Sex-based differences in clearance of chronic Plasmodium falciparum infection
Abstract
Multiple studies have reported a male bias in incidence and/or prevalence of malaria infection in males compared to females. To test the hypothesis that sex-based differences in host-parasite interactions affect the epidemiology of malaria, we intensively followed Plasmodium falciparum infections in a cohort in a malaria endemic area of eastern Uganda and estimated both force of infection (FOI) and rate of clearance using amplicon deep-sequencing. We found no evidence of differences in behavioral risk factors, incidence of malaria, or FOI by sex. In contrast, females cleared asymptomatic infections at a faster rate than males (hazard ratio [HR]=1.82, 95% CI 1.20 to 2.75 by clone and HR = 2.07, 95% CI 1.24 to 3.47 by infection event) in multivariate models adjusted for age, timing of infection onset, and parasite density. These findings implicate biological sex-based differences as an important factor in the host response to this globally important pathogen.
Introduction
Malaria, a protozoan infection of the red blood cells, remains one of the greatest global health challenges (World malaria report, 2019). Infection with malaria parasites results in a wide range of clinical disease presentations, from severe to uncomplicated; in addition, in hyperendemic areas, asymptomatic infections are common (Bousema et al., 2014). It is well established that chronic asymptomatic infection with Plasmodium falciparum, the most common and fatal malaria parasite, can lead to morbidity for those infected and contribute to ongoing transmission (Bousema et al., 2014; Okell et al., 2012; Tadesse et al., 2018; Slater et al., 2019). Characterization of these asymptomatic infections is paramount as they represent a major obstacle for malaria elimination efforts. Therefore, an understanding of how host immunity and parasite factors interact to cause disease tolerance is required. While age-specific immunity to malaria in hyperendemic areas is well-characterized, less attention has been paid to the possibility of a sex bias in malarial susceptibility despite evidence for a male bias in malaria infections in non-human animals and a male bias in the prevalence of other human parasitic infections (Zuk and McKean, 1996; Klein, 2004; Roberts et al., 2001).
The clearest evidence for sexual dimorphism in malaria susceptibility is in pregnant women, who are at greater risk of malaria infection and also experience more severe disease and higher mortality (Desai et al., 2007). However, multiple studies performed in different contexts have demonstrated a male bias in incidence and/or prevalence of malaria infection in school-aged children and adults (Molineaux et al., 1980; Pathak et al., 2012; Landgraf et al., 1994; Camargo et al., 1996; Abdalla et al., 2007); this bias is more well established in hypoendemic areas, but has also been observed in hyperendemic regions (Houngbedji et al., 2015; Mulu et al., 2013). Where there is a male bias in malaria infection, it has often been postulated that these differences in malaria incidence or prevalence stem from an increased risk of males acquiring infection due to socio-behavioral factors (Pathak et al., 2012; Camargo et al., 1996; Moon and Cho, 2001; Finda et al., 2019). However, because biological sex itself has been demonstrated to affect responses to other pathogens, an alternative hypothesis is that the sexes may have different responses to the malaria parasite once infected (Nhamoyebonde and Leslie, 2014; Bernin and Lotter, 2014; Fischer et al., 2015; Fish, 2008).
Estimating the host response to P. falciparum infection requires close follow-up of infected individuals, sensitive detection of parasites, and the ability to distinguish superinfection, which is common in endemic areas, from persistent infection. To test the hypothesis that sex-based differences in host–parasite interactions affect the epidemiology of malaria, we intensively followed a representative cohort of individuals living in a malaria endemic area of eastern Uganda. Using frequent sampling, ultrasensitive quantitative PCR (qPCR), and amplicon deep-sequencing to genotype parasite clones, we were able to accurately detect the onset of new infections and follow all infections over time to estimate their duration. Using these data, we show that females cleared their asymptomatic infections more rapidly than males, implicating biological sex-based differences as important in the host response to this globally important pathogen.
Results
Cohort participants and P. falciparum infections
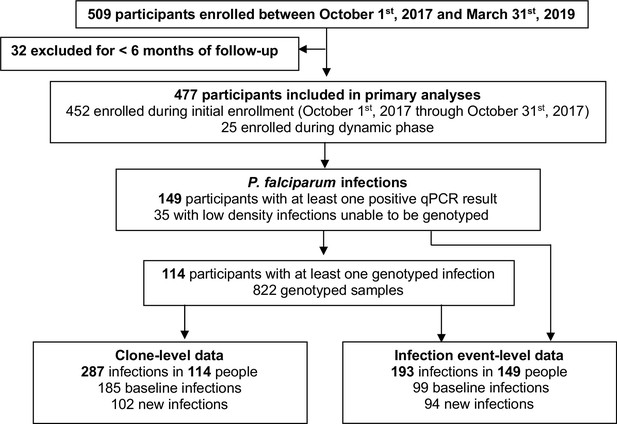
This analysis involved data from 477 children and adults (233 males and 244 females) that were followed for a total of 669.6 person-years (Table 1). 25 participants, 10 males and 15 females, were enrolled after initial enrollment (Figure 1); median duration of follow-up in those who were dynamically enrolled was 0.84 years (IQR 0.69–1.23) compared to 1.45 years (IQR 1.43–1.47) in those enrolled during initial enrollment. 149 of 477 participants (31.2%) included in the analysis had at least one P. falciparum infection detected (Figure 1). 114 participants had 822 successfully genotyped samples and had infections characterized by clone and by infection event. 35 samples (from 35 unique participants) had very low-density infections (<1 parasite/µL) that could not be genotyped; these infections were characterized at the infection event level only. We achieved a read count of >10,000 for 92% of genotyped samples, identifying 45 unique AMA-1 clones in our population (frequencies and sequences in Supplementary file 1e). At the clone level, the proportion of baseline infections out of all infections in males was 117/171 (68.4%), compared to 68/116 (58.8%) baseline infections in females (p=0.10). At the infection event level, there were 54/104 (51.9%) baseline infections in males and 45/89 (50.6%) baseline infections in females (p=0.89).
