Philosophy of Biology: The analysis of living systems can generate both knowledge and illusions
Abstract
Life relies on phenomena that range from changes in molecules that occur within nanoseconds to changes in populations that occur over millions of years. Researchers have developed a vast range of experimental techniques to analyze living systems, but a given technique usually only works over a limited range of length or time scales. Therefore, gaining a full understanding of a living system usually requires the integration of information obtained at multiple different scales by two or more techniques. This approach has undoubtedly led to a much better understanding of living systems but, equally, the staggering complexity of these systems, the sophistication and limitations of the techniques available in modern biology, and the need to use two or more techniques, can lead to persistent illusions of knowledge. Here, in an effort to make better use of the experimental techniques we have at our disposal, I propose a broad classification of techniques into six complementary approaches: perturbation, visualization, substitution, characterization, reconstitution, and simulation. Such a taxonomy might also help increase the reproducibility of inferences and improve peer review.
Introduction
Scientific knowledge relies on discoveries that continually extend, modify, or negate prior discoveries, making any inference from any study provisional. All practicing scientists intuitively appreciate this fact. Yet, collective blindness to alternative inferences could promote the acceptance of knowledge that later turns out to be unreliable. Such illusions are easily recognized in hindsight, but are difficult to anticipate. At an extreme, these illusions can provide a false sense of security about the foundations of a discipline, and also obscure the boundaries between the known and the unknown.
The pace of discovery and the array of techniques used in modern science make it difficult to evaluate new findings and place them in context. Modern biology is a good example: all biologists want to understand living systems, be they cells or populations or something in between, but the complexity of living systems and the sophistication of modern scientific enquiry can make it challenging to see the frontiers of the subject. Is it even possible to evaluate progress towards the overarching goals of any area of biology? A common framework for thinking about a wide range of experiments in biology could help us assess discoveries and their potential impact.
Six approaches for analyzing living systems on multiple scales
One feature that many experimental techniques have in common, no matter how powerful they are, is that they are only useful over a limited range of length and time scales. Yet, the integration of information across different length and time scales is necessary to understand living systems because the outcome of events that occur at the level of molecules within nanoseconds and that unfold in populations over millions of years can depend on each other. For example, a mutation in DNA that disrupts molecular interactions could alter the cell, the tissue, the organism, and even populations. Conversely, mechanisms that have been honed over evolutionary time in populations can repair the mutation in DNA or compensate for its consequence.
Here I argue that it is possible to classify the many different experimental techniques used in biology into six broad approaches (Figure 1): perturbation, visualization, substitution, characterization, reconstitution, and simulation. Each approach is applicable across all scales, ranging from molecules within an organism to populations of organisms within an ecosystem, which means that this framework can be used to evaluate any experiment and place it within the wider array of possible experiments. The six approaches are discussed in greater detail below, with an emphasis on analysis at the molecular scale, along with some examples that illustrate application to other scales. Possible uses of this classification for improved inference and peer review are also discussed.
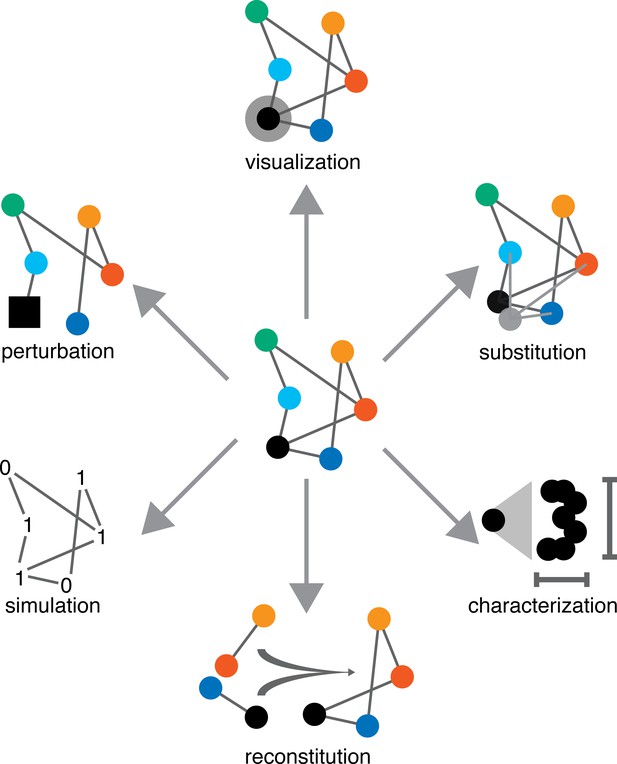
Six approaches for the analysis of living systems.
In this schematic diagram, a living system is represented as an abstract network (center), with the colored nodes (circles) representing the different parts of the system, and the grey edges representing the interactions between the parts. Four of the six approaches described in this article involve doing something to one part of the system (shown here in black); the fifth approach involves combining multiple parts; and the sixth approach involves simulating some or all of the parts.
1. Perturbation
This approach involves changing something in a system and then observing it. What happens after a perturbation can depend not only on the part of the system being perturbed but also on how this part interacts with the rest of the system. Simplistic inference based on the presumed change in the part alone can be misleading. For example, loss of the pha-1 gene in the worm C. elegans results in complete loss of the pharynx (throat) because all the cells of the pharynx fail to undergo morphogenesis and terminal differentiation (Schnabel and Schnabel, 1990). The initial simple model was that the PHA-1 protein is a 'master regulator' of pharynx differentiation. But subsequent analyses revealed that it is an antidote to a maternal toxin (Ben-David et al., 2017). Take away both the maternal toxin and PHA-1, and a pharynx is made just fine. Perturbation can be permanent or transient and can disrupt gene sequence (genetics), gene regulators (epigenetics), or the environment ('environomics'). In every case, the impact of the change depends on the changed part and the rest of the interacting system.
