A novel motif of Rad51 serves as an interaction hub for recombination auxiliary factors
Figures
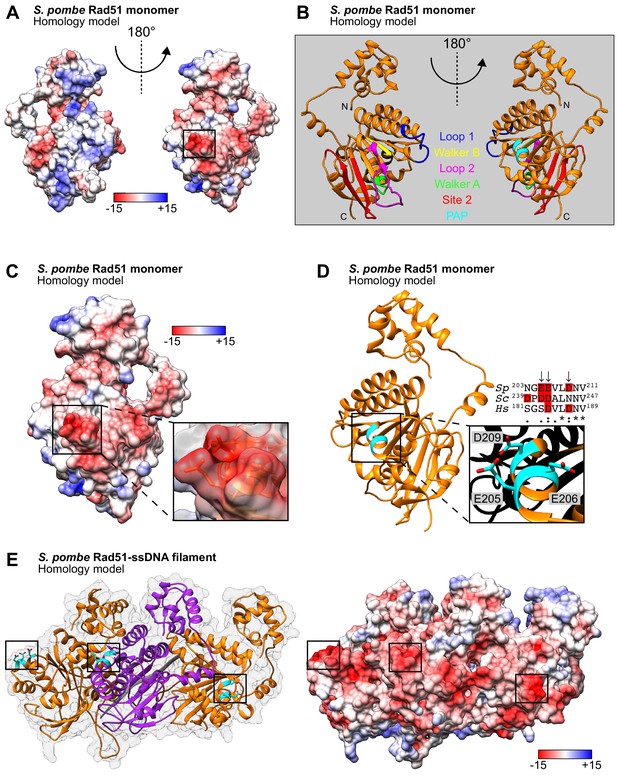
E205, E206, and D209 form a protruding acidic patch (PAP) on the exterior of the Rad51 presynaptic filament.
(A) Homology model of an SpRad51 monomer (residues 42–360). Surface representation colored according to Coulombic surface charge. The molecule on the left is rotated 180° to visualize the PAP on the right, with the region of interest squared. (B) Ribbon depiction of an SpRad51 monomer with relevant motifs highlighted. The molecules are oriented as in (A). (C) Surface representation colored according to Coulombic surface charge. The PAP is enlarged with a semi-transparent surface revealing residues E205, E206, and D209, which have their side-chains shown. (D) Ribbon depiction of SpRad51 with the α-helix containing the PAP enlarged to illustrate the respective positions of each residue (colored in cyan) with their side-chains revealed (O atoms in red). Sequence alignment shows the corresponding region in S. pombe, S. cerevisiae and H. sapiens (Sp, Sc, and Hs, respectively), with arrows indicating PAP residues in S. pombe and acidic residues highlighted in red. (E) Ribbon depiction of three SpRad51 monomers (alternating orange and purple) bound to ssDNA (9-mer poly-dT in gray) with a near-transparent surface for visualization (left). The side-chains of E205, E206, and D209 are revealed in cyan (O atoms in red) and their positions are squared. The surface is made opaque and colored according to Coulombic surface charge to demonstrate that the PAP constitutes dense, negatively charged regions on the exterior of the ssDNA filament (right). Numbers in the legends for (A,C,E) are in units of kcal/(mol•e).
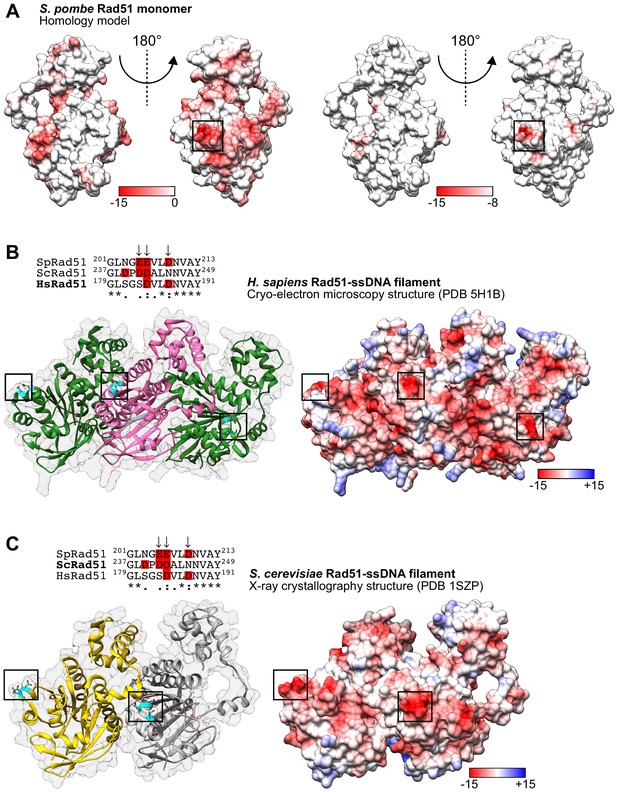
The PAP is conserved in the HsRad51 and ScRad51 presynaptic filaments.
(A) Surface representation of an SpRad51 monomer colored according to Coulombic surface charge. The squared region forms a protruding acidic patch (PAP) that is among the most negatively charged regions on the surface of SpRad51 and corresponds to residues E205, E206, and D209. (B) Ribbon depiction of three HsRad51 monomers (alternating green and pink) bound to ssDNA (9-mer poly-dT in gray) with a near-transparent surface (left). The side-chains of residues indicated in the sequence alignment are revealed in cyan (O atoms in red) and their positions are highlighted by squares. The surface is made opaque and colored according to Coulombic surface charge to demonstrate that these residues constitute negatively charged patches on the exterior of the ssDNA filament (right). In the sequence alignment, arrows indicate PAP residues in S. pombe and acidic residues are highlighted in red. (C) Ribbon depiction of two ScRad51 monomers (yellow and gray) with a near-transparent surface (left) bound to ssDNA (not shown). The side-chains of residues indicated in the sequence alignment are revealed in cyan (O atoms in red), and their positions are highlighted by squares. The surface is made opaque and colored according to Coulombic surface charge to demonstrate that these residues constitute negatively charged patches on the exterior of the ssDNA filament (right). In the sequence alignment, arrows indicate PAP residues in S. pombe and acidic residues are highlighted in red. Numbers in the legends of (A–C) are in units of kcal/(mol•e).