Behavioral risk factors for malaria infection and measures of malaria burden in study population, stratified by age and sex.
| Metric | Age and gender categories | |||||||
|---|---|---|---|---|---|---|---|---|
| All | <5 years old | 5–15 years old | 16 years and older | |||||
| Male | Female | Male | Female | Male | Female | Male | Female | |
| Number of participants, n | 233 | 244 | 73 | 84 | 101 | 71 | 59 | 89 |
| Median days of follow-up per participant | 530.0 | 530.0 | 525.0 | 524.5 | 530.0 | 531.0 | 530.0 | 530.0 |
| Slept under LLIN the previous night | 53.6% | 56.3% | 54.1% | 56.3% | 47.9% | 47.8% | 63.7% | 64.3% |
| Person-years of follow-up | 324.2 | 345.4 | 87.3 | 96.7 | 152.6 | 118.5 | 84.22 | 130.1 |
| Number of overnight trips | 44 | 107 | 21 | 19 | 9 | 10 | 14 | 78 |
| Incidence of overnight trips*, (95% CI) | 0.14 (0.09–0.20) | 0.31 (0.20–0.49) | 0.24 (0.14–0.42) | 0.20 (0.09–0.42) | 0.06 (0.03–0.13) | 0.08 (0.03–0.24) | 0.17 (0.09–0.31) | 0.60 (0.30–1.18) |
| Episodes of malaria* | 11 | 13 | 5 | 2 | 5 | 9 | 1 | 2 |
| Incidence of malaria**, (95% CI) | 0.03 (0.02–0.06) | 0.04 (0.02–0.09) | 0.06 (0.02—0.16) | 0.02 (0.00–0.11) | 0.03 (0.01–0.08) | 0.08 (0.03–0.23) | 0.01 (0.00–0.08) | 0.02 (0.00–0.17) |
| Number of routine visits, n | 4316 | 4583 | 1164 | 1293 | 2034 | 1568 | 1118 | 1722 |
| Prevalence of microscopic parasitemia*** | 2.9% | 1.4% | 1.8% | 1.1% | 4.4% | 2.7% | 1.3% | 0.5% |
| Prevalence of parasitemia by qPCR | 14.4% | 9.2% | 5.8% | 3.7% | 17.0% | 15.3% | 18.5% | 7.7% |
| Geometric mean parasite density**** | 3.41 | 3.06 | 4.86 | 13.06 | 6.31 | 4.20 | 1.09 | 1.02 |
| Median complexity of infection, (IQR) | 3 (1–7) | 2 (1–2) | 1 (1–2.5) | 2 (1–2.3) | 4 (2–9) | 2 (1–2) | 1 (1–2) | 1 (1–2) |
-
*Malaria includes one episode (female,<5 years old), due to non-falciparum species (P. malariae).
**per person-year.
-
***Parasitemia by light microscopy includes one episode (female, 5–15 years old) due to non-falciparum species (P. ovale).
****Geometric mean parasite density in parasites/µL of all qPCR-positive routine visits.
Behavioral malaria risk factors and measures of malaria burden
There was no difference in reported rates of LLIN use the previous night by sex (Table 1). Women over the age of 16 traveled overnight outside of the study area more than men (incidence rate ratio [IRR] for females vs. males = 3.61, 95% confidence interval [CI] 1.83 to 7.13), a potential risk factor for malaria exposure. Antimalarial use outside the study clinic was reported only four times (3 females and one male, all under the age of 5). In this region receiving regular rounds of IRS, the incidence of symptomatic malaria was low in all age categories, and there was no evidence of a difference in incidence of symptomatic malaria by sex overall (IRR for females vs. males = 1.11, 95% CI 0.49 to 2.52) or when adjusted for age (IRR = 1.26, 95% CI 0.54 to 2.96). In contrast, prevalence ratio (PR) of P. falciparum parasitemia by microscopy in females versus males across all age categories was 0.49 (95% CI 0.36 to 0.65), with relative differences in prevalence most pronounced in the oldest age group. Similar findings were seen when prevalence was assessed by ultrasensitive qPCR, with PR = 0.64 in females vs. males (95% CI 0.43 to 0.96), again with the largest differences seen in the oldest age group. Adjusting for age as a categorical variable, LLIN use, and travel did not qualitatively change prevalence ratios for microscopic parasitemia (PR in females vs. males = 0.57, 95% CI 0.42 to 0.77) or for qPCR-positive parasitemia (PR in females vs. males = 0.67, 95% CI 0.60 to 0.76). We also found no evidence for a difference in parasite density as determined by qPCR between males and females after adjusting for age as a continuous variable (p=0.47). Median complexity of infection (COI) was higher in males than in females overall, driven primarily by a higher COI in male school-aged children.