Prior conceptions about the ability of living systems to accommodate change can influence selection of perturbed systems for further analysis or indeed recognition that a perturbation has occurred. When performing grafting experiments on embryos to discover regions that cause changes when moved (Mangold and Spemann, 1924; see Spemann and Mangold, 2001 for a translation), interpreting the results requires the idea that moving a piece of embryonic tissue could perturb patterning. Such perturbation is not guaranteed because of processes that can recover anatomy and function. For example, the entire organism can be generated from half the number of cells during regulative development (Driesch, 1891; see Willier and Oppenheimer, 1964 for a translation), new cells can be made to regain pattern during regeneration (Browne, 1909), and cells can move to regain pattern during reorganization (Abrams et al., 2015). When using mutagens or chemicals to induce a particular change, unanticipated compensatory mechanisms can confound inference (El-Brolosy and Stainier, 2017). A recent case that highlights this problem is the disagreement between morpholino-based perturbations and Cas-9-mediated genome editing in Zebrafish (Kok et al., 2015), some of which is currently thought to be explained by transcriptional adaptation in response to mutations (Ma et al., 2019; El-Brolosy et al., 2019). Compensation of any kind can blunt the impact of perturbations.
2. Visualization
Describing what can be seen in one or more parts of a system is often a first step towards understanding systems at any scale. Yet, limitations on what can be seen complicate inference. For example, describing an ecosystem requires accurate counts of organisms, which can be difficult to achieve and can distort deduced trophic relationships (reviewed in Elphick, 2008). Techniques for observing animal behavior without perturbation needed to be developed to reveal increases in the nocturnal activity of land animals (Gaynor et al., 2018) and light-based communication in deep-sea organisms (Burford and Robison, 2020). The advance of methods for seeing progressively smaller tumors illustrate how functionally important anatomical features of an organism could remain invisible until methods improve.
At the molecular scale, all visualization requires specialized instruments and techniques. The part to be seen can be either directly tagged with a label or indirectly through physical interactions with a label. Inference using these approaches requires the label to be similarly detectable in all parts of the system and the labeled parts to be used by the system in the same way as the unlabeled parts. A common tagging approach is the fusion of fluorescent protein or fluorescent RNA to a protein or RNA of interest (Rodriguez et al., 2017; Truong and Ferré‐D'Amaré, 2019). However, because different cellular compartments can distort the signal from such fluorescent tags, specific optimization is needed for reliably detecting fluorescence in parts of a cell that differ in characteristics such as pH or oxidation state (Costantini et al., 2015). In addition, the fluorescent tag needs to be added to particular places on the part being visualized to avoid interfering with its functions. For example, the Gα proteins of heterotrimeric G proteins interact with receptors, the beta-gamma subunits, regulators of G-protein signaling, and effectors, which leaves only a few places where a Gα protein can be tagged without interfering with one of its functions (Hughes et al., 2001). Both how the system impacts the approach and how the approach impacts the system matter.
3. Substitution
This approach involves adding measurable proxies for parts of a system to infer how those parts are used by the system. For inference based on this approach to be successful, the proxy should not interfere with any aspect of the system while, at the same time, interacting with the system in the same way as the part, effectively substituting for the part in regulatory interactions. For example, one species can be used as a sentinel to report on the health of the ecosystem under the assumption that impacts on the sentinel species reflects impact on the ecosystem (Popkin, 2020). An inert tracer can be used as a proxy for other particles to understand how they move through a circulatory system (Errico et al., 2015). A reporter gene with the regulatory surroundings of a protein-coding gene can produce a visible proxy that can reveal how the system regulates the gene (Chalfie et al., 1994). In every case, if the proxy interferes with any aspect of the living system, a mix of perturbation and substitution has occurred, thus complicating inference.
Creating perfect proxies is not trivial and unexpected behaviors of proxies have revealed new aspects of how living systems function. For example, attempts to overexpress a gene for making a pigment in plants using many copies of the gene led instead to the silencing of the same gene expressed from the genome of the plant (Napoli et al., 1990; van der Krol et al., 1990) and the eventual discovery of RNA interference (Fire et al., 1998) – a potent method of silencing genes. Now we can assemble reporter genes with surgical precision using genome-editing approaches such as CRISPR (Knott and Doudna, 2018). Yet, permanent silencing of an apparently perfectly assembled proxy can occur through germline small RNAs in C. elegans for reasons that are not yet clear (reviewed in Almeida et al., 2019). Similarly, unexpected behaviors of proxies for parts involved in protein sorting, RNA splicing, or any other process of interest have the potential to reveal new aspects of the system rather than reliably reflecting the behavior of the parts. Sometimes a failed proxy teaches a new way to perturb the system.
4. Characterization
The purpose of this approach is to infer the properties of parts of the system. Organisms can be characterized as predator or prey based on their dietary habits to understand their influence on an ecosystem. However, some features of organisms can be less obvious and their eventual detection could warrant revisions of our understanding of the ecosystem (such as the recent detection of widespread biofluorescence in amphibians; Lamb and Davis, 2020). While the in vitro growth of organs (such as the optic cup of the eye; Eiraku et al., 2011) and cells (since the time of Harrison, 1906) can facilitate their detailed characterization, crucial aspects of their functions in vivo could be changed in vitro (for example, see Bissell and Labarge, 2005) for a discussion of the profound influence of in vivo microenvironments).
Similar considerations also plague methods for analyzing the properties of molecules. The shape of a protein can be inferred by solving its structure and the sequence of a genome can be inferred by interrogating its bases. These approaches typically require isolation of the parts or subsets of interacting parts and therefore do not reveal how the characterized parts interact with the rest of the system or whether aspects characterized in isolation are relevant for the system. For example, X-ray crystallography cannot be used to deduce the structures of regions of molecules that do not take on a rigid conformation (Acharya and Lloyd, 2005). Sometimes a characterization can turn out to be incomplete or wrong after a long period of time. For example, it took ~17 years to discover that a technique for determining if a cytosine on DNA is methylated (Frommer et al., 1992) does not discriminate 5-methyl cytosine from 5-hydroxymethyl cytosine (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). What we thought was one kind of modified base on DNA turned out to be an unknown mixture of two kinds of modified bases.