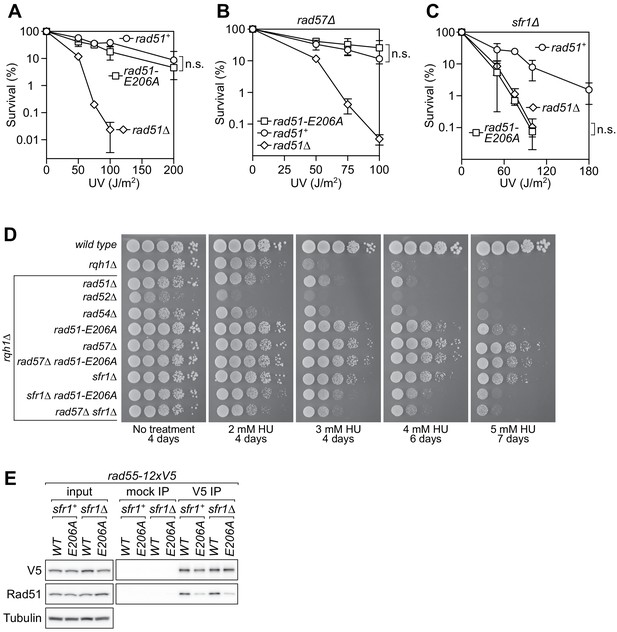
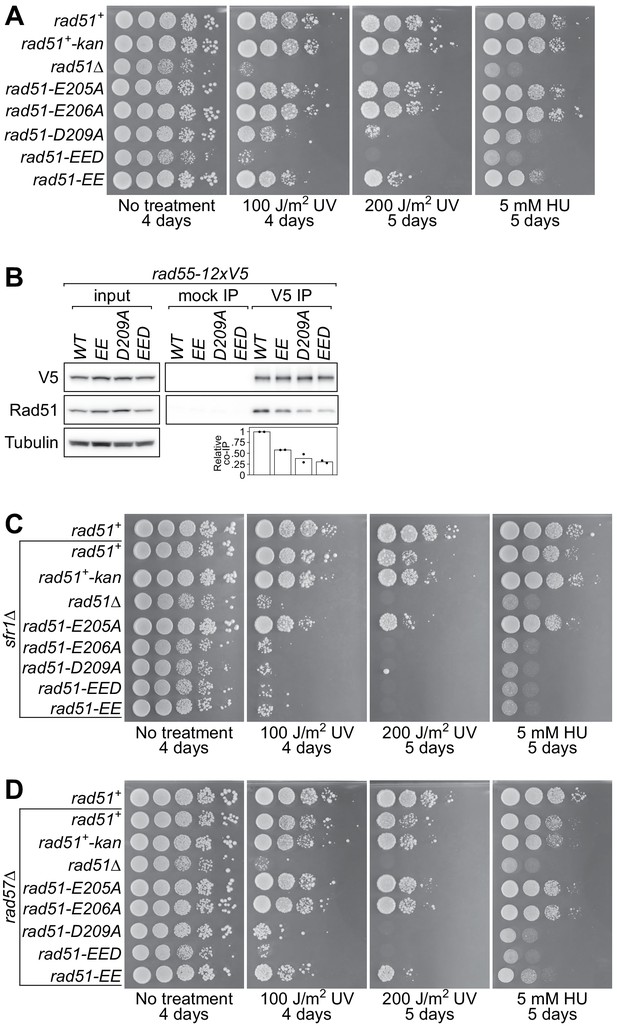
The rad51-E206A mutant is specifically defective in the interaction with Rad55-Rad57.
(A–C) Following acute UV irradiation, a clonogenic assay was employed to test the survival of rad51+, rad51-E206A, and rad51Δ in the wild-type background (A), the rad57Δ background (B), and the sfr1Δ background (C). Statistical significance at the highest dose of UV was assessed by unpaired two-tailed t-test. n.s., not significant (A, p=0.506; B, p=0.242; C, p=0.593). (D) Tenfold serial dilutions of the indicated strains were spotted onto standard media without treatment or containing hydroxyurea (HU). Following growth for the indicated time at 30°C, plates were imaged. (E) Soluble cell extracts treated with a benzonase-like nuclease were prepared from each strain under native conditions (input). Immunoprecipitation (IP) was then performed with mock (human IgG from non-immunized animal) or anti-V5 antibodies. Tubulin serves as a loading control. Data in (A–C) are means of three independent experiments and error bars depict standard deviation.
-
Figure 2—source data 1
Survival (%) following UV irradiation for data in Figure 2A–C.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig2-data1-v1.xlsx
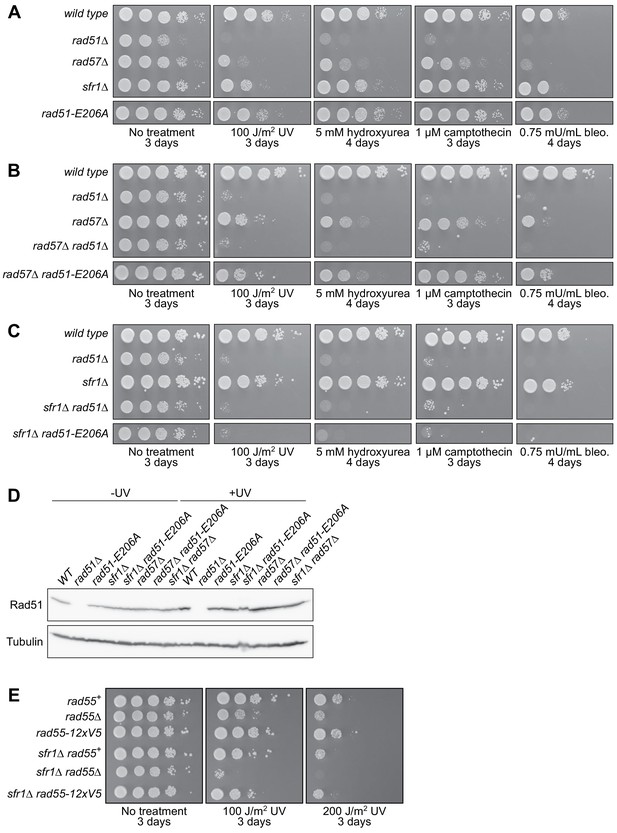
Specificity of the DNA damage sensitivity of rad51-E206A.
(A–C, E) Tenfold serial dilutions of each strain were spotted onto standard media containing DNA damaging agents, or onto standard media with or without acute UV exposure, as indicated. Following growth at 30°C for the indicated time, plates were imaged. Although cropped separately from other strains, the respective rad51-E206A strains were spotted onto the same plate in each case. Bleo., bleomycin. (D) The indicated strains were grown to log-phase, at which point cultures were split into two. One sub-culture was left untreated, and the other was UV-irradiated (200 J/m2). Following recovery for 3 hr at 30°C, cellular levels of Rad51 were examined by immunoblotting. Tubulin serves as a loading control.