Force of infection by age and sex
To determine whether higher infection prevalence in males was due to an increased rate of infection, we used longitudinal genotyping to calculate the force of infection (FOI, number of new blood stage infections per unit time). Overall, the FOI was low, with new infections occurring on average less than once every 5 years (Table 2). There was no evidence for a significant difference in FOI by sex overall (IRR for females vs. males = 0.88, 95% CI 0.48 to 1.62 by clone and IRR = 0.83, 95% CI 0.52 to 1.33 by infection event). There was also no evidence for a significant difference in FOI by sex when adjusted for age category (IRR = 0.88, 95% CI 0.47 to 1.63 by clone and IRR = 0.83, 95% CI 0.53 to 1.31 by infection event). For analysis both by clone and by infection event, there was a trend toward higher FOI in males compared to females. We performed a sensitivity analysis by decreasing the number of skips necessary to declare an infection cleared (Supplementary file 1f). Performing the same analysis using one skip for clearance instead of 3 skips increased the FOI in all groups and increased the trend toward higher FOI in males, with IRR for females vs. males when adjusted for age category = 0.71, 95% CI 0.41 to 1.24 by clone and IRR = 0.75, 95% CI 0.47 to 1.22 by infection event.
Molecular force of infection (FOI) by clone and by infection event, stratified by age and sex.
| Molecular force of infection (FOI) | Sex | Age category | |||
|---|---|---|---|---|---|
| All | <5 years | 5–15 years | 16 years or older | ||
| By clone, ppy* (95% CI) | All | 0.18 (0.13–0.24) | 0.14 (0.07–0.28) | 0.19 (0.08–0.43) | 0.20 (0.08–0.46) |
| Male | 0.19 (0.12–0.30) | 0.16 (0.07–0.39) | 0.19 (0.10–0.37) | 0.22 (0.09–0.54) | |
| Female | 0.17 (0.09–0.31) | 0.12 (0.03–0.49) | 0.19 (0.08–0.45) | 0.18 (0.06–0.54) | |
| By event, ppy* (95% CI) | All | 0.14 (0.11–0.18) | 0.09 (0.06–0.16) | 0.16 (0.09–0.30) | 0.16 (0.08–0.32) |
| Male | 0.16 (0.11–0.22) | 0.13 (0.07–0.27) | 0.18 (0.11–0.28) | 0.15 (0.07–0.29) | |
| Female | 0.13 (0.08–0.21) | 0.06 (0.02–0.18) | 0.14 (0.07–0.27) | 0.17 (0.07–0.41) | |
-
*per person-year.
Rate of clearance of infection and duration of infection by sex
Since females had a lower prevalence of infection but similar rate of acquiring infections compared to males, we evaluated whether there was a difference between sexes in the rate at which infections were cleared. Asymptomatic infections were included in this analysis if they were not censored as stated in the methods. At the clone level, 105 baseline infections and 53 new infections were included; there was a slightly higher proportion of baseline infections in males (68/99, 68.7%), compared to the proportion of baseline infections in females (37/59, 62.7%) (p=0.49). At the infection event level, 58 baseline infections and 51 new infections were included and there was no difference in the proportion of baseline infections by sex, with 32/60 (53.3%) baseline infections in males and 26/49 (53.1%) baseline infections in females (p=1.0).
Unadjusted hazard ratios for clearing infecting clones showed that asymptomatic infections cleared naturally (i.e., when not treated by antimalarials) at nearly twice the rate in females vs. males (hazard ratio (HR) 1.92, 95% CI 1.19 to 3.11, Table 3). In addition, new infections cleared faster than baseline infections and monoclonal infections cleared faster than polyclonal infections. Unadjusted hazard ratios for clearance of infection events (as opposed to clones) also showed faster clearance in females vs. males (HR = 2.30, 95% CI 1.20 to 4.42). Results were similar in multivariate models including age, gender, the period during which the infection was first observed, and parasite density, demonstrating faster clearance in females vs. males (HR = 1.82, 95% CI 1.20 to 2.75 by clone and HR = 2.07, 95% CI 1.24 to 3.47 by infection event). Complexity of infection was not included in the final adjusted model because the model fit the data less well when COI was included as a predictor. In both adjusted models, new infections cleared faster than baseline infections. Higher parasite densities were associated with slower clearance by clone and by infection event, but the effect size was larger when data were analyzed by infection event (HR = 0.44, 95% CI 0.35 to 0.54). There was no evidence for interaction between age and sex in either adjusted model. We performed a sensitivity analysis by decreasing the number of skips necessary to declare an infection cleared. Regardless of whether three skips, two skips, or one skip was used to determine infection clearance, females cleared their infections faster than males both by clone and by infection event (Supplementary file 1g).
Hazard ratios for rates of clearance of infection, by clone and by infection event.
| Predictors | Categories | Hazard ratio by clone (95% CI) | Hazard ratio by infection event (95% CI) | ||
|---|---|---|---|---|---|
| Unadjusted | Adjusted | Unadjusted | Adjusted | ||
| Sex | Male | ref | ref | ref | ref |
| Female | 1.92 (1.19–3.11) | 1.82 (1.20–2.75) | 2.30 (1.20–4.42) | 2.07 (1.24–3.47) | |
| Age | 16 years or greater | ref | ref | ref | ref |
| 5–15 years | 0.66 (0.39–1.10) | 0.81 (0.49–1.36) | 0.82 (0.39–1.74) | 1.27 (0.72–2.25) | |
| <5 years | 1.64 (0.79–3.41) | 1.55 (0.76–3.17) | 2.01 (0.80–5.00) | 1.75 (0.87–3.53) | |
| Complexity of infection (COI) | Polyclonal (COI > 1) | ref | -- | ref | -- |
| Monoclonal (COI = 1) | 1.63 (1.03–2.57) | -- | 0.95 (0.38–2.34) | -- | |
| Infection status | Present at baseline | ref | ref | ref | ref |
| New infection | 1.94 (1.22–3.07) | 1.75 (1.05–2.94) | 4.66 (2.58–8.42) | 4.32 (2.59–7.20) | |
| Parasite density * | 0.85 (0.69–1.06) | 0.81 (0.65–1.00) | 0.41 (0.32–0.51) | 0.44 (0.35–0.54) | |
-
*Increasing parasite density (log10) in parasites/microliter, as measured by qPCR.