5. Reconstitution
This approach involves recovering particular properties of the system or subsystem by combining parts. Reconstitution can be performed at many different scales – from individual molecules being combined to recreate properties exhibited by collections of molecules, through cells being combined to recreate properties of organs or developing organisms, to organisms being combined to capture aspects of ecosystems. Recovery of a particular property of the system or subsystem by combining parts can provide compelling evidence for how the parts can function in the system. For example, the direct observation of F1-ATPase proteins rotating attached glowing actin filaments provided vivid support for a model for ATP production whereby the protein acts as a rotary motor (Noji et al., 1997). Interacting regulators can be combined to recreate behaviors in vitro, as in the case of the oscillations in phosphorylation recreated using three proteins and ATP (Rust et al., 2011). Similar assembly within cells can inform plausible regulatory logic of oscillatory gene expression (Elowitz and Leibler, 2000) – potentially a reconstitution of defined components (regulators) within poorly defined environments (cells).
Parts that are themselves living systems – such as cells – can be combined into tissues that develop into complex organs like the human brain when induced in vitro (Lancaster et al., 2013), highlighting the ability of cells to form highly structured communities. Likewise, the formation of human embryos from sperm and egg in vitro (Edwards et al., 1969) establishes the sufficiency of these two cell types. On a larger scale, multiple organisms can be grown together to recreate symbioses (such as Vibrio fischeri and its squid host Euprymna scolopes; McFall-Ngai and Ruby, 1991). However, in every case differences between the reconstituted system and the living system could persist either because of missing parts or erroneous arrangement of the parts. The progressive addition of such missing factors can recover increasingly sophisticated properties of the system (see Ganzinger and Schwille, 2019) for a review of this progression in understanding the cytoskeleton). Each missing property in a reconstitution experiment drives the discovery of additional factors to add towards the distant goal of reconstituting entire living systems of varying complexity from molecules.
6. Simulation
This approach is used to represent or abstract features of the system for analysis. Examples of simulations include: sketching ideas on a sheet of paper (such as Darwin's 1837 sketch of the branching diagram of descent with modification; Barrett, 1959); building models of molecular structures (such as the metal model of DNA; Crick and Watson, 1954); analyzing a real problem in abstract systems (such as the exploration of self-replication in cellular automata; reviewed in Sipper, 1998); using computers to make quantitative or qualitative predictions of observed phenomena (such as computational models of cellular and molecular behaviors; reviewed in Mogilner et al., 2006). All simulations include some features and omit others.
Computer-aided simulations can explore the possible behaviors of systems under conditions that are experimentally unattainable using current technology to guide future experiments. Such calculations can span from sub-second molecular dynamics to multi-year experimental evolution. For example, the simultaneous behavior of multiple molecular complexes that regulate gene expression can be examined to test the validity of models aggregated from analyses on each complex. Such 'whole-cell' models can predict cell behavior despite being incomplete (Karr et al., 2012) and can be used to guide the experimental analysis of cells (Sanghvi et al., 2013). Further efforts in this direction hope to connect molecular detail to cell behavior, but require tremendous computational resources (see review by Feig and Sugita, 2019). At the other end of the scale, artificial 'organisms' that follow certain rules of evolution can be simulated in silico to gain intuition about the ways in which complexity could evolve (Lenski et al., 2003). However, recent surprises (Phillips et al., 2019) in the workings of the lac operon of E. coli, which has been the subject of more than half a century of experimentation and modeling (Vilar et al., 2003), underscore the need for iterative refinements of all in silico models for processes in living systems.
Potential applications
This brief overview provides a glimpse into how the modern biologist seeks tentative conclusions using multiple powerful but imperfect approaches. Appreciation of the strengths and weaknesses of each approach could improve the reproducibility of inferences presented in a study, and could also increase fairness during peer review.
Towards reproducible inference
Progress in science results from both exploratory and confirmatory analyses (Tukey, 1980), but exploratory analyses are often not reported (or are misleadingly reported as being confirmatory). Some advocate their disclosure as supplements to the main 'story' of the paper (Thompson et al., 2020) and others advocate obfuscating them if needed in service of improved story telling (Sanes, 2019). These opposing opinions about how science should be communicated are also relevant for establishing the desired reproducibility and rigor in science because problems can occur at the level of methods, results, or inferences (Goodman et al., 2016). Ways to improve the reproducibility of methods and results are being widely implemented, such as the inclusion of more detailed descriptions of experimental methods used and the statistical analyses performed. For inferences to be reproducible, qualitatively similar conclusions have to be drawn when a study is replicated or re-analysed (Goodman et al., 2016). However, it is not clear how the reproducibility of inferences can be improved, although the need for improvement is underscored by the results of a recent study which found that different teams of researchers drew different conclusions from the same neuroimaging dataset (Botvinik-Nezer et al., 2020).
Published inferences can be preceded by continuous interplay of exploratory and confirmatory research. As a result, the number of possible inferences explored in a study is typically not carefully documented. Furthermore, there can always be unimagined additional inferences of varying probabilities. This difficulty in deciding 'N' for plausible inferences makes it impossible for a practicing scientist to know what 'statistical test' they could use to communicate the reasonableness of their collective inferences. Furthermore, language could exacerbate this difficulty because persuasive story telling can make it difficult to spot gaps in logic. Although critical linguistic analysis has been proposed as a way to ameliorate this problem (see Segal, 1993; Horton, 1995) and the ensuing discussions), it has not been widely implemented. In short, the expected reproducibility of inferences is difficult to quantify because of the way science is done and is obscured because of the way science is communicated.
Inferences that are drawn from a study are influenced by prevailing paradigms. Erroneous paradigms can drive the accumulation of wrong inferences for long periods, as famously occurred with the Ptolemaic model of our solar system. Such episodes illustrate how knowledge derived from perfectly reproducible observations can nevertheless be illusory. The scientific community could collectively decide that such irreproducible inferences are part of progress – each one a potential impetus for a paradigm shift (Kuhn, 1962) to be eventually realized by some discerning scientist(s). Alternatively, we could explore ways to reduce the time and resources spent taking such missteps. In either case, having an explicit overview of the basis for the overall inferences and conclusions drawn in a study could clarify the next experiments and stimulate exploration of alternative inferences.
