HBO1-MLL interaction promotes AF4/ENL/P-TEFb-mediated leukemogenesis
Figures
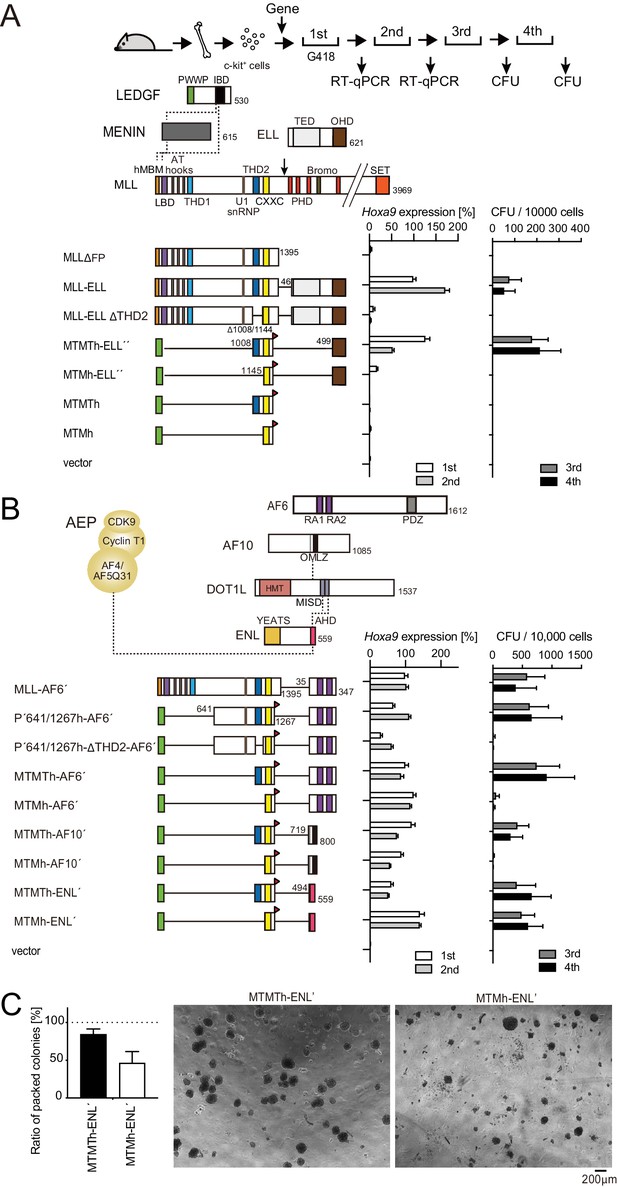
Trithorax homology domain 2 (THD2)-mediated functions promote MLL-fusion-dependent leukemic transformation.
(A) Structure/function analysis of MLL-ELL. Various MLL-ELL constructs were examined for the transformation of myeloid progenitors. HA-tag (h: indicated as a red triangle) was fused to MTM and MTMT constructs. A schema of myeloid progenitor transformation assay is shown on top. Hoxa9 expression normalized to Gapdh in first-round and second-round colonies (left) is shown as the relative value of MLL-ELL (arbitrarily set at 100%) with error bars (mean ± SD of PCR triplicates). Colony-forming ability at third- and fourth-round passages (right) is shown with error bars (mean ± SD of ≥3 biological replicates). IBD: integrase-binding domain; hMBM: high-affinity MENIN-binding motif; LBD: LEDGF-binding domain; THD1/2: trithorax homology domains 1 and 2; SET: Suver3-9/enhancer-of-zeste/trithorax motif; TED: transcription elongation domain; OHD: occludin homology domain. (B) Requirement of THD2 for various MLL fusion proteins in leukemic transformation. Various MLL fusion constructs were examined for the transformation of myeloid progenitors as in (A). RA1/2: RAS association domains 1/2; PDZ: PSD-95/Dlg/ZO-1 domain; OM: octapeptide motif; LZ: leucine zipper. MISD: minimum interaction site of DOT1L: YEATS: Yaf9 ENL AF9 Taf14 Sas5 domain; AHD: ANC1 homology domain. (C) Colony morphologies of MTMTh or MTMh-ENL′ transformed cells. The colonies on day 5 of fourth passage are shown with a scale bar. The ratio of compact colonies (≥100 total colonies were counted in each experiment) is shown on the left (mean ± SD of six biological replicates). Representative images are shown on the right.