We next estimated durations of asymptomatic infection by age and sex using results from a model that included these covariates (Figure 2). Durations of infection ranged from 103 days to 447 days by clone, and from 87 to 536 days by infection event. Males had a longer duration of infection across all age categories. Children aged 5–15 years had the longest duration of infection, followed by adults. Therefore, overall, males aged 5–15 years had the longest estimated duration of infection by either clone (447 days) or infection event (526 days).
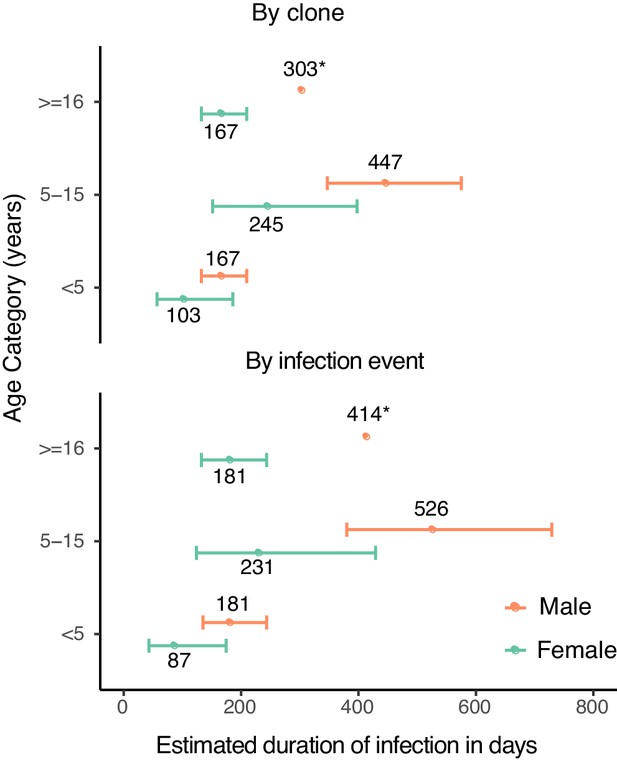
Estimates of duration of infection from sex- and age-adjusted model.
Estimated duration of infection in days, calculated by adjusting the point estimate of the baseline hazard by the coefficients of the sex- and age-adjusted model. Error bars represent standard errors of duration obtained from variance in the model coefficients. Point estimates of duration are labeled (*).
Discussion
Previous studies have reported a higher prevalence of malaria infection in males compared to females, with the difference often ascribed to differences in exposure (Molineaux et al., 1980; Pathak et al., 2012; Landgraf et al., 1994; Camargo et al., 1996; Abdalla et al., 2007; Houngbedji et al., 2015; Mulu et al., 2013). In a cohort study in eastern Uganda, we noted higher prevalence of malaria infection in males compared to females. By closely following a cohort of children and adults and genotyping every detected infection with sensitive amplicon deep-sequencing, we were able to estimate both the rate of infection (FOI) and duration of infection and to compare these measures by sex. We found that lower prevalence in females did not appear to be due to lower rates of infection but rather due to faster clearance of asymptomatic infections. To our knowledge, this is the first study to report a sex-based difference in the duration of malaria infection.
Though there are some conflicting reports in the literature, the majority of studies of malaria incidence and/or prevalence that evaluated associations with sex in late childhood, adolescence and adulthood have found a male bias in the observed measure of burden (Molineaux et al., 1980; Pathak et al., 2012; Landgraf et al., 1994; Camargo et al., 1996; Abdalla et al., 2007; Houngbedji et al., 2015; Mulu et al., 2013). We note that this is more often observed in hypoendemic settings and may be confounded by factors such as treatment-seeking behavior; however, this male bias has been reported in studies of both P. vivax and P. falciparum. Overall, these studies consistently suggest that males exhibit higher incidence and/or prevalence of malaria that begins during late childhood, persisting through puberty and the majority of adulthood (excepting the years when pregnancy puts women at higher risk). One possible explanation put forward for the sex-specific difference in burden has been that males are more frequently bitten by malaria-carrying mosquitos due to behavioral differences such as working outside, not sleeping under a net, or traveling for work (Pathak et al., 2012; Camargo et al., 1996; Moon and Cho, 2001; Finda et al., 2019). In our study, however, there were no statistically significant differences in malaria incidence or FOI by sex, though there was a trend toward higher FOI in males. We also saw no evidence of behavioral trends that would result in more infections in males; in fact, older women in our study did most of the traveling outside the study area (an area of low transmission compared to surrounding areas). We did not assess work habits as part of our study questionnaire, but the fact that we observe similar patterns in prevalence by sex in all age categories makes this a less likely explanation. Therefore, in this particular cohort, it appears that the observed male bias in parasite prevalence is best explained by slower clearance of infection in males. We cannot rule out that males were differentially exposed in the recent past (e.g., had a higher force of infection when transmission was higher, which could have affected immunity), and we plan to genotype samples from a prior cohort from 2011 to 2017 to test this hypothesis.