Currently, inferences are discussed in the introduction and discussion sections of papers: however, it would be much better if papers included explicit statements about the rationales for the inferences drawn by the authors (as is currently done for experimental methods and statistical analyses). This could be done in a relatively straightforward manner by adding two elements to the methods section of a paper: i) an explicit statement of the prevailing paradigm or prior model as understood by the authors; ii) a summary of the basis for any proposed modification of the model in plain language (for example, 'Inferences for the proposed model were drawn based on a perturbation approach using two techniques with distinct caveats and a visualization approach using one technique'). Finally, a common taxonomy of approaches with acknowledged limitations, refined through periodic debates, could help outsiders develop their own critical overviews of progress in a field and help the next generation of scientists decide on their future research directions.
Towards fair peer review
Authors and reviewers debate about methods, results, and inferences during peer review. In such debates, the approaches outlined in this article could provide a common context for articulating strengths and weaknesses. Such structured evaluation could benefit both peer review of a particular study and review articles that appraise a field to identify areas of future development. Since all doubt about the conclusions of any study cannot be eliminated, a commonly agreed upon level of support for claims is needed. For example, it would be unreasonable to expect any study to include experiments from all six approaches. Moreover, since a study that uses a single experimental approach well could be better than a study that uses two or more approaches poorly, statements about quantity and quality need to be developed. Such explicit statements about the acceptable level of doubt for publication in a particular journal, or for receiving a grant from a particular funding agency, could guide reviewers and authors through the process of peer review.
Peer review by journals or funding agencies also involves estimating the impact of the study. Since the ultimate value of any study is established through follow up work by the community at large, any estimate of potential impact made during peer review is at best an educated guess influenced by prevailing paradigms (Park et al., 2014; Siler et al., 2015). Furthermore, despite agreeing upon a common level of support for claims, there can be disagreements because of subjective interpretations (for example, see Keeling et al., 2019 for a discussion of the different meanings of the word 'function' in biology). This subjectivity is underscored by studies on grant review that document low agreement when different panels judge the same proposal (Pier et al., 2018). Given all this, being more specific about the paradigms involved in a paper or grant application – both in the paper or application itself, and during the review and assessment of the paper or application – should lead to better decisions being made.
Experiments are underway on the process of peer review (Rennie, 2016; Rodgers, 2017; Polka et al., 2018), with a number of journals opting to publish all exchanges between authors and reviewers to improve transparency (see, for example, Nature, 2020). These exchanges are typically between experts and can be more abstruse than the paper being reviewed, making it difficult for the interested reader to decipher. One solution is to provide a summary of the discussion as is already done by some funding agencies. The editor of a manuscript could guide readers by summarizing the major points of debate during its review: ideally, this summary would be written in plain language (e.g., 'The need for a visualization approach, caveats of two experimental techniques, and plausibility of alternative inferences were debated'). eLife recently took a step in this direction with the introduction of 'acceptance explanations'. The exchanges that take place between authors and reviewers during peer review provide a genuine opportunity for readers to learn about the process of science.
Conclusion
Biologists are engaged in understanding living systems through experimental and theoretical efforts in many subdisciplines. Placing new advances in the context of overall progress in biology can be challenging because of the dazzling variety and sophistication of techniques used. The six complementary approaches presented here provide a scaffold for classifying techniques and contextualizing discoveries. Appreciating the scope and limitations of each approach could improve how inferences are drawn and how studies are reviewed. Together, better inference and better review could promote the realistic appraisal of discoveries and limit persistent illusions of knowledge.
Data availability
No data were generated for this work.
References
-
The advantages and limitations of protein crystal structuresTrends in Pharmacological Sciences 26:10–14.https://doi.org/10.1016/j.tips.2004.10.011
-
BookA transcription of Darwin's first notebook [B] on 'Transmutation of species'In: Barrett P. H, editors. Bulletin of the Museum of Comparative Zoology, 122. Michigan State University. pp. 245–296.
-
The production of new hydranths in Hydra by the insertion of small graftsJournal of Experimental Zoology 7:1–23.https://doi.org/10.1002/jez.1400070102
-
A palette of fluorescent proteins optimized for diverse cellular environmentsNature Communications 6:7670.https://doi.org/10.1038/ncomms8670
-
The complementary structure of deoxyribonucleic acidProceedings of the Royal Society A 223:80–97.https://doi.org/10.1098/rspa.1954.0101
-
Entwicklungsmechanische studien I&IIZeitschrift Für Wissenschaftliche Zoologie 53:160–184.
-
How you count counts: the importance of methods research in applied ecologyJournal of Applied Ecology 45:1313–1320.https://doi.org/10.1111/j.1365-2664.2008.01545.x
-
Whole-cell models and simulations in molecular detailAnnual Review of Cell and Developmental Biology 35:191–211.https://doi.org/10.1146/annurev-cellbio-100617-062542
-
More from less – bottom-up reconstitution of cell biologyJournal of Cell Science 132:jcs227488.https://doi.org/10.1242/jcs.227488
-
What does research reproducibility mean?Science Translational Medicine 8:341ps12.https://doi.org/10.1126/scitranslmed.aaf5027
-
Observations on the living developing nerve fiberExperimental Biology and Medicine 4:140–143.https://doi.org/10.3181/00379727-4-98
-
Visualization of the functional Gαq-green fluorescent protein fusion in living cellsJournal of Biological Chemistry 276:4227–4235.https://doi.org/10.1074/jbc.M007608200
-
BookThe Structure of Scientific RevolutionsChicago: University of Chicago Press.https://doi.org/10.5897/PPR2013.0102
-
Über induktion von embryonalanlagen durch implantation artfremder organisatorenArchiv Für Mikroskopische Anatomie Und Entwicklungsmechanik 100:599–638.https://doi.org/10.1007/BF02108133
-
Quantitative modeling in cell biology: what is it good for?Developmental Cell 11:279–287.https://doi.org/10.1016/j.devcel.2006.08.004
-
The growing and glowing toolbox of fluorescent and photoactive proteinsTrends in Biochemical Sciences 42:111–129.https://doi.org/10.1016/j.tibs.2016.09.010
-
Accelerated discovery via a whole-cell modelNature Methods 10:1192–1195.https://doi.org/10.1038/nmeth.2724
-
Strategies of influence in medical authorshipSocial Science & Medicine 37:521–530.https://doi.org/10.1016/0277-9536(93)90287-E
-
Fifty years of research on self-replication: an overviewArtificial Life 4:237–257.https://doi.org/10.1162/106454698568576
-
Induction of embryonic primordia by implantation of organizers from a different speciesThe International Journal of Developmental Biology 45:13–38.