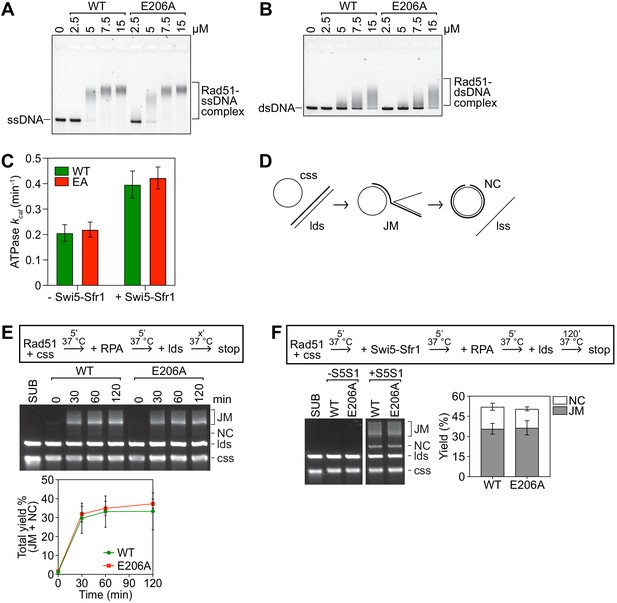
Rad51-E206A retains normal DNA binding and recombinase activity.
(A,B) The indicated concentrations of Rad51 (WT) or Rad51-E206A (E206A) were incubated with 30 micromolar nucleotide (µM nt) PhiX174 virion DNA (ssDNA; A) or 20 µM nt of linearized PhiX174 RF I DNA (dsDNA; B), protein-DNA complexes were crosslinked with glutaraldehyde and then resolved by agarose gel electrophoresis. (C) The ATPase activity of 5 µM Rad51 (WT) and Rad51-E206A (EA) was measured in the presence of ssDNA (10 µM nt), with or without Swi5-Sfr1 (0.5 µM), and kcat was calculated. (D) Schematic of the strand exchange assay with full-length DNA substrates (PhiX174 virion ssDNA and ApaLI-linearized PhiX174 RF I dsDNA). (E) Strand exchange reactions were conducted according to the scheme outlined above the gel. Rad51 (WT or E206A), 15 µM. RPA, 1 µM. cssDNA, 30 µM nt. ldsDNA, 20 µM nt. (F) Strand exchange reactions were conducted according to the scheme outlined above the gel. Rad51 (WT or E206A), 5 µM. Swi5-Sfr1 (S5S1), 0.5 µM. RPA, 1 µM. cssDNA, 10 µM nt. ldsDNA, 10 µM nt. Data in (C,E,F) are means of three independent experiments and error bars depict standard deviation.
-
Figure 3—source data 1
kcat values in Figure 3C and strand exchange yield (%) in Figure 3E,F.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig3-data1-v1.xlsx
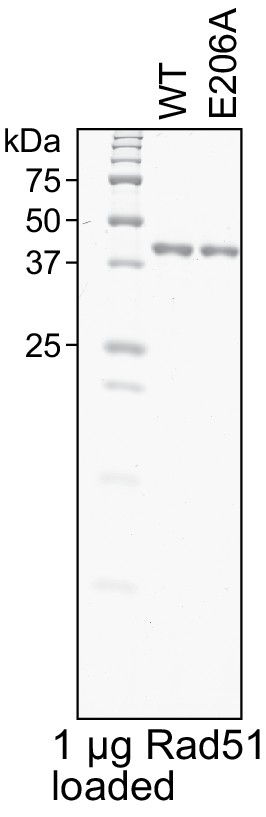
Purity analysis of Rad51 and Rad51-E206A.
Purified Rad51 (WT) and Rad51-E206A (E206A) were examined by SDS-PAGE and coomassie staining.
The PAP is essential for Rad51-dependent DNA repair.
(A,C,D) Tenfold serial dilutions of the indicated strains were spotted onto standard media with or without acute UV irradiation, or standard media containing 5 mM hydroxyurea (HU). Following growth at 30°C for the indicated time, plates were imaged. (B) Soluble cell extracts treated with a benzonase-like nuclease were prepared from each strain under native conditions (input). Immunoprecipitation (IP) was then performed with mock (human IgG from non-immunized animal) or anti-V5 antibodies. Tubulin serves as a loading control. For quantification, Rad51 signal was normalized to V5 signal and expressed relative to wild type. Data in (B) are the means of two independent biological replicates with individual values shown.
-
Figure 4—source data 1
Relative co-IP of Rad51 (%) for Figure 4B.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig4-data1-v1.xlsx
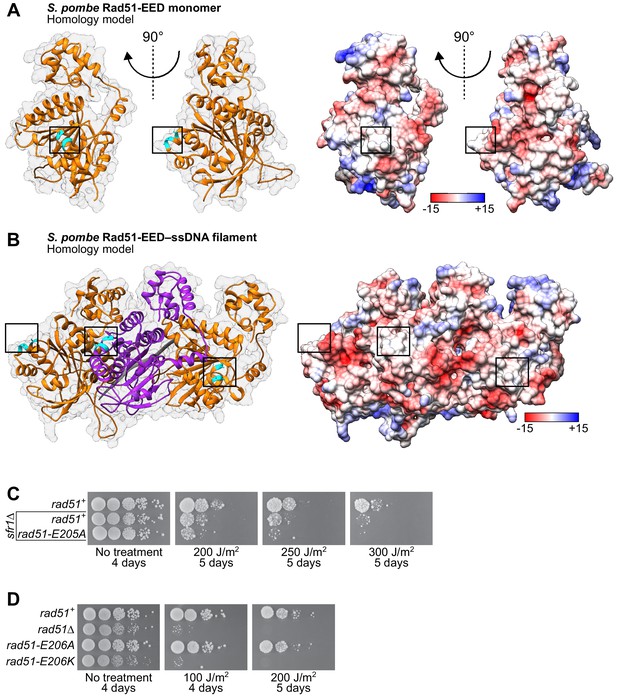
The EED mutation neutralizes the PAP of SpRad51 and is phenocopied by the E206K mutation.
(A) Ribbon depiction of an SpRad51-EED monomer or (B) three SpRad51-EED monomers (alternating orange and purple) bound to ssDNA (9-mer poly-dT in gray), both with a near-transparent surface (left) with the side-chains of residues EED (E205A, E206A, D209A) revealed in cyan. The surface is made opaque and colored according to Coulombic surface charge (right) to demonstrate that the EED mutation neutralizes the negative charge of the PAP. (C,D) Tenfold serial dilutions of the indicated strains were spotted onto standard media with or without acute UV irradiation. Following growth at 30°C for the indicated time, plates were imaged. Numbers in the legends of (A) and (B) are in units of kcal/(mol•e).