Very few studies have been conducted to explore immunological differences between males and females in their response to the malaria parasite. RTS,S vaccination is associated with higher all-cause mortality in girls compared to boys, and a trend toward higher risk of fatal malaria has been noted in vaccinated girls compared to boys, suggesting possible sexual dimorphism in immunological responses to malaria (Klein et al., 2016). The comprehensive Garki project found a male bias in the prevalence of P. falciparum infection after the age of 5 years that was noted to increase after control measures including IRS and mass drug administration. They also found that females had higher levels of certain antibodies against P. falciparum compared to males, but saw no difference in these levels after control measures (Molineaux et al., 1980). Hormonal differences have been posited to play a role in these sex-based immunological differences to malaria infection; for example, studies in mice show that testosterone appears to downregulate the immune response to malaria (Delić et al., 2011; Wunderlich et al., 1991). One study of irradiated sporozoite vaccination in mice showed that female mice vaccinated after puberty were better protected than males following parasite challenge, but this sex difference was not seen if the mice were vaccinated prior to puberty, suggesting a role for sex hormones in the development of immunity (Vom Steeg et al., 2019). Testosterone was again associated with decreased protection from malaria in that study (Vom Steeg et al., 2019). In addition, in Kenya, two studies showed that dehydroepiandrosterone sulfate (DHEAS) levels were significantly associated with decreased parasite density in both males and females, even after adjustment for age, but neither study directly compared males to females (Kurtis et al., 2001; Leenstra et al., 2003). Given that levels and types of sex hormones differ between males and females, as does the age of onset of puberty, the interaction between these hormones, age, and protection against malaria is likely to be complicated. We did not detect an interaction between age and sex in our model. This could be due to a lack of statistical power or imperfect detectability, as adults have lower density infections than children due to improved anti-parasite immunity (Bousema et al., 2014; Okell et al., 2012). In addition, the adult age group was comprised of a wide age range, which included post-menopausal women and might have blunted the effect of sex in that age group if there is a hormonal basis for some of this effect. Repeating our analysis using different age categories (<8 years, 8–13 years, and 13+ years) did not significantly change our findings. It is also possible that sex-based differences are explained by a different mechanism, such as differences in innate immunity, Y-linked chromosomal factors in males, increased X-linked gene expression in females, or by a combination of factors. More studies are needed to elucidate the relationship between sex-based biological differences between males and females and their impact on the development of effective antimalarial immunity in humans.
Of the variables we evaluated in addition to sex, baseline infection status and parasite density were most strongly associated with the rate of clearance of infection. Infections that were already present at the beginning of the study and persisted past the left censoring date may have been a non-random selection of well-established asymptomatic infections that were present at baseline at a higher frequency than average because they had a fundamentally different trajectory than newly established asymptomatic infections. Higher parasite densities were also associated with slower clearance of infection in both adjusted models, but the effect was most pronounced when the data were analyzed by infection event. This may be because low-density infections that we were unable to successfully genotype were only included in the infection event analysis, and these events tended to have short durations. The inclusion of parasite density in our multivariate models did not meaningfully alter associations between sex and duration of infection, providing evidence that the sex-based differences in duration were not mediated primarily by differences in parasite density in our cohort.
A limitation of our study is the statistical model’s assumption that all infections clear at the same rate, which was necessary because we did not observe the beginning of most infections. To rely less on this assumption, we adjusted for baseline infections, allowing them to have a different rate of clearance than new infections; this did not change our primary finding of a difference in rate of clearance by sex. We also acknowledge there may be other unmeasured confounders, such as genetic hemoglobinopathies or sex-based differences in unreported outside antimalarial use, that could affect our results. Another caveat is that our findings may not be generalizable to areas with different transmission intensity given that our study was conducted in a very specific setting: an area with previously very high transmission intensity that has been greatly reduced in recent years by repeated rounds of IRS.
Additionally, because it is difficult to genotype low-density infections and because parasite densities fluctuate over time, we allowed several ‘skips’ in detection before declaring an infection cleared or the same clone in an individual a new infection with the same clone. This requires the assumption that re-infection with the same clone is relatively rare, which is reasonable given the high genetic diversity and low force of infection seen in this setting; this assumption has been made in other longitudinal genotyping studies (Felger et al., 2012; Smith et al., 1999). Our method may have biased us toward longer durations of infection and fewer new infections overall, but this is unlikely to have introduced any significant bias in the observed associations by sex. We performed a sensitivity analysis to ensure that changing the number of skips allowed did not significantly change the main finding regarding longer duration of infection in males and our findings were consistent. Given decreasing parasite densities over time and a setting with low force of infection and low EIR, assuming perfect recovery of clones is likely to artificially inflate the force of infection in this cohort. There were not enough infections to perform a rigorous analysis of the distribution of clones within and between households, but given that the overall force of infection was quite low, the probability of re-infection with the same clone already present in a participant from another member in the household (which could bias toward longer duration of infection) was low and unlikely to have introduced any significant bias by sex.
In summary, we estimated the clearance of asymptomatic P. falciparum infections by genotyping longitudinal samples from a cohort in Nagongera, Uganda, using sensitive amplicon deep-sequencing and found that females cleared their infections at a faster rate than males; this finding remained consistent when adjusting for age, baseline infection status, and parasite density. Furthermore, we found no conclusive evidence for a sex-based difference in exposure to infection, either behaviorally or by FOI. Though there have long been observed differences in malaria burden between the sexes, there is still little known about sex-based biological differences that may mediate immunity to malaria. Unfortunately, much reported malaria data is still sex-disaggregated. Our findings should encourage epidemiologists to better characterize sexual dimorphism in malaria and motivate increased research into biological explanations for sex-based differences in the host response to the malaria parasite.
Materials and methods
Study setting and population-level malaria control interventions
Request a detailed protocolThis cohort study was carried out in Nagongera sub-county, Tororo district, eastern Uganda, an area with historically high malaria transmission. However, 7 rounds of indoor residual spraying (IRS) from 2014 to 2019 have resulted in a significant decline in the burden of malaria (Nankabirwa et al., 2019). Pre-IRS, the daily human biting rate (HBR) was 34.3 and the annual entomological inoculation rate (EIR) was 238; after 5 years of IRS, in 2019 the daily HBR was 2.07 and overall annual EIR was 0.43 as reported by Nankabirwa et. al (Nankabirwa et al., 2020).