-
We need both exploratory and confirmatoryAmerican Statistician 34:23–25.https://doi.org/10.2307/2682991
-
Modeling network dynamics: the lac operon, a case studyThe Journal of Cell Biology 161:471–476.https://doi.org/10.1083/jcb.200301125
-
Experimental Production of Partial and Double Formations38–50, The potency of the first two cleavage cells in echinoderm development. Experimental production of partial and double formations, Experimental Production of Partial and Double Formations, New York, Hafner Press.
Decision letter
-
Peter RodgersSenior and Reviewing Editor; eLife, United Kingdom
In the interests of transparency, eLife publishes the most substantive revision requests and the accompanying author responses.
Thank you for submitting your article "Knowledge and illusions when using the complementary approaches needed to understand living systems" to eLife for consideration as a Features Article. Your article has been reviewed by three peer reviewers, and I am writing to invite you to revise the article in response to the comments of these reviewers.
The points that need to be addressed in the report of reviewer #1 are clear to see. In the report from reviewer #2, please address the points in the second and fourth paragraphs; please also take on board the comments in the third paragraph about making the article more readable (but also note that if the article is accepted for publication, it will be edited for readability and clarity). As regards the report from reviewer #3, it may be difficult to answer the questions in the second paragraph, but addressing the points raised by reviewer #1 will help to address some of these concerns.
Reviewer #1
This Essay addresses methodological and philosophical aspects of Biology broadly conceived. The author asks important questions, including "How is an outsider to evaluate [biologists'] progress?" (although I'm not sure a solid answer is proposed here), and gives a classification (systematization) of the major approaches in the field. This and a number of other important meta-scientific questions, including reproducibility and fairness of review, are discussed. Overall this is an important approach and I think will be valuable, but it seems a little thin on actionable suggestions (except for the good and novel idea to be explicit in papers about inference and method with respect to a common taxonomy), or on deep insights that hadn't been made in longer pieces in the past (admittedly, in journals that few biologists read). I suggest some points which will increase value for readers.
The title might use a little work - I'm not sure everyone will understand the meaning of "illusions" (without having read the paper first), but it may draw people in so perhaps it's fine. Overall I find the arguments convincing. A few minor comments:
- "Inference using [perturbation] approaches requires prior models of how the living system is built." I am not sure this is right - many perturbation experiments (e.g., classic work in developmental biology using transplantation, teratology, etc.) were done with absolutely no model of mechanism, and even modern screening (high-throughput) approaches are often done in a model-agnostic manner. Overall I suggest that the Perturbation section not be exclusively focused on genetics, as this is just one of the perturbational tools in the biologists' toolbox. It's too narrowly conceived as written, to match the title of the piece which refers to "living systems" (i.e., not just the molecular level).
- "Visualization" is also focused entirely on molecular tagging. I think if the author wants to stick to this level of analysis, the title might need to be made more specific - right now, it promises a more comprehensive view of biology. Lots of aspects of visualizing systems on the anatomical, metabolic, neurobiological, behavioral, and population levels are not even mentioned here.
- "Reconstitution" is likewise envisioned purely molecularly, not including for example important reconstitution tools like grafts/chimeras, organoids, biobots, etc. and other examples of synthetic bioengineering.
- Applications. The author is right to focus some spotlight on exploratory analyses, which are often hidden in favor of 'hard inference' storytelling, but not much is said here about what to do about this.
Overall, the task of trying to systematize biology in these ways is valuable and I applaud the author for taking this on. However, as written, it's very narrowly construed and I'm not sure it provides the insights for all of Biology that the introductory text suggests, or that eLife readers will want. One way to resolve this is to put "Molecular Biology" or similar in the title, and simply decide that this will be only about that level of investigation. On the other hand, if there is room, I would be happier to see a bigger piece that lived up to the much more fundamental and inclusive aim suggested in the Abstract: "acquire knowledge about living systems". As it is now, it's far too narrow for that (as large a goal as that is).
Reviewer #2
I have found this a very difficult paper to review. Normally as a reviewer I try to assess the extent to which the methodology and results are rigorous and whether the underpinning literature is treated with appropriate scholarship, then whether the conclusions are justified (or at least the speculations reasonable) based on the information presented. But this manuscript is not a scientific paper but an essay on epistemology, therefore an "opinion" piece. Therefore I cannot use my usual method to assess it. I can only answer with more "opinion" and I generally shy away from doing this as this is the realm of religion and politics, not Science.
Overall I do feel that this piece does raise some interesting points that are worthy of further discussion and consideration by many scientists. I am not sure I agree that this is a definitive classification of approaches (for example simple "description" of the system at a chosen level or levels is not obviously included and many studies start this way, and there are various types of theoretical model building that are not easily included under "simulation") but the six chosen ones for functional analysis are common ones.
Whether this is the type of paper that eLife wants to publish, and under what heading, is a matter for the editors. I think it belongs more as an editorial in a journal like Nature or Science, or perhaps as an Essay in BioEssays or similar. In either case it needs to be written much more fluidly to make it more attractive and accessible to experimental scientists who are not otherwise inclined to think about these bigger philosophical and epistemological issues. I find this quite difficult to read even as someone who has been thinking about these issues for some time.
I don't like the proscriptive tone in relation to what reviewers should do - it gives an unnecessarily arrogant tone to the essay. While it is true that being aware of the strengths and limitations of particular types of approaches is essential to determine whether the conclusions of a set of experiments are justified, there are many different ways for reviewers to achieve this and this six-part classification is too rigid a framework to advocate for general use. I think the paper would gain if the comments about peer review were removed. Where this classification (or other ways of evaluating evidence and conclusions) is probably particularly important is in the writing of reviews that synthesize the work of a number of papers in a field - many reviews are not sufficiently analytical or integrative and therefore often fail to undertake a critical evaluation that could allow readers to see the strengths and limitations of different studies more clearly.