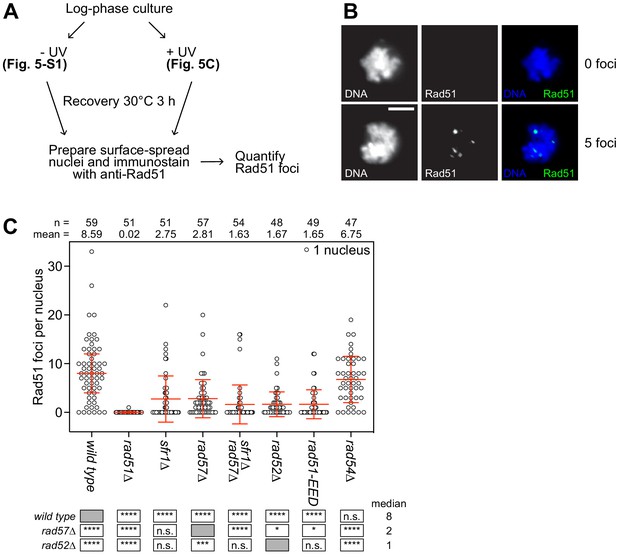
The PAP is critical for the recruitment of Rad51 to sites of DNA damage.
(A) Schematic of cytological analysis. (B) Representative images of UV-irradiated nuclear spreads containing 0 or 5 Rad51 foci. Scale bar, 5 µm. (C) The indicated strains were grown to log phase and cultures were split into two. One sub-culture was UV-irradiated (200 J/m2; shown here) and the other was not (shown in Figure 5—figure supplement 1). Following 3 hr of recovery, nuclei were surface-spread and immunostained with a polyclonal PVDF-purified anti-Rad51 antibody. The indicated number of nuclei were manually picked, images in the DAPI and Rad51 channels were captured, and semi-automated quantification of foci was performed using FIJI software. Bars depict the median and interquartile range. Mean number of foci are shown for each strain. Statistical analysis was by Wilcoxon ranked sum test with the indicated median values. n.s., not significant (p>0.05). * p<0.05. ***p<0.001. ****p<0.0001.
-
Figure 5—source data 1
Number of foci per nucleus following UV irradiation for Figure 5C.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig5-data1-v1.xlsx
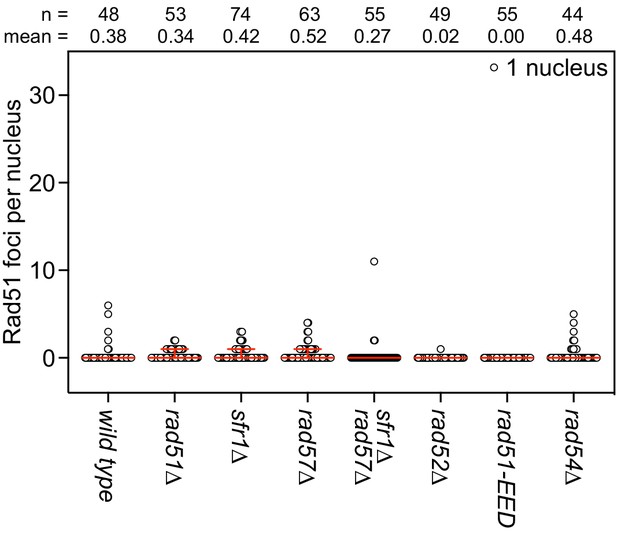
DNA damage dependency of Rad51 foci.
The indicated strains were grown to log phase and cultures were split into two. One sub-culture was UV-irradiated (200 J/m2; shown in Figure 5C) and the other was not (shown here). Following 3 hr of recovery, nuclei were surface-spread and immunostained with an affinity purified anti-Rad51 antibody. The indicated number of nuclei were manually picked, images in the DAPI and Rad51 channels were captured, and semi-automated quantification of foci was performed using FIJI software. Bars depict the median and interquartile range. Mean number of foci are shown for each strain.
-
Figure 5—figure supplement 1—source data 1
Number of foci per nucleus without UV irradiation.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig5-figsupp1-data1-v1.xlsx
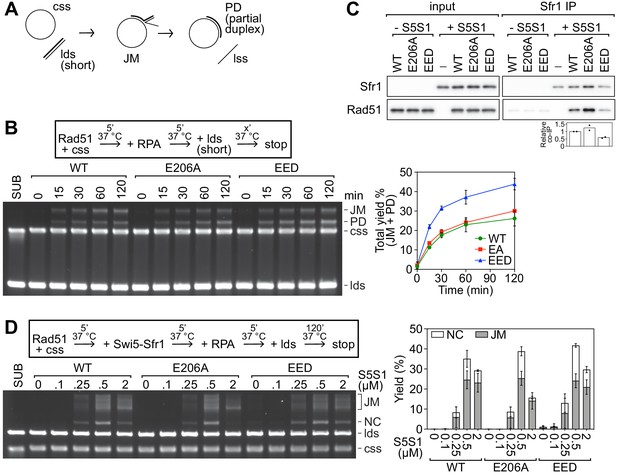
Neutralization of the PAP does not impair the intrinsic activity of Rad51 or the stimulation by Swi5-Sfr1.
(A) Schematic of the strand exchange assay with shortened DNA substrates (PhiX174 virion ssDNA and a 1.6 kb fragment of PhiX RF I dsDNA). (B) Strand exchange reactions were conducted according to the scheme outlined above the gel. Rad51 (WT, E206A, or EED), 15 µM. RPA, 1 µM. cssDNA, 30 µM nt. ldsDNA, 20 µM nt. (C) Purified Rad51 (WT, E206A, or EED) was incubated with purified Swi5-Sfr1 (or the equivalent volume of protein storage buffer) and subjected to immunoprecipitation with an anti-Sfr1 antibody. For quantification, Rad51 signal was normalized to Sfr1 signal and expressed relative to wild type. (D) Strand exchange reactions were conducted according to the scheme outlined above the gel. Rad51 (WT, E206A, or EED), 5 µM. Swi5-Sfr1 (S5S1), indicated. RPA, 1 µM. cssDNA, 10 µM nt. ldsDNA, 10 µM nt. Data in (B,D) are means of three independent experiments and error bars depict standard deviation. Data in (C) are the means of two independent experiments with individual values shown.
