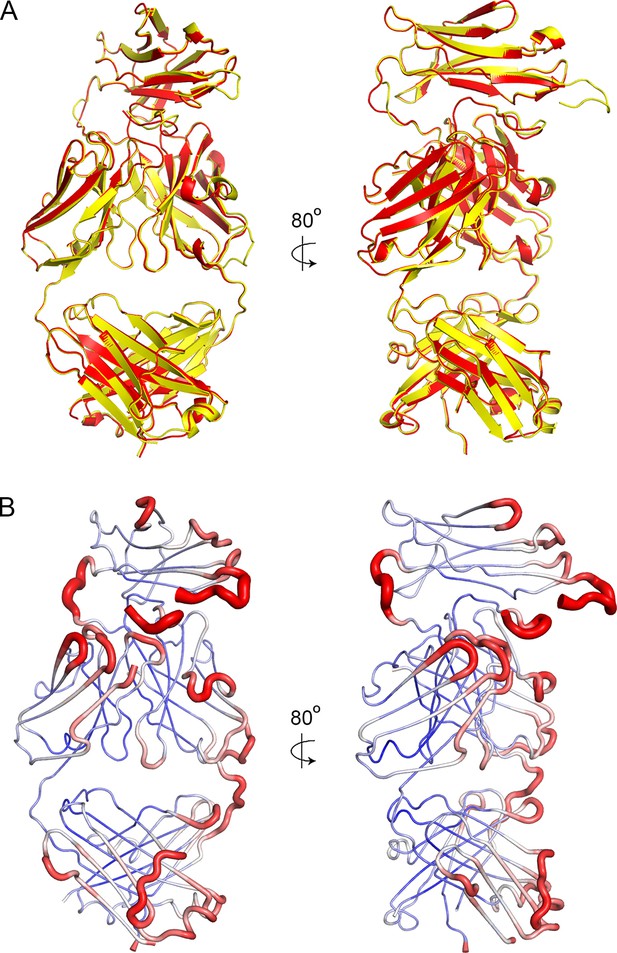
Crystal packing and order.
(A) A superimposition of the two antibody molecules in the asymmetric unit, with chains A and B in yellow and C and D in red. (B) The structure of chains A and B shown in putty representation with putty thickness determined by B factor. The colour scale for blue to red is from B factors of 30 to 100.
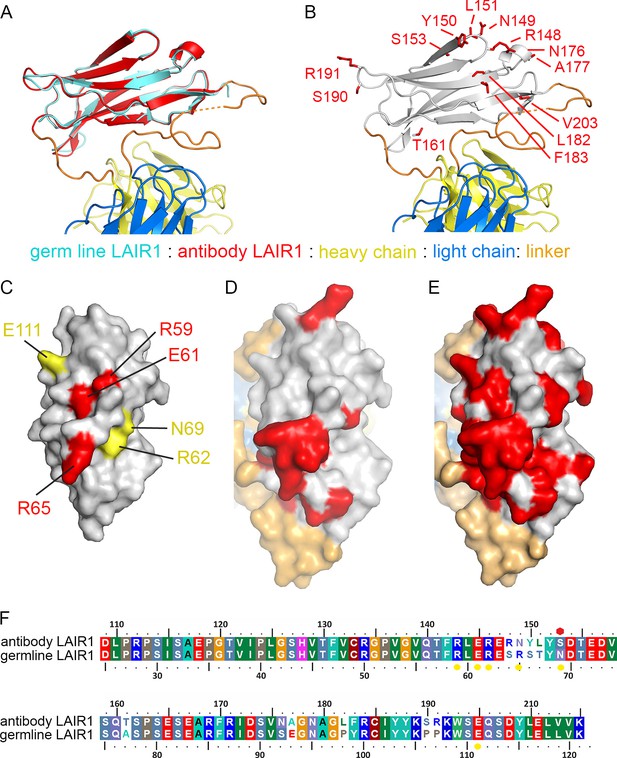
Structure and polymorphism in the LAIR1 insertion.