Reviewer #3
This is an unusual paper for me to review and I offer my reactions to it – I am not sure whether what I have to say is in any objective way right or wrong. The author makes some interesting observations about how scientists use perturbation, visualization, substitution, characterization, reconstitution, and simulation of some aspect of a living system in an effort try to understand how it works. The author emphasizes how each of these approaches in isolation, coupled with sociological norms of scientists have contributed to erroneous or misleading conclusions, usually using one spectacular example to make the point for each approach in this essay.
Its not clear to what degree these examples are representative of the broader life science enterprise, though its clear they have played a role in sustaining some misconceptions for a long time. Is there some way in which the significance and frequency of examples used can be put into perspective?
The issues the author raises are certainly worth considering, and an essay that brings them to the reader's attention is important. However, it is not clear how effective the suggested solution of explicitly spelling out the assumed paradigm will be. Other suggestions for the editor in capturing the discussions and making this a more transparent process are good – and already standard practice for eLife!
Communicating more of the backstory behind a study including the blind alleys, alternate hypotheses and sometimes accidental discovery of the appropriate framework would also be helpful and interesting. Most scientists are quite aware of the contrived nature of the narrative that is often created to make a concise slick paper with few unresolved open questions. Have space limitations and the perceived desires of some "premier" journals (at least in the past) played some role in encouraging the evolution of this contrived narrative? Perhaps digital publishing provides the opportunity to more easily respond to the issues emphasized in this essay, with opportunities to provide transparency and multiple opportunities to dig deeper through supplementary material into the back story, alternative hypotheses, statistical analysis etc.
https://doi.org/10.7554/eLife.56354.sa1Author response
[We repeat the reviewers’ points here in italic, and include our replies point by point, as well as a description of the changes made, in Roman.]
Reviewer #1
This Essay addresses methodological and philosophical aspects of Biology broadly conceived. The author asks important questions, including "How is an outsider to evaluate [biologists'] progress?" (although I'm not sure a solid answer is proposed here), and gives a classification (systematization) of the major approaches in the field. This and a number of other important meta-scientific questions, including reproducibility and fairness of review, are discussed. Overall this is an important approach and I think will be valuable, but it seems a little thin on actionable suggestions (except for the good and novel idea to be explicit in papers about inference and method with respect to a common taxonomy), or on deep insights that hadn't been made in longer pieces in the past (admittedly, in journals that few biologists read). I suggest some points which will increase value for readers.
I thank the reviewer for the positive words and for endorsing explicit inclusion of statements about inferences in the Methods section. Where possible, I have made suggestions for how one might address problems. However, it is possible that some problems are not easily solvable at the present time potentially because of underdeveloped methods (e.g., evaluating the impact of conflated exploratory and confirmatory research). In such cases, I have merely drawn attention to the problem and highlighted the current difficulty.
The title might use a little work - I'm not sure everyone will understand the meaning of "illusions" (without having read the paper first), but it may draw people in so perhaps it's fine. Overall I find the arguments convincing.
I have simplified the title to be “Knowledge and illusions acquired during the analysis of living systems”.
Editor's note: The title was later changed to: "The analysis of living systems can generate both knowledge and illusions"
A few minor comments:
Collectively, the comments below suggest an expansion of the scope of the article to include the analysis of living systems on multiple scales. Although the taxonomy can apply across multiple scales (reason for representing living systems as an abstract network in Figure 1), the previous version did not highlight this aspect and was focused on a single scale - molecular - for simplicity. I have now highlighted the multi-scale nature of the proposed taxonomy during the early part of the article and provided specific examples to address other scales throughout the article as suggested by the reviewer.
- "Inference using [perturbation] approaches requires prior models of how the living system is built." I am not sure this is right - many perturbation experiments (e.g., classic work in developmental biology using transplantation, teratology, etc.) were done with absolutely no model of mechanism, and even modern screening (high-throughput) approaches are often done in a model-agnostic manner. Overall I suggest that the Perturbation section not be exclusively focused on genetics, as this is just one of the perturbational tools in the biologists' toolbox. It's too narrowly conceived as written, to match the title of the piece which refers to "living systems" (i.e., not just the molecular level).
I thank the reviewer for pointing out this error. This statement was meant to draw attention to the prior conception of the living system that is necessary to infer what a perturbed outcome means or indeed that a perturbation has occurred. I agree that ‘model’ is not a good word for conveying this idea because it connotes molecular mechanism in the context of the rest of the article (previous version). I have now expanded upon the statement to make the intended meaning clearer. I have also included some examples of the different kinds of perturbation suggested by the reviewer and illustrated the particular prior conceptions that impact the inference after a perturbation experiment. For example, in the famous Spemann and Mangold experiments on the organizer in embryology, interpreting the results requires the idea that a piece of embryonic tissue could be the source of unknown factors that help pattern the entire embryo. Modern screening selects for particular perturbed outcomes based on the prior expectation of what a perturbed system of interest would look like or that a particular manipulation is a perturbation, which sometimes lead to unexpected outcomes. For example, genetic compensation can confound inference (El-Brolosy and Stainier, PLoS Genetics, 2017). A prominent recent case that highlights this problem is the disagreement between morpholino-based perturbations and Cas-9-mediated genome editing in Zebrafish (Kok et al., Dev. Cell, 2015), some of which is currently thought to be explained by transcriptional adaptation (Ma et al., Nature, 2019, El-Brolosy et al., Nature, 2019).
- "Visualization" is also focused entirely on molecular tagging. I think if the author wants to stick to this level of analysis, the title might need to be made more specific - right now, it promises a more comprehensive view of biology. Lots of aspects of visualizing systems on the anatomical, metabolic, neurobiological, behavioral, and population levels are not even mentioned here.
I have now used examples from multiple scales to illustrate the kinds of visualizations that are possible and the kinds of limitation that can confound inference. For example, at the anatomical scale, limitations in the resolution of measurement combined with preconceptions promoted by prevailing theories can result in missed features of the system – e.g., consider estimations of tumor size and the developments that have been necessary to improve their visualization. At the population scale, difficulties in counting a particular species can distort the deduced composition and trophic relationships in an ecosystem. At the behavioral level, inability to visualize aspects of behavior can result in profound misconceptions about an organism. For example, recent ability to see animal behavior at night in the wild without perturbation is changing our understanding of land animals. In the deep ocean, recent ability to image without perturbation has revealed light-based communication in benthic organisms.