-
Figure 6—source data 1
Strand exchange yield (%) in Figure 6B,D and relative co-IP of Rad51 (%) for Figure 6C.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig6-data1-v1.xlsx
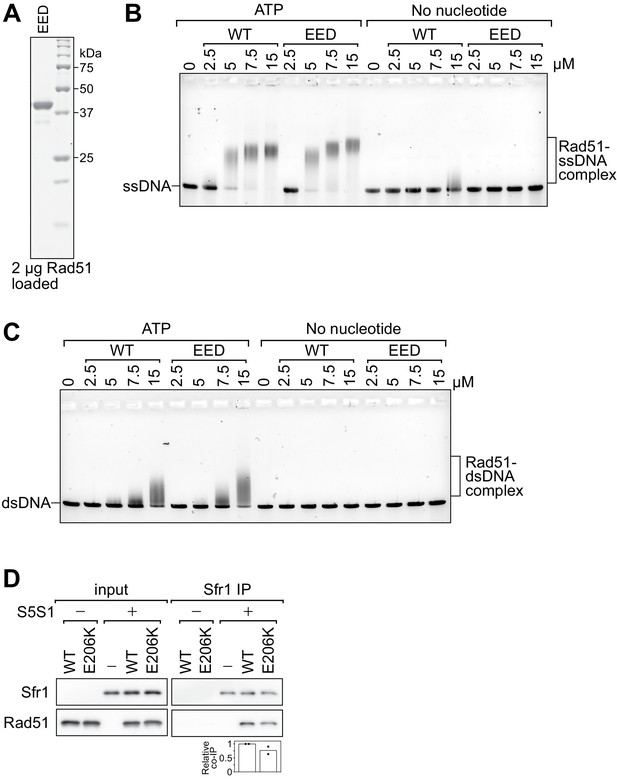
Rad51-EED binds both ssDNA and dsDNA like wild-type Rad51.
(A) Purified Rad51-EED was examined by SDS-PAGE and coomassie staining. (B, C) The indicated concentrations of Rad51 (WT) or Rad51-EED (EED) were incubated with 30 µM nt PhiX174 virion DNA (ssDNA; B) or 20 µM nt of linearized PhiX174 RF I DNA (dsDNA; C) in the presence or absence of ATP. Protein-DNA complexes were crosslinked with glutaraldehyde and then resolved by agarose gel electrophoresis. (D) Purified Rad51 (WT or E206K) was incubated with purified Swi5-Sfr1 (or the equivalent volume of protein storage buffer) and subjected to immunoprecipitation with an anti-Sfr1 antibody. For quantification, Rad51 signal was normalized to Sfr1 signal and expressed relative to wild type. Data in (D) are the means of two independent experiments with individual values shown.
-
Figure 6—figure supplement 1—source data 1
Relative co-IP of Rad51 (%) for Figure 6—figure supplement 1D.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig6-figsupp1-data1-v1.xlsx
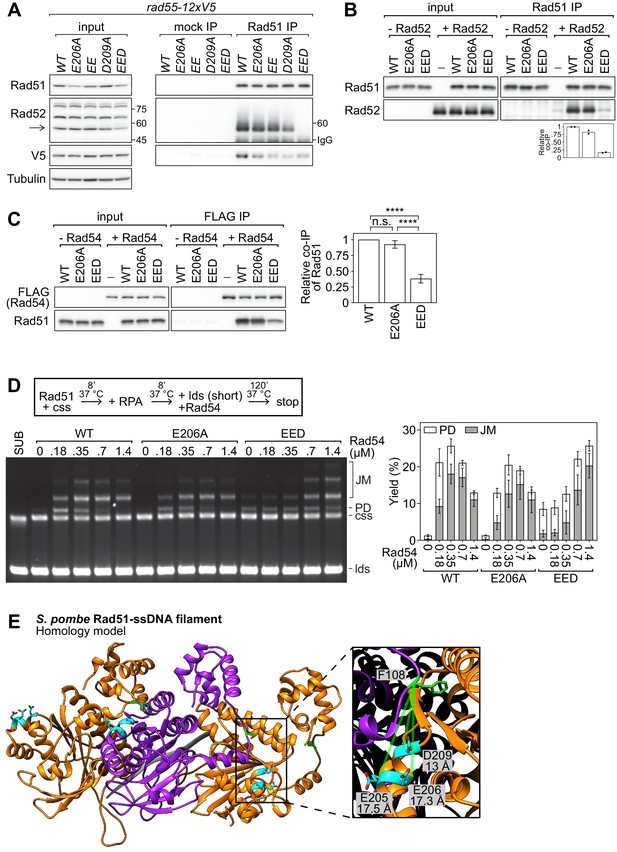
The PAP facilitates the interaction of Rad51 with Rad52 and Rad54.
(A) Soluble cell extracts treated with a benzonase-like nuclease were prepared from each strain under native conditions (input). Immunoprecipitation (IP) was then performed with mock (human IgG from non-immunized animal) or anti-Rad51 antibodies. Arrow denotes the Rad52 band (other bands were still observed in a rad52∆ strain, see Figure 7—figure supplement 1A) and numbers denote the position of the size markers in kDa. To ensure separation of Rad52 from the IgG heavy chain band, the IP samples were separated by 7% SDS-PAGE instead of the 12% SDS-PAGE employed for the input samples. As a result, the 45 kDa and 75 kDa bands of the size marker are not within the cropped area of the anti-Rad52 immunoblot for the IP sample. Tubulin serves as a loading control. (B) Purified Rad51 (WT, E206A, or EED) was incubated with purified Rad52 (or the equivalent volume of protein storage buffer) and subjected to immunoprecipitation with anti-Rad51 antibody. For quantification, Rad51 signal was normalized to Sfr1 signal and expressed relative to wild type. (C) Purified Rad51 (WT, E206A, or EED) was incubated with purified FLAG-Rad54 (or the equivalent volume of protein storage buffer) and subjected to immunoprecipitation with FLAG-agarose resin. For quantification, Rad51 signal was normalized to FLAG-Rad54 signal and expressed relative to wild type. Statistical significance was assessed by one-way ANOVA with Tukey’s multiple comparisons test. n.s., not significant (p=0.2538). ****p<0.0001. (D) Strand exchange reactions were conducted according to the scheme outlined above the gel. Rad51 (WT, E206A, or EED), 7 µM. Rad54, indicated. RPA, 0.7 µM. cssDNA, 7 µM nt. ldsDNA, 14 µM nt. (E) Ribbon depiction of three SpRad51 monomers (alternating orange and purple) bound to ssDNA (9-mer poly-dT in gray). Residues E205, E206, and D209 are colored in cyan with their side-chains shown (O atoms in red), and F108 is colored in green with its side-chain shown. The α-helix of the rightmost monomer containing E205, E206, and D209 is enlarged, along with the F108 residue of the central monomer, to illustrate the close proximity of these residues at the subunit interface. Dotted-green lines show the Cα-Cα distance between each residue and F108. The Rad51 filament is employed as a proxy to demonstrate that insertion of Phe in the FxxA motif of auxiliary factors could be facilitated by interactions with the PAP. Data in (B) are means of two independent experiments with individual values shown. Data in (C,D) are means of three independent experiments and error bars depict standard deviation.