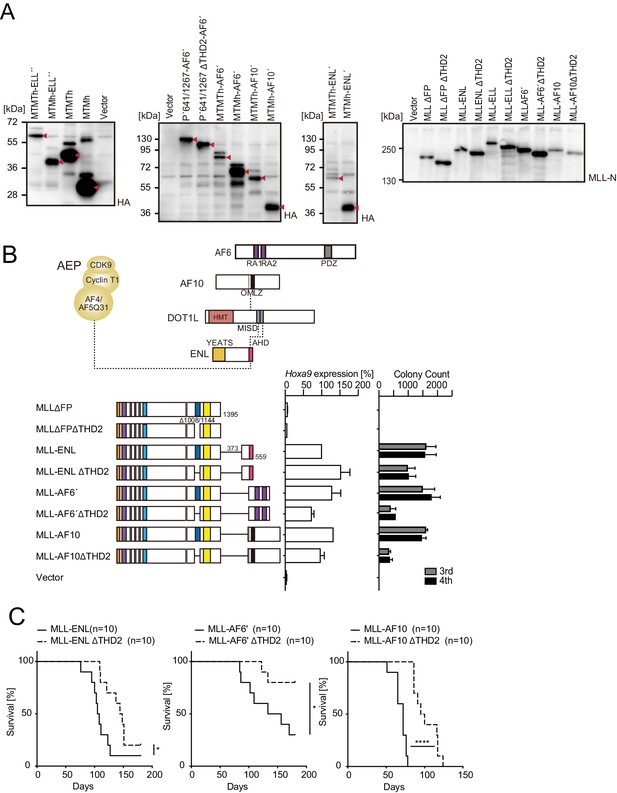
Trithorax homology domain 2 (THD2) promotes MLL fusion-mediated leukemogenesis.
(A) Protein expression from various MLL fusion constructs. Virus-packaging cells transiently expressing each MLL fusion construct shown in Figure 1A and B and (B) were analyzed by western blotting with the indicated antibodies. The expected bands corresponding to the transgene products are indicated by a red arrow head. (B) Negative effects imposed by the loss of THD2 domain on transformation ex vivo. Various MLL fusion constructs lacking the THD2 domains were examined for the transformation of myeloid progenitors as in Figure 1A. (C) Negative effects imposed by the loss of THD2 domain on transformation in vivo. Leukemogenic activity of MLL fusion constructs was examined. Each MLL fusion construct was transduced to c-Kit-positive hematopoietic progenitors and transplanted into syngeneic mice (n=10). Statistical analysis was performed using the log-rank test and Bonferroni correction with the wild-type control. *p ≤ 0.05, ****p ≤ 0.0001.
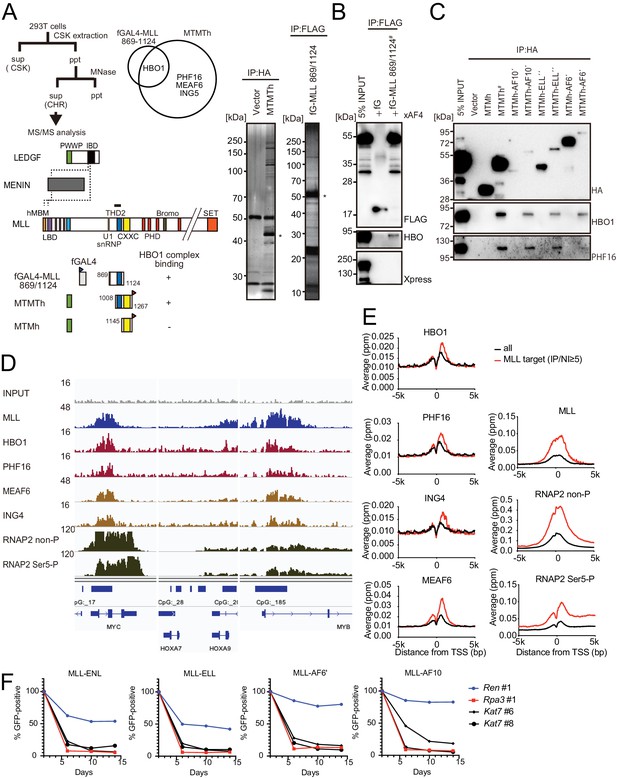
HBO1 complex associates with MLL proteins at promoters.