- "Reconstitution" is likewise envisioned purely molecularly, not including for example important reconstitution tools like grafts/chimeras, organoids, biobots, etc. and other examples of synthetic bioengineering.
I have now expanded this section to include reconstitution on additional scales. For example, human embryos can be formed from sperm and egg in vitro (Edwards, et al., 1969), establishing the sufficiency of these two cell types for the formation of human embryos.
- Applications. The author is right to focus some spotlight on exploratory analyses, which are often hidden in favor of 'hard inference' storytelling, but not much is said here about what to do about this.
The complications introduced by the conflation of exploratory and confirmatory research when story telling is emphasized require a deeper analysis and debate by the scientific community that are beyond the scope of this article. Unfortunately, a facile solution for this problem may not be available. Nevertheless, widespread awareness of the problem is the starting point for collectively working towards a solution. Therefore, the primary goals of this article remain to introduce a unified classification of approaches to understand living systems and invite the scientific community to debate the ways to implement such system-level and meta-scientific thinking to improve the practice of science.
Overall, the task of trying to systematize biology in these ways is valuable and I applaud the author for taking this on. However, as written, it's very narrowly construed and I'm not sure it provides the insights for all of Biology that the introductory text suggests, or that eLife readers will want. One way to resolve this is to put "Molecular Biology" or similar in the title, and simply decide that this will be only about that level of investigation. On the other hand, if there is room, I would be happier to see a bigger piece that lived up to the much more fundamental and inclusive aim suggested in the Abstract: "acquire knowledge about living systems". As it is now, it's far too narrow for that (as large a goal as that is).
I have now expanded the scope of the article to highlight the application of the taxonomy to additional scales and included illustrative examples across scales throughout the article.
Reviewer #2
I have found this a very difficult paper to review. Normally as a reviewer I try to assess the extent to which the methodology and results are rigorous and whether the underpinning literature is treated with appropriate scholarship, then whether the conclusions are justified (or at least the speculations reasonable) based on the information presented. But this manuscript is not a scientific paper but an essay on epistemology, therefore an "opinion" piece. Therefore I cannot use my usual method to assess it. I can only answer with more "opinion" and I generally shy away from doing this as this is the realm of religion and politics, not Science.
As noted by reviewer #1, this article addresses methodological and philosophical aspects of science, particularly as it applies to understanding living systems. The spirit of the article is in keeping with the belief that philosophical considerations can have practical uses for science (e.g., Laplane et al., PNAS, 2019). Methodologically, this article develops an overarching classification and invites future debates about such overall classifications. Philosophically, this article highlights the ever present unknown unknown when using any approach that can cast doubt over the most ardently held knowns.
Overall I do feel that this piece does raise some interesting points that are worthy of further discussion and consideration by many scientists. I am not sure I agree that this is a definitive classification of approaches (for example simple "description" of the system at a chosen level or levels is not obviously included and many studies start this way, and there are various types of theoretical model building that are not easily included under "simulation") but the six chosen ones for functional analysis are common ones.
As stated in the article, future periodic debates about the classification would help enrich and sharpen our understanding of the methods we have and will acquire. The previous version of the article did not expand upon application of the taxonomy across multiple scales, which makes it unclear where “description” fits within the classification. As suggested by reviewer #1, I have now expanded upon the multi-scale application, and “description” at any scale is essentially “visualization”, i.e., seeing what is there and reporting on it. For example, when describing an ecosystem, we count the number of organisms, look at their shape, size and any other characteristics that we can see (using the naked eye or using additional instruments). In the current expanded section on “simulation”, methods ranging from the sketching of ideas on a sheet of paper to simulate how a system might be working to running in silico computations to explore a virtual recreation of a system are included.
Whether this is the type of paper that eLife wants to publish, and under what heading, is a matter for the editors. I think it belongs more as an editorial in a journal like Nature or Science, or perhaps as an Essay in BioEssays or similar. In either case it needs to be written much more fluidly to make it more attractive and accessible to experimental scientists who are not otherwise inclined to think about these bigger philosophical and epistemological issues. I find this quite difficult to read even as someone who has been thinking about these issues for some time.
I have strived to simplify sentences throughout the manuscript and appreciate the editorial help offered by the journal for improving clarity and readability. I hope that these efforts make the article more engaging.
I don't like the proscriptive tone in relation to what reviewers should do - it gives an unnecessarily arrogant tone to the essay. While it is true that being aware of the strengths and limitations of particular types of approaches is essential to determine whether the conclusions of a set of experiments are justified, there are many different ways for reviewers to achieve this and this six-part classification is too rigid a framework to advocate for general use. I think the paper would gain if the comments about peer review were removed. Where this classification (or other ways of evaluating evidence and conclusions) is probably particularly important is in the writing of reviews that synthesize the work of a number of papers in a field - many reviews are not sufficiently analytical or integrative and therefore often fail to undertake a critical evaluation that could allow readers to see the strengths and limitations of different studies more clearly.
The intent of the suggestions is not to be proscriptive, but rather to add a specific suggestion that can either be taken on or ignored after debating its merits. I have now reworded the section to mitigate any perceived arrogance in tone. As highlighted in the article, the process of peer review is currently undergoing a lot of experimentation and this suggestion adds to the debate on the best way to achieve peer review, which in my opinion is an invaluable part of science. I believe that constructive peer review can improve an author’s work as it certainly has for this article.
I agree with the excellent opinion of the reviewer that this classification would help with the writing of more analytical and integrative reviews, and the revised manuscript now includes a sentence to highlight this use in the peer review section.
Reviewer #3
This is an unusual paper for me to review and I offer my reactions to it-I not sure whether what I have to say is in any objective way right or wrong. The author makes some interesting observations about how scientists use perturbation, visualization, substitution, characterization, reconstitution, and simulation of some aspect of a living system in an effort try to understand how it works. The author emphasizes how each of these approaches in isolation, coupled with sociological norms of scientists have contributed to erroneous or misleading conclusions, usually using one spectacular example to make the point for each approach in this essay.