Study design, enrollment, and follow-up
Request a detailed protocolAll members of 80 randomly selected households with at least two children were enrolled in October 2017 using a list generated by enumerating and mapping all households in Nagongera sub-county (Nankabirwa et al., 2020). The cohort was dynamic such that residents joining the household were enrolled and residents leaving the household were withdrawn. Data for this analysis was collected from October 1st, 2017 through March 31st, 2019; participants were included if they had at least 6 months of contiguous follow-up. Data from the 25 dynamically enrolled participants contributed to all analyses, including that of baseline infections. Participants were followed at a designated study clinic open daily from 8 AM to 5 PM. Participants were encouraged to seek all medical care at the study clinic and avoid the use of antimalarial medications outside of the study. Routine visits were conducted every 28 days and included a standardized clinical evaluation, assessment of overnight travel outside of Nagongera sub-county, and collection of blood by phlebotomy for detection of malaria parasites by microscopy and molecular studies. Participants came in for non-routine visits in the setting of illness. Blood smears were performed at enrollment, at all routine visits and at non-routine visits if the participant presented with fever or history of fever in the previous 24 hr. Participants with fever (>38.0°C tympanic) or history of fever in the previous 24 hr had a thick blood smear read urgently. If the smear was positive, the patient was diagnosed with malaria and treated with artemether-lumefantrine. Participants with asymptomatic parasitemia as detected by qPCR or microscopy were not treated with antimalarials, consistent with Uganda national guidelines. Study participants were visited at home every 2 weeks to assess use of long-lasting insecticidal nets (LLINs) the previous night.
Laboratory methods
Thick blood smears were stained with 2% Giemsa for 30 min and evaluated for the presence of asexual parasites and gametocytes. Parasite densities were calculated by counting the number of asexual parasites per 200 leukocytes (or per 500, if the count was less than 10 parasites per 200 leukocytes), assuming a leukocyte count of 8,000/μL. A thick blood smear was considered negative if examination of 100 high power fields revealed no asexual parasites. For quality control, all slides were read by a second microscopist, and a third reviewer settled any discrepant readings. In our experienced microscopists’ hands, the lower limit of detection is approximately 20–50 parasites/µL.
For qPCR and genotyping, we collected 200 µL of blood at enrollment, at each routine visit, and at the time of malaria diagnosis. DNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen) and parasitemia was quantified using an ultrasensitive varATS qPCR assay with a lower limit of detection of 0.05 parasites/µL (Hofmann et al., 2015). Samples with a parasite density >= 0.1 parasites/µL blood were genotyped via amplicon deep-sequencing. All samples positive for asexual parasites by microscopy but negative for P. falciparum by qPCR were tested for the presence of non-falciparum species using nested PCR (Snounou et al., 1993).
Sequencing library preparation
Request a detailed protocolHemi-nested PCR was used to amplify a 236 base-pair segment of apical membrane antigen 1 (AMA-1) using a published protocol (Miller et al., 2017), with modifications (Supplementary file 1a). Samples were amplified in duplicate, indexed, pooled, and purified by bead cleaning. Sequencing was performed on an Illumina MiSeq platform (250 bp paired-end).
Bioinformatics methods
Request a detailed protocolData extraction, processing, and haplotype clustering were performed using SeekDeep (Hathaway et al., 2018), followed by additional filtering (Lerch et al., 2019). Supplementary file 1b shows the full bioinformatics workflow.
Data analysis
Request a detailed protocolA clone was defined as a genetically identical group of parasites, for example with identical haplotypes. Because polyclonal infections can occur due to co-infection (one mosquito bite transmitting multiple clones) or superinfection (multiple bites), we analyzed the infection data both by clone and by infection event. For analysis by clone, each unique clone was counted as an infection and each clone’s disappearance as a clearance event. For analysis by infection event, any new clones seen within 3 visits of the date of the first newly detected clone(s) were grouped together and considered one new ‘infection event.’ Clearance of infection for these events required that all clones in the group be absent. A baseline infection was defined as a clone or infection event (group of clones) detected in the first 60 days of observation of a participant. New infections were defined as a new clone or infection event (group of clones) detected in a participant after day 60.
Imperfect detection of P. falciparum clones is known to be a limitation of all PCR-based genotyping methods; both biological factors, such as deep organ sequestration of clones in the lifecycle of the parasite, and methodological factors, such as difficulty amplifying minority clones (especially at lower parasite densities), results in imperfect detectability of all clones present in a single blood sample drawn on a single day (Felger et al., 2012; Koepfli and Mueller, 2017; Nguyen et al., 2018; Koepfli et al., 2011; Ross et al., 2012). Though amplicon deep-sequencing is less likely than other methods to miss minority clones (Lerch et al., 2017), detectability was a particular concern in our study, in which the majority of infections were submicroscopic and parasite densities declined over time [Supplementary file 1c]. To account for the fact that a clone might be missed in a single sample due to fluctuations in parasite density and/or methodological limitations, we allowed 3 ‘skips’ in detection and classified an infection as cleared only if it was not identified in four contiguous samples from routine visits. If the infection (as determined by clone or by infection event) was absent for four routine samples in a row, the last date of detection would be the end date of that infection. Therefore, for a clone that had previously infected an individual to be classified as a new infection, it had to have been absent from that individual for at least four routine visits. Additional details are found in Supplementary file 1d. This decision was supported by the fact that the diversity of the genotyped AMA-1 amplicon was quite high (expected heterozygosity = 0.949), resulting in a low probability (5.1%) for infection with the same clone by chance.