(A) An alignment of germ line LAIR1 (cyan) with the antibody LAIR1 insertion (red). (B) The residues that differ between the LAIR1 insertion in antibody MGD21 and germ line LAIR1 are shown as red sticks. (C) A surface representation of the structure of LAIR1 (grey) with residues whose mutation has a major (red) or minor (yellow) effect on collagen binding highlighted (Brondijk et al., 2010). (D) A surface view of the LAIR1 insert in antibody MGD21 (grey) with residues that differ from germ line LAIR1 highlighted (red). (E) A surface view of the LAIR1 insert (grey) with residues that differ from germ line LAIR1 in all 27 antibodies tested to date (Tan et al., 2016) highlighted (red). (F) A sequence alignment of germ line LAIR1 and the LAIR1 insert in the MGD21 antibody. Yellow circles are sites residues shown to play a role in collagen binding while a red hexagon represents a potential N-linked glycosylation site mutated in the LAIR1 insert.
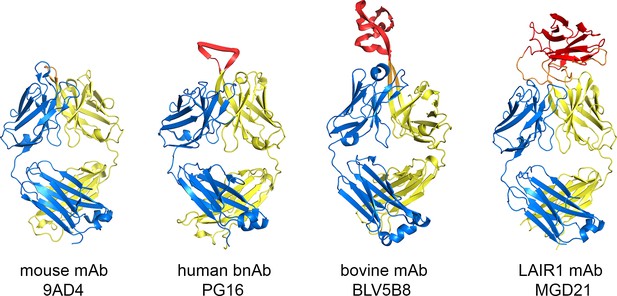
Comparison of the LAIR1-containing antibody with other unusual antibodies.
The structure of the LAIR1-containing monoclonal antibody is compared with a classical mouse monoclonal antibody (9AD4; PDB code 4U0R), a human monoclonal antibody with broadly neutralizing potential against HIV (PG16; PDB code 4DQ0) and a bovine monoclonal antibody (BLV5B8; PDB code 4K3E). In each case, the light chain is blue and the two immunoglobulin domains of the heavy chain are yellow. Inserted domains are shown in red with linker regions in orange.
Tables
Data collection and refinement statistics. The structure was determined from a single crystal. Values in parentheses are for highest-resolution shell. Rfree was determined using 1968 reflections (4.8%) The structure is deposited with pdb code 5NST.
| Fab-MGD21 | |
|---|---|
| Data collection | |
| Space group | C121 |
| Cell dimensions | |
| a, b, c (Å) | 169.8, 86.5, 104.0 |
| α β γ (°) | 90.0, 126.7, 90.0 |
| Wavelength | 0.92819 |
| Resolution (Å) | 81.90–2.52 (2.56–2.52) |
| Total Observations | 131833 (5451) |
| Total Unique | 40946 (2031) |
| Rpim (%) | 5.4 (67.8) |
| Rmerge (%) | 8.3 (88.5) |
| Rmeas (%) | 9.9 (112.1) |
| CC1/2 | 0.992 (0.571) |
| I/σ(I) | 7.4 (1.0) |
| Completeness (%) | 99.8 (98.3) |
| Multiplicity | 3.2 (2.7) |
| Wilson B factor | 55.216 |
| Refinement | |
| Number of reflections | 40946 |
| Rwork / Rfree | 21.9/26.7 |
| Number of residues | |
| Protein | 1076 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.25 |
| All Atom clash score | 5 |
| B factors | |
| All atoms | 71.53 |
| Solvent | 63.12 |
| Variable domains | 65.17 |
| Constant domains | 74.29 |
| LAIR1 insert | 73.70 |
| Linkers | 94.71 |
| Ramachandran plot | |
| Favored (%) | 95.2% |
| Allowed (%) | 4.8% |
| Disallowed (%) | 0.0% |
A list of interactions between the LAIR1 insert and linkers that occupies the heavy chain CDR3 loop and the other five CDR loops of the antibody.
| CDR loop | Residue | Group | LAIR1 region | Residue | Group | Interaction |
|---|---|---|---|---|---|---|
| Light chain CDR1 | Q27 | Side chain | Linker 2 | A222 | Main Chain | Hydrogen Bond |
| Light chain CDR2 | Y49 | Side chain | Linker 1 | L102 | Side chain | Hydrophobic Packing |
| Light chain CDR2 | N53 | Side chain | Linker 1 | S104 | Side chain | Hydrogen Bond |
| Light chain CDR3 | C93 | Side chain | Linker 2 | C223 | Side chain | Disulphide Bond |
| Light chain CDR3 | F94 | Main Chain | Linker 2 | E227 | Side Chain | Hydrogen Bond |
| Heavy chain CDR1 | N32 | Side chain | LAIR1 | R134 | Side chain | Hydrogen Bond |
| Heavy chain CDR2 | R57 | Side Chain | LAIR1 | P109 | Main Chain | Hydrogen Bond |