-
Figure 7—source data 1
Relative co-IP of Rad52 and Rad51 in Figure 7B,C, and strand exchange yield (%) in Figure 7D.
- https://cdn.elifesciences.org/articles/64131/elife-64131-fig7-data1-v1.xlsx
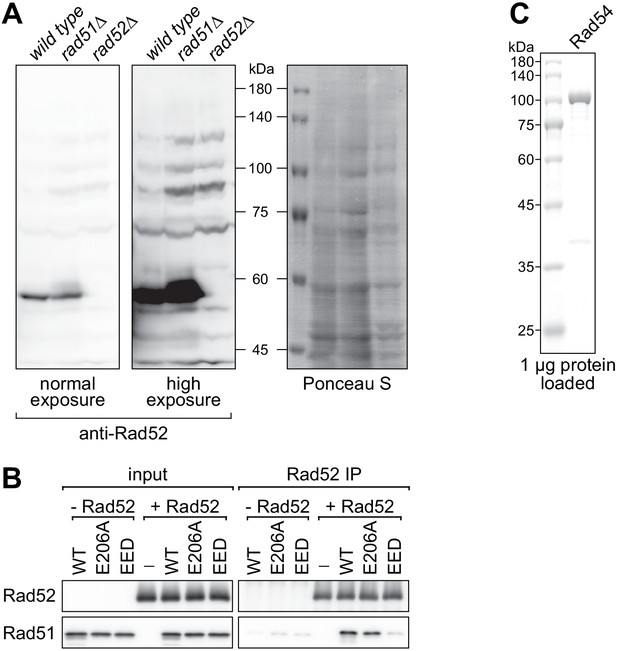
Anti-Rad52 antibody verification and Rad54 purity analysis.
(A) Cellular proteins were processed by the TCA method and subjected to immunoblotting with a polyclonal anti-Rad52 antibody to identify the Rad52 band. Ponceau S serves as a loading control. (B) Purified Rad51 (WT, E206A, or EED) was incubated with purified Rad52 (or the equivalent volume of protein storage buffer) and subjected to immunoprecipitation with anti-Rad52 antibody. (C) Purified FLAG-Rad54 was examined by SDS-PAGE and coomassie staining.
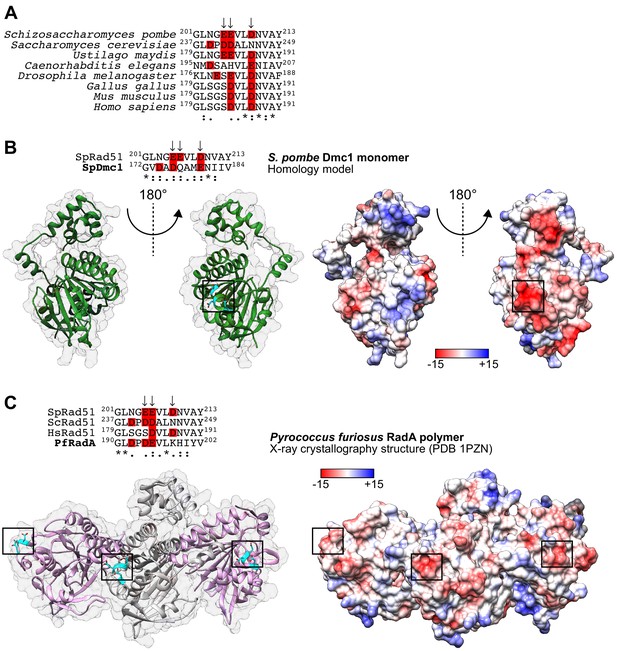
The PAP is conserved in eukaryotic Dmc1 and archaeal RadA.
(A) Sequence alignment of Rad51 from the indicated eukaryotic species. Arrows indicate PAP residues in S. pombe and acidic residues are highlighted in red. (B) Homology model of an SpDmc1 monomer (residues 15–332). Ribbon depictions (left) are in the same orientation as the surface representation colored according to Coulombic surface charge (right). In each case, the molecule on the left is rotated 180° to visualize the protruding acidic patch (PAP) on the right, with the region of interest squared. The side-chains of residues indicated in the sequence alignment are revealed in cyan (O atoms in red) and their positions are highlighted by squares. In the sequence alignment, arrows indicate PAP residues in S. pombe and acidic residues are highlighted in red. (C) Ribbon depiction of three Pyrococcus fusiosus RadA monomers (alternating pink and gray) with a near-transparent surface (left). The side-chains of residues indicated in the sequence alignment are revealed in cyan (O atoms in red) and their positions are highlighted in squares. The surface is made opaque and colored according to Coulombic surface charge to demonstrate that these residues constitute negatively charged patches on the exterior of the ssDNA filament (right). In the sequence alignment, arrows indicate PAP residues in S. pombe and acidic residues are highlighted in red. Numbers in the legends of (B,C) are in units of kcal/(mol•e).
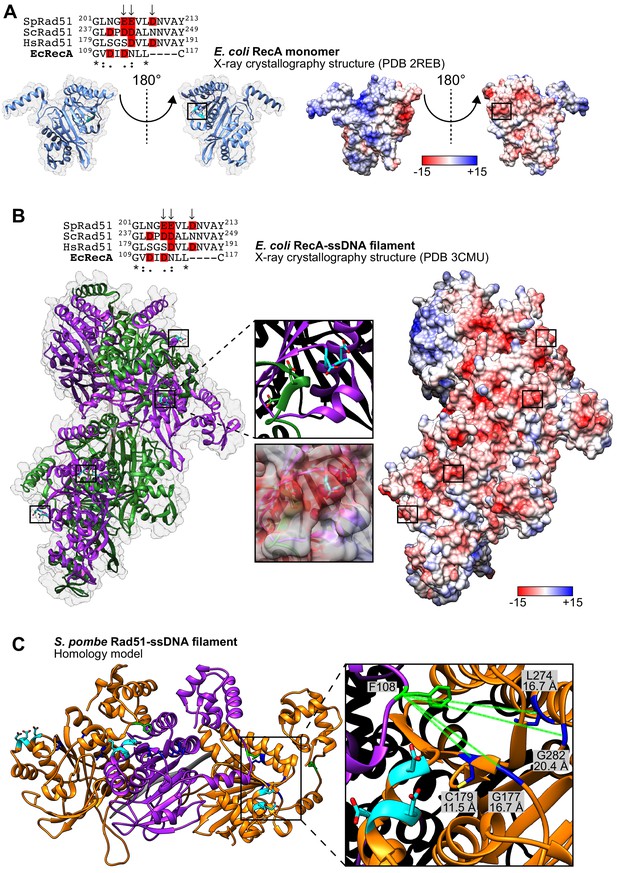
The PAP is not conserved in bacterial RecA.