(A) Purification of trithorax homology domain 2 (THD2) domain-associating factors. A FLAG-tagged (f: indicated as a blue triangle) GAL4 DNA-binding domain fused with the MLL fragment containing the residues 869–1124 or HA-tagged MTMT fragment was transiently expressed in HEK293T cells. A schema of the fractionation-assisted chromatin immunoprecipitation (fanChIP) method is shown on top. The transgene products were purified from the chromatin fraction and analyzed by mass spectrometry. Silver-stained images (right) of the purified materials are shown. Asterisk indicates the position of the transgene products. A Venn diagram of identified THD2 domain-associating factors by mass spectrometry is shown. (B) Association of GAL4-THD2 fusion with HBO1. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293T cells transiently expressing FLAG-tagged GAL4-MLL 869/1124 construct and Xpress-tagged AF4 (xAF4) was performed. Co-purification of HBO1, but not xAF4, was confirmed. #: the sample used for the input. (C) THD2-dependent association with the HBO1 complex. IP-western blotting of the chromatin fraction of virus-packaging cells, transiently expressing various HA-tagged MTMT (or MTM) fusion constructs, was performed. (D) Genomic localization of MLL and the HBO1 complex components in HB1119 cells. The ChIP followed by deep sequencing (ChIP-seq) profiles were visualized using the Integrative Genomics Viewer (The Broad Institute). The minimum value of the y-axis was set at 0, while the maximum value for each sample is indicated. (E) Average distribution of proteins near the transcription start sites (TSSs) of HB1119 cells. Genes whose MLL ChIP signal/input ratio at the promoter proximal transcribed region was ≥5 were defined as MLL target genes. Average ChIP signal distribution of indicated proteins at the MLL target genes (red) or all genes (black) is shown. The y-axis indicates the frequency of the ChIP-seq tag count (ppm) in 25 bp increments. (F) Requirement of HBO1 for myeloid progenitors immortalized by various oncogenes. sgRNA competition assays for Hbo1 were performed on immortalized myeloid progenitors. The ratio of GFP-positive cells co-expressing sgRNA was measured by flow cytometry. sgRNA for Renilla luciferase (Ren) was used as a negative control, which does not affect proliferation. sgRNA for Rpa3 was used as a positive control, which impairs proliferation.
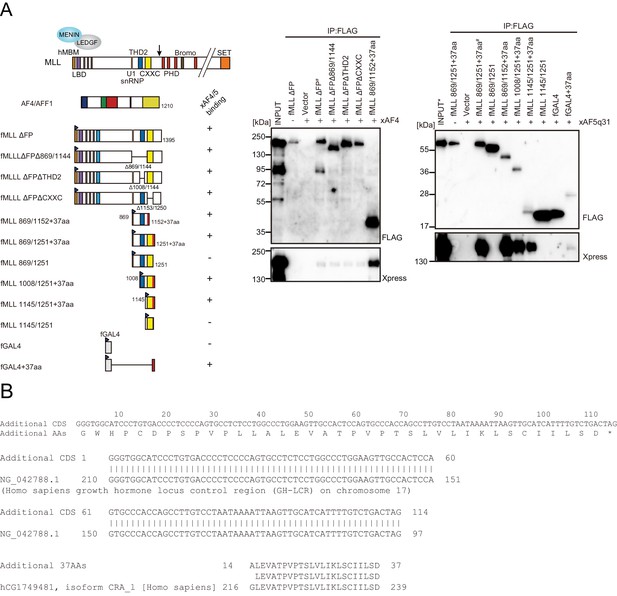
Trithorax homology domain 2 (THD2) does not mediate association with AF4 family proteins.
(A) AF4 family proteins bind the additional 37 residues derived from the expression vector. Immunoprecipitation-western blotting of the chromatin fraction of HEK293T cells transiently expressing various FLAG-tagged MLL constructs along with xAF4/xAF5Q31 was performed using fractionation-assisted chromatin immunoprecipitation (fanChIP) method. (B) Sequence of the additional residues derived from the pCMV5 vector. The coding sequences for the additional 37 amino acids were tethered to MLL in frame. A part of it corresponded to the chorionic somatomammotropin hormone 1 gene.