I thank the reviewer for the positive words.
Its not clear to what degree these examples are representative of the broader life science enterprise, though its clear they have played a role in sustaining some misconceptions for a long time. Is there some way in which the significance and frequency of examples used can be put into perspective?
The reviewer raises an excellent and difficult question, answering which requires extensive study that is beyond the scope of this article. Specifically, the answer needs a comprehensive historical (and predictive?) survey of the science that has been done (and will be done?). One source that presents a partial historical survey of examples where misconceptions arose, persisted for a long time and then were overthrown is the book ‘Structure of Scientific Revolutions’ by Thomas Kuhn. However, even such a study doesn’t allow evaluation of the frequency. Evaluation of the frequency is being made all the more difficult by the growing volume of scientific literature.
In the revised version of the article, application of the taxonomy to additional scales and subdisciplines are highlighted in an effort to underscore the broad use of such a taxonomy of methods across the life sciences (and perhaps all sciences).
The issues the author raises are certainly worth considering, and an essay that brings them to the reader's attention is important. However, it is not clear how effective the suggested solution of explicitly spelling out the assumed paradigm will be. Other suggestions for the editor in capturing the discussions and making this a more transparent process are good -and already standard practice for eLife!
Explicitly spelling out the assumed paradigm can help authors and readers identify the assumptions/frameworks that are necessary for the inferences to be valid. Often these remain unstated and could result in quests for concepts and entities that may not exist – historical examples include the search for luminiferous ether as the medium transmitting light and phlogiston as the carrier of heat. Nevertheless, the suggestions presented in the article are starting points for future debate. The primary goal of the article is to bring attention to these issues that impact how science is done, presented, and evaluated.
Unlike eLife, many journals do not yet have active summaries of the review process by the editors. I hope the arguments presented in this article add to those presented elsewhere to aid wider adoption of this practice.
Communicating more of the backstory behind a study including the blind alleys, alternate hypotheses and sometimes accidental discovery of the appropriate framework would also be helpful and interesting. Most scientists are quite aware of the contrived nature of the narrative that is often created to make a concise slick paper with few unresolved open questions. Have space limitations and the perceived desires of some "premier" journals (at least in the past) played some role in encouraging the evolution of this contrived narrative? Perhaps digital publishing provides the opportunity to more easily respond to the issues emphasized in this essay, with opportunities to provide transparency and multiple opportunities to dig deeper through supplementary material into the back story, alternative hypotheses, statistical analysis etc.
I agree with the reviewer that digital publishing can facilitate the clear declaration of the often tortuous route to discovery. However, our collective inability to evaluate the statistical rigor of the inferences in the study would remain. Furthermore, pursuit of today’s ‘complete’ story while ignoring or obfuscating incompleteness, deliberately makes scientific understanding more saltatory than it needs to be – proceeding from one false picture to the next and reducing confidence in published models. The article therefore highlights this difficulty to invite debate on the most productive courses of action for the scientific community.
https://doi.org/10.7554/eLife.56354.sa2Article and author information
Author details
Funding
National Institutes of Health (R01GM111457)
- Antony M Jose
National Institutes of Health (R01GM124356)
- Antony M Jose
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
I thank Tom Kocher, Karen Carleton, Charles Delwiche, and members of the Jose lab for long discussions; and Tom Kocher, Karen Carleton, members of the Jose lab, the editor and the reviewers for their comments on the manuscript.
Publication history
- Received:
- Accepted:
- Version of Record published:
Copyright
© 2020, Jose
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 10,919
- views
-
- 871
- downloads
-
- 10
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cancer Biology
- Cell Biology
The endoplasmic reticulum (ER) is an essential sensing organelle responsible for the folding and secretion of almost one-third of eukaryotic cells' total proteins. However, environmental, chemical, and genetic insults often lead to protein misfolding in the ER, accumulating misfolded proteins, and causing ER stress. To solve this, several mechanisms were reported to relieve ER stress by decreasing the ER protein load. Recently, we reported a novel ER surveillance mechanism by which proteins from the secretory pathway are refluxed to the cytosol to relieve the ER of its content. The refluxed proteins gain new prosurvival functions in cancer cells, thereby increasing cancer cell fitness. We termed this phenomenon ER to CYtosol Signaling (or ‘ERCYS’). Here, we found that in mammalian cells, ERCYS is regulated by DNAJB12, DNAJB14, and the HSC70 cochaperone SGTA. Mechanistically, DNAJB12 and DNAJB14 bind HSC70 and SGTA - through their cytosolically localized J-domains to facilitate ER-protein reflux. DNAJB12 is necessary and sufficient to drive this phenomenon to increase AGR2 reflux and inhibit wt-p53 during ER stress. Mutations in DNAJB12/14 J-domain prevent the inhibitory interaction between AGR2-wt-p53. Thus, targeting the DNAJB12/14-HSC70/SGTA axis is a promising strategy to inhibit ERCYS and impair cancer cell fitness.
-
- Cancer Biology
- Cell Biology
DNA base lesions, such as incorporation of uracil into DNA or base mismatches, can be mutagenic and toxic to replicating cells. To discover factors in repair of genomic uracil, we performed a CRISPR knockout screen in the presence of floxuridine, a chemotherapeutic agent that incorporates uracil and fluorouracil into DNA. We identified known factors, such as uracil DNA N-glycosylase (UNG), and unknown factors, such as the N6-adenosine methyltransferase, METTL3, as required to overcome floxuridine-driven cytotoxicity. Visualized with immunofluorescence, the product of METTL3 activity, N6-methyladenosine, formed nuclear foci in cells treated with floxuridine. The observed N6-methyladenosine was embedded in DNA, called 6mA, and these results were confirmed using an orthogonal approach, liquid chromatography coupled to tandem mass spectrometry. METTL3 and 6mA were required for repair of lesions driven by additional base-damaging agents, including raltitrexed, gemcitabine, and hydroxyurea. Our results establish a role for METTL3 and 6mA in promoting genome stability in mammalian cells, especially in response to base damage.