Data analysis was conducted in R (R Development Core Team, 2019) and Python (Python Language Reference, 2020). Travel was self-reported at routine visits, and LLIN use on the previous night was reported by field entomology teams who surveyed households every two weeks. Microscopic parasite prevalence was defined by the number of smear-positive routine visits over all routine visits. Parasite prevalence by qPCR was defined as the number of qPCR-positive visits over all routine visits. Thus, these measures represent the average prevalence during routine visits. Prevalence ratios were computed using Poisson regression with generalized estimating equations to adjust for repeated measures. Comparison of parasite density by sex was made using linear regression with generalized estimating equations to adjust for repeated measures. Force of infection (FOI) was defined as the number of new infections, including malaria episodes, divided by person time. Since the start of baseline infections was not observed, baseline infections were not included in calculating FOI. Poisson regression with generalized estimating equations to account for repeated measures was used to estimate malaria incidence, FOI, and to calculate incident rate ratios (IRR) for malaria and FOI. Hazards for clearance of untreated, asymptomatic infections were estimated using time-to-event models (shared frailty models fit using R package ‘frailtypack,’ version 2.12.2) (Rondeau and Gonzalez, 2005; Rondeau et al., 2012). Infections were censored for this analysis if they were only observed in the first three months or the last three months (before January 01, 2018 and after January 01, 2019) because they were not observed for long enough to determine whether clearance occurred. If an infection was observed for only one timepoint, it was assigned a duration of 14 days. These models assumed a constant hazard of clearance and included random effects to account for repeated measures in individuals. Parasite density was included in the model as a time-varying covariate. Duration of infection in days was calculated as 1/adjusted hazard.
Data accessibility
Request a detailed protocolData from the cohort study is available through an open-access clinical epidemiology database resource, ClinEpiDB at https://clinepidb.org/ce/app/record/dataset/DS_51b40fe2e2. Genotyping data and code used to generate tables and figures is available on GitHub (Briggs, 2020; copy archived at swh:1:rev:cf6c3256e609f4f136fc8d90f9cae1d61d6d8d63).
Data availability
Data from the PRISM2 cohort study is available through a novel open-access clinical epidemiology database resource here: https://clinepidb.org/ce/app/record/dataset/DS_51b40fe2e2. Sequencing data is available on Github at https://github.com/EPPIcenter/sex_based_differences as referenced in the paper. In addition. all sequences of haplotypes are included in Supplementary file 1.
-
ClinEpiDBID DS_51b40fe2e2. PRISM2 ICEMR Cohort.
References
-
Sex Bias in the outcome of human tropical infectious diseases: influence of steroid hormonesJournal of Infectious Diseases 209 Suppl 3:S107–S113.https://doi.org/10.1093/infdis/jit610
-
Asymptomatic malaria infections: detectability, transmissibility and public health relevanceNature Reviews Microbiology 12:833–840.https://doi.org/10.1038/nrmicro3364
-
Hypoendemic malaria in Rondonia (Brazil, western Amazon region): seasonal variation and risk groups in an urban localityThe American Journal of Tropical Medicine and Hygiene 55:32–38.https://doi.org/10.4269/ajtmh.1996.55.32
-
Epidemiology and burden of malaria in pregnancyThe Lancet Infectious Diseases 7:93–104.https://doi.org/10.1016/S1473-3099(07)70021-X
-
The X-files in immunity: sex-based differences predispose immune responsesNature Reviews Immunology 8:737–744.https://doi.org/10.1038/nri2394
-
SeekDeep: single-base resolution de novo clustering for amplicon deep sequencingNucleic Acids Research 46:e21.https://doi.org/10.1093/nar/gkx1201
-
Malaria epidemiology at the clone levelTrends in Parasitology 33:974–985.https://doi.org/10.1016/j.pt.2017.08.013
-
Parasite density of Plasmodium falciparum malaria in Ghanaian schoolchildren: evidence for influence of sex hormones?Transactions of the Royal Society of Tropical Medicine and Hygiene 88:73–74.https://doi.org/10.1016/0035-9203(94)90505-3
-
WebsiteThe garki project : research on the epidemiology and control of malaria in the Sudan savanna of west africaWorld Health Organization. Accessed February 18, 2020.
-
Incidence patterns of vivax malaria in civilians residing in a high-risk county of Kyonggi-do (province), Republic of koreaThe Korean Journal of Parasitology 39:293–299.https://doi.org/10.3347/kjp.2001.39.4.293
-
Persistent parasitemia despite dramatic reduction in malaria incidence after 3 rounds of indoor residual spraying in Tororo, UgandaThe Journal of Infectious Diseases 219:1104–1111.https://doi.org/10.1093/infdis/jiy628
-
Malaria transmission, infection, and disease following sustained indoor residual spraying of insecticide in Tororo, UgandaThe American Journal of Tropical Medicine and Hygiene 103:1525–1533.https://doi.org/10.4269/ajtmh.20-0250
-
Biological differences between the sexes and susceptibility to tuberculosisJournal of Infectious Diseases 209 Suppl 3:S100–S106.https://doi.org/10.1093/infdis/jiu147
-
SoftwareR: A language and environment for statistical computingR Foundation for Statistical Computing, Vienna, Austria.