(A) Ribbon depictions of the E. coli RecA monomer (left) are in the same orientation as the surface representation colored according to Coulombic surface charge (right). In each case, the molecule on the left is rotated 180° to visualize the equivalent of the protruding acidic patch (PAP) on the right, with the region of interest squared. The side-chains of residues indicated in the sequence alignment are revealed in cyan (O atoms in red) and their positions are highlighted by squares. In the sequence alignment, arrows indicate PAP residues in S. pombe and acidic residues are highlighted in red. (B) Ribbon depiction of six E. coli RecA monomers (alternating purple and green) with a near-transparent surface (left) bound to ssDNA (15-mer poly-dT in gray). The side-chains of residues indicated in the sequence alignment are revealed in cyan (O atoms in red) and their positions in four monomers are highlighted in squares. One such region is enlarged to demonstrate that the indicated residues form a loop rather than the short α-helix observed in the monomer structure in (A). The surface is made opaque and colored according to Coulombic surface charge to demonstrate that these residues nevertheless constitute negatively charged patches on the exterior of the ssDNA filament (right), although closer examination reveals that this is partly due to two acidic residues in the adjacent monomer (shown enlarged). In the sequence alignment, arrows indicate PAP residues in S. pombe and acidic residues are highlighted in red. (C) Ribbon depiction of three SpRad51 monomers (alternating orange and purple) bound to ssDNA (9-mer poly-dT in gray). Residues E205, E206, and D209 are colored in cyan with their side-chains shown (O atoms in red), and F108 is colored in green with its side-chain shown. Residues G177, C179, G282, and L274 (Kim et al., 2002) and their side-chains are shown in blue (S atom in yellow), with dotted-green lines depicting the Cα-Cα distances from F108. Numbers in the legends of (A,B) are in units of kcal/(mol−1•e).
Videos
A model of the SpRad51 monomer with motifs labeled.
A homology model of the SpRad51 monomer is depicted in ribbon form and labeled as follows: N and C, N- and C-termini; L1 and L2, loop 1 and loop 2 of DNA binding site 1; A and B, Walker A and Walker B motifs; Site 2, DNA binding site 2. Residues E205, E206, and D209, which constitute the protruding acidic patch (PAP), are shown in cyan following 180° rotation of the model in the y-axis.
Visualization of the protruding acidic patch (PAP) in monomer form.
A homology model of the SpRad51 monomer is depicted in ribbon form. The side-chains of residues E205, E206, and D209 are shown, intermittently highlighted in cyan (O atoms in red). The surface is made opaque and colored according to Coulombic surface charge to demonstrate that these residues constitute a dense negatively charged patch that we refer to as the PAP.
Visualization of the protruding acidic patch (PAP) in filament form.
A homology model of the SpRad51-ssDNA filament. Ribbon depiction of three monomers (alternating orange and purple) bound to ssDNA (9-mer poly-dT in gray). The side-chains of residues E205, E206, and D209 are shown, intermittently highlighted in cyan. The surface is made opaque and colored according to Coulombic surface charge to demonstrate that these residues constitute dense negatively charged patches on the exterior of the ssDNA filament.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Schizosaccharomyces pombe; Msmt-0 | Wild type | Akamatsu et al., 2003 | NA310 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-E206A-kanMX6 | This study | NA211 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51::hphMX6 | This study | BA263 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-E206A- kanMX6 rad57::hphMX6 | This study | NA253 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad57::hphMX6 | This study | BA268 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad57::hphMX6 rad51::natMX6 | This study | NA383 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::kanMX6 | This study | NA138 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::kanMX6 rad51::hphMX6 | This study | NA233 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-E206A-kanMX6 sfr1::kanMX6 | This study | NA236 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | Wild type | This study | BA396 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 | This study | NA324 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 rad51::hphMX6 | This study | NA394 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 rad52::arg3 | This study | BA546 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 rad54::ura4 | This study | BA548 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 rad51-E206A-kanMX6 | This study | NA387 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 rad57::hphMX6 | This study | NA396 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 rad57::hphMX6 rad51-E206A-kanMX6 | This study | NA389 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 sfr1::kanMX6 | This study | NA386 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 sfr1::kanMX6 rad51-E206A-kanMX6 | This study | NA398 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; mat1PD17::LEU2 | rqh1::natMX6 rad57::hphMX6 sfr1::kanMX6 | This study | NA391 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad52::arg3 | Hiroshi Iwasaki (lab stock) | BA481 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad54::ura4 | Hiroshi Iwasaki (lab stock) | BA518 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-E205A-E206A-D209A-kanMX6 | This study | BA499 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51+-kanMX6 | This study | NA209 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-E205A-kanMX6 | This study | BA445 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-D209A-kanMX6 | This study | BA493 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-E205A-E206A-kanMX6 | This study | BA449 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 | This study | BA172 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 rad51-E206A-kanMX6 | This study | NA238 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 sfr1::kanMX6 | This study | BA170 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 rad51-E206A-kanMX6 sfr1::kanMX6 | This study | NA243 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 rad51-E205A-E206A-kanMX6 | This study | BA529 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 rad51 D209A-kanMX6 | This study | BA521 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 rad51-E205A-E206A-D209A-kanMX6 | This study | BA523 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad57::hphMX6 rad51+-kanMX6 | This study | NA249 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad57::hphMX6 rad51-E205A-kanMX6 | This study | BA443 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad57::hphMX6 rad51-D209A-kanMX6 | This study | BA491 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad57::hphMX6 rad51-E205A-E206A-D209A-kanMX6 | This study | BA501 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad57::hphMX6 rad51-E205A-E206A-kanMX6 | This study | BA447 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::kanMX6 rad51+-kanMX6 | This study | NA222 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::natMX6 rad51-E205A-kanMX6 | This study | BA461 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::natMX6 rad51-E206A-kanMX6 | This study | BA476 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::natMX6 rad51-D209A-kanMX6 | This study | BA485 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::nathMX6 rad51-E205A-E206A-kanMX6 | This study | BA463 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | sfr1::natMX6 rad51-E205A-E206A-D209A-kanMX6 | This study | BA497 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55::natMX6 | Argunhan et al., 2020 | BA150 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55-12xV5-natMX6 | This study | BA165 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad55::arg3 sfr1::kanMX6 | Argunhan et al., 2020 | BA126 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| S. pombe; Msmt-0 | rad51-E206K-kanMX6 | This study | BA552 | leu-1–32 ura4-D18 his3-D1 arg3-D1 |
| Antibody | Rat polyclonal anti-Rad51 | Argunhan et al., 2020 | 1:10,000 | |
| Antibody | Mouse monoclonal anti-tubulin | Sigma-Aldrich | T5168 | 1:10,000 |
| Antibody | Mouse monoclonal anti-V5 | Bio-Rad | MCA1360 | 1:10,000 4 µL / IP |
| Antibody | Rabbit polyclonal anti-Rad51 | Haruta et al., 2006 | 10 µL / IP | |
| Antibody | ChromPure Human IgG, whole molecule polyclonal | Jackson Immunoresearch Laboratories | 009-000-003 | 0.336 µL / IP |
| Antibody | Rabbit monoclonal Alexa fluor 488 | Thermo Fisher Scientific | A11034 | 1:300 |
| Antibody | Mouse monoclonal anti-Sfr1 | Argunhan et al., 2020 | 1:200 | |
| Antibody | Mouse monoclonal anti-FLAG-tag | Sigma-Aldrich | F3165 | 1:10,000 |
| Antibody | Rabbit polyclonal anti-Rad52 | Kurokawa et al., 2008 | 1:10,000 | |
| Antibody | Anti-mouse IgG (HRP-conjugated) | GE Healthcare | NA931 | 1:5000 |
| Antibody | Anti-rabbit IgG (HRP-conjugated) | GE Healthcare | NA934 | 1:5000 |
| Antibody | Anti-rat IgG (HRP-conjugated) | Jackson Immunoresearch Laboratories | 712-035-153 | 1:10,000 |
| Recombinant DNA reagent | PhiX174 virion DNA (ssDNA plasmid) | NEB | N3023L | |
| Recombinant DNA reagent | PhiX174 RF I DNA (dsDNA plasmid) | NEB | N3021L | |
| Peptide, recombinant protein | S. pombe Rad51 | Kurokawa et al., 2008 | ||
| Peptide, recombinant protein | S. pombe Rad51-E206A | This study | ||
| Peptide, recombinant protein | S. pombe Rad51 E205A-E206A-D209A | This study | ||
| Peptide, recombinant protein | S. pombe RPA | Haruta et al., 2006 | ||
| Peptide, recombinant protein | S. pombe Swi5-Sfr1 | Haruta et al., 2006 | ||
| Peptide, recombinant protein | S. pombe Rad52 | Kurokawa et al., 2008 | ||
| Peptide, recombinant protein | S. pombe Rad54 | This study | ||
| Commercial assay or kit | Amicon Ultra-15, 10K MWCO | Merck | UFC901096 | |
| Commercial assay or kit | Dynabeads Protein A | ThermoFisher | 10002D | |
| Commercial assay or kit | ImmunoStar Zeta chemiluminescence solution | FujiFilm Wako | 297–72403 | |
| Commercial assay or kit | Malachite Green Phosphate Assay Kit | BioAssay Systems | POMG-25H | |
| Commercial assay or kit | TurboNuclease | Accelagen | N0103P | |
| Commercial assay or kit | Lysing enzyme | Sigma-Aldrich | L1412 | |
| Commercial assay or kit | In-Fusion HD Cloning | TaKaRa | 639650 | |
| Chemical compound, drug | ATP | Sigma-Aldrich | A2383 | |
| Chemical compound, drug | Phosphocreatine di(tris) salt | Sigma-Aldrich | P1937 | |
| Chemical compound, drug | Bio-Safe Coomassie Stain | Bio-Rad | 1610786 | |
| Chemical compound, drug | Proteinase K | TaKaRa | 9034 | |
| Chemical compound, drug | Creatine Kinase | Sigma-Aldrich | 10127566001 | |
| Sequenced-based reagent | 398_F | This study | PCR primers | CCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATA |
| Sequenced-based reagent | 400_R | This study | PCR primers | ATCCAAAACTGCCTCACCATTTAAGCCATA |
| Sequenced-based reagent | 399_F | This study | PCR primers | AATGGTGAGGCAGTTTTGGATAACGTTGCA |
| Sequenced-based reagent | 405_R | This study | PCR primers | CTTTGTTAGCAGCCGGATCC |
| Sequenced-based reagent | 350_F | This study | PCR primers | GATTGGAAATCGTAAGGATCCGCGAATTTCTTATGATTTATGATTTTTATT |
| Sequenced-based reagent | 351_R | This study | PCR primers | ATAGTAAGGAGTCCTCAGTATAGCGACCAGCATTC |
| Sequenced-based reagent | 409_F | This study | PCR primers | CGGTATCGATAAGCTTGATATCCATAAGACTGAGGCAGAGGT |
| Sequenced-based reagent | 412_R | This study | PCR primers | CTCTGTATCTGCCATTATAACTTGTTAAGCACGAAATTATCAC |
| Sequenced-based reagent | 417_F | This study | PCR primers | CTGGTCGCTATACTGATTATTTTAGTATCGTTTCATTTTTATTTATTATTTTGCA |
| Sequenced-based reagent | 419_R | This study | PCR primers | GCGGTGGCGGCCGCTCTAGAGCATGCTTGGAAGGCTTT |
| Sequenced-based reagent | 200_F | This study | PCR primers | CTACTGGTATTCAGGATTATCAAAGTATTCCTACCAATAGCTCACAACGACGTAAGAGATCCATTTTGGAATGTGAGTCCCGGATCCCCGGGTTAATTAA |
| Sequenced-based reagent | 201_R | This study | PCR primers | TAGGTATAATAAATTAATAGATATGGGCAAAACAACCATCACTATGCTAAAAAATTCGTAACTGAAGCCACCATTTTTACGAATTCGAGCTCGTTTAAAC |
| Software, algorithm | FIJI | Schindelin et al., 2012 | ||
| Software, algorithm | Prism version 8 | GraphPad | ||
| Other | DAPI stain | Sigma-Aldrich | D9542 | (1 mg/mL) |