-
Sex-associated hormones and immunity to protozoan parasitesClinical Microbiology Reviews 14:476–488.https://doi.org/10.1128/CMR.14.3.476-488.2001
-
Frailtypack: a computer program for the analysis of correlated failure time data using penalized likelihood estimationComputer Methods and Programs in Biomedicine 80:154–164.https://doi.org/10.1016/j.cmpb.2005.06.010
-
Effect of insecticide-treated bed nets on the dynamics of multiple Plasmodium falciparum infectionsTransactions of the Royal Society of Tropical Medicine and Hygiene 93 Suppl 1:53–57.https://doi.org/10.1016/S0035-9203(99)90328-0
-
High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reactionMolecular and Biochemical Parasitology 61:315–320.https://doi.org/10.1016/0166-6851(93)90077-B
-
Sex differences in parasite infections: patterns and processesInternational Journal for Parasitology 26:1009–1024.https://doi.org/10.1016/S0020-7519(96)80001-4
Article and author information
Author details
Funding
National Institute of Allergy and Infectious Diseases (T32 AI007641-16)
- Jessica Briggs
National Institute of Allergy and Infectious Diseases (U19AI089674)
- Grant Dorsey
Fogarty International Center (K43TW010365)
- Joaniter I Nankabirwa
Fogarty International Center (D43TW010526)
- Emmanuel Arinaitwe
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Ethics
Human subjects: IRB approval for the PRISM2 cohort study was obtained in Uganda (UNCST HS-119ES, SOMREC: 2017-099), UK (LSHTM: 14266), and US (UCSF IRB: 17-22544, Stanford: UCSF IRB reliance). Informed consent was obtained from all participants prior to enrollment in the study per IRB guidelines.
Copyright
© 2020, Briggs et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,677
- views
-
- 302
- downloads
-
- 62
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Epidemiology and Global Health
Background:
Biological aging exhibits heterogeneity across multi-organ systems. However, it remains unclear how is lifestyle associated with overall and organ-specific aging and which factors contribute most in Southwest China.
Methods:
This study involved 8396 participants who completed two surveys from the China Multi-Ethnic Cohort (CMEC) study. The healthy lifestyle index (HLI) was developed using five lifestyle factors: smoking, alcohol, diet, exercise, and sleep. The comprehensive and organ-specific biological ages (BAs) were calculated using the Klemera–Doubal method based on longitudinal clinical laboratory measurements, and validation were conducted to select BA reflecting related diseases. Fixed effects model was used to examine the associations between HLI or its components and the acceleration of validated BAs. We further evaluated the relative contribution of lifestyle components to comprehension and organ systems BAs using quantile G-computation.
Results:
About two-thirds of participants changed HLI scores between surveys. After validation, three organ-specific BAs (the cardiopulmonary, metabolic, and liver BAs) were identified as reflective of specific diseases and included in further analyses with the comprehensive BA. The health alterations in HLI showed a protective association with the acceleration of all BAs, with a mean shift of –0.19 (95% CI −0.34, –0.03) in the comprehensive BA acceleration. Diet and smoking were the major contributors to overall negative associations of five lifestyle factors, with the comprehensive BA and metabolic BA accounting for 24% and 55% respectively.
Conclusions:
Healthy lifestyle changes were inversely related to comprehensive and organ-specific biological aging in Southwest China, with diet and smoking contributing most to comprehensive and metabolic BA separately. Our findings highlight the potential of lifestyle interventions to decelerate aging and identify intervention targets to limit organ-specific aging in less-developed regions.
Funding:
This work was primarily supported by the National Natural Science Foundation of China (Grant No. 82273740) and Sichuan Science and Technology Program (Natural Science Foundation of Sichuan Province, Grant No. 2024NSFSC0552). The CMEC study was funded by the National Key Research and Development Program of China (Grant No. 2017YFC0907305, 2017YFC0907300). The sponsors had no role in the design, analysis, interpretation, or writing of this article.
-
- Epidemiology and Global Health
- Microbiology and Infectious Disease
Background:
In many settings, a large fraction of the population has both been vaccinated against and infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Hence, quantifying the protection provided by post-infection vaccination has become critical for policy. We aimed to estimate the protective effect against SARS-CoV-2 reinfection of an additional vaccine dose after an initial Omicron variant infection.
Methods:
We report a retrospective, population-based cohort study performed in Shanghai, China, using electronic databases with information on SARS-CoV-2 infections and vaccination history. We compared reinfection incidence by post-infection vaccination status in individuals initially infected during the April–May 2022 Omicron variant surge in Shanghai and who had been vaccinated before that period. Cox models were fit to estimate adjusted hazard ratios (aHRs).
Results:
275,896 individuals were diagnosed with real-time polymerase chain reaction-confirmed SARS-CoV-2 infection in April–May 2022; 199,312/275,896 were included in analyses on the effect of a post-infection vaccine dose. Post-infection vaccination provided protection against reinfection (aHR 0.82; 95% confidence interval 0.79–0.85). For patients who had received one, two, or three vaccine doses before their first infection, hazard ratios for the post-infection vaccination effect were 0.84 (0.76–0.93), 0.87 (0.83–0.90), and 0.96 (0.74–1.23), respectively. Post-infection vaccination within 30 and 90 days before the second Omicron wave provided different degrees of protection (in aHR): 0.51 (0.44–0.58) and 0.67 (0.61–0.74), respectively. Moreover, for all vaccine types, but to different extents, a post-infection dose given to individuals who were fully vaccinated before first infection was protective.
Conclusions:
In previously vaccinated and infected individuals, an additional vaccine dose provided protection against Omicron variant reinfection. These observations will inform future policy decisions on COVID-19 vaccination in China and other countries.
Funding:
This study was funded the Key Discipline Program of Pudong New Area Health System (PWZxk2022-25), the Development and Application of Intelligent Epidemic Surveillance and AI Analysis System (21002411400), the Shanghai Public Health System Construction (GWVI-11.2-XD08), the Shanghai Health Commission Key Disciplines (GWVI-11.1-02), the Shanghai Health Commission Clinical Research Program (20214Y0020), the Shanghai Natural Science Foundation (22ZR1414600), and the Shanghai Young Health Talents Program (2022YQ076).