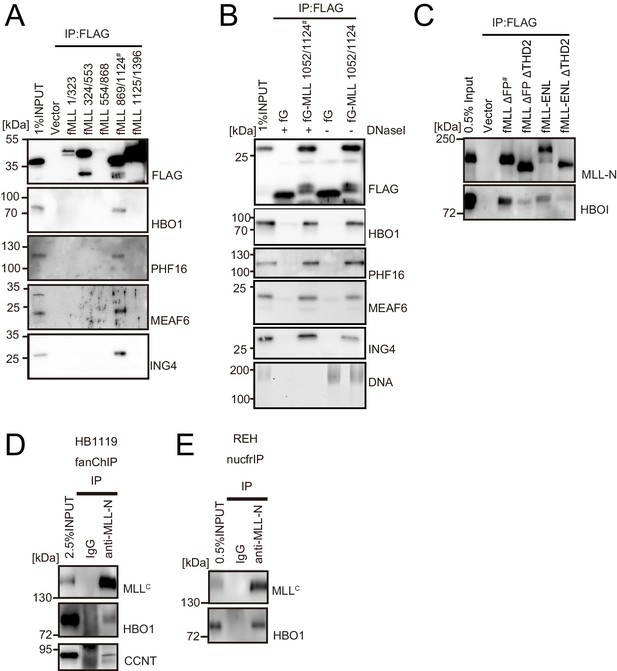
HBO1 complex binds to MLL in various contexts.
(A) Domain mapping of MLL for interaction with the HBO1 complex. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293T cells transiently expressing various FLAG-tagged MLL constructs was performed as in Figure 2B. (B) Effects of DNase treatment on the interaction between MLL and the HBO complex. Co-precipitates were treated with DNaseI, washed multiple times, and analyzed by western blotting. DNA was detected by SYBR green. (C) Interaction with HBO1 through multiple contacts. IP-western blotting of the chromatin fraction of HEK293T cells transiently expressing various FLAG-tagged MLL constructs was performed as in Figure 2B. (D) Interaction between MLL proteins and HBO1 in HB1119 leukemia cells. IP-western blotting of the chromatin fraction of HB1119 cells was performed using an anti-MLL antibody. (E) Interaction between MLL and HBO1 in REH leukemia cells. IP-western blotting of the nucleosome fraction of REH leukemia cells was performed (using anti-MLL antibody) by the nucfrIP method.
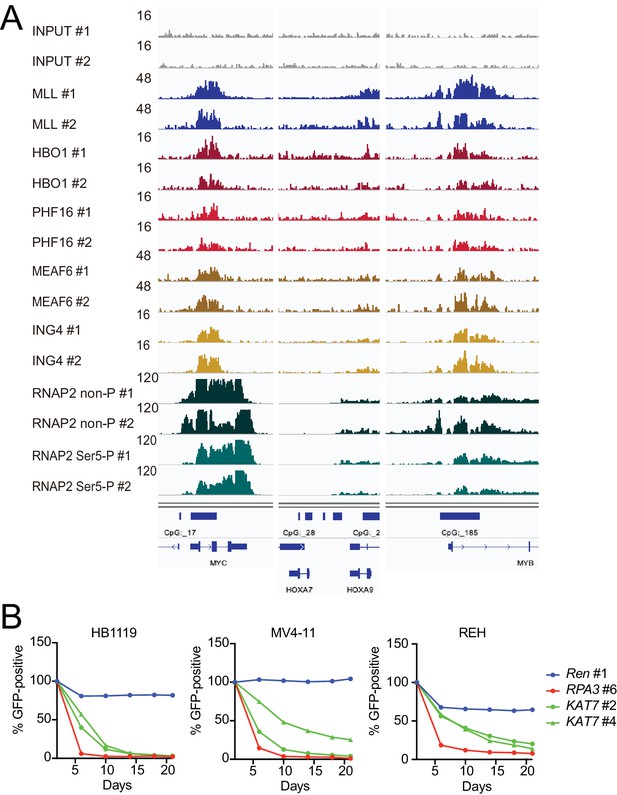
HBO1 is required for proliferation of MLL leukemia cells.
(A) Confirmation of chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) data by repeated ChIP-seq analyses. Duplicated ChIP-seq analyses in HB1119 cells were performed on each target proteins individually. (B) Requirement of HBO1 for various human leukemia cell lines. sgRNA competition assays for KAT7 were performed on Cas9-expressing leukemia cell lines including HB1119, MV4-11, and REH. The ratio of GFP-positive cells co-expressing sgRNA was measured by flow cytometry. sgRNA for Renilla luciferase (Ren) was used as a negative control, which does not affect proliferation. sgRNA for RPA3 was used as a positive control, which impairs proliferation.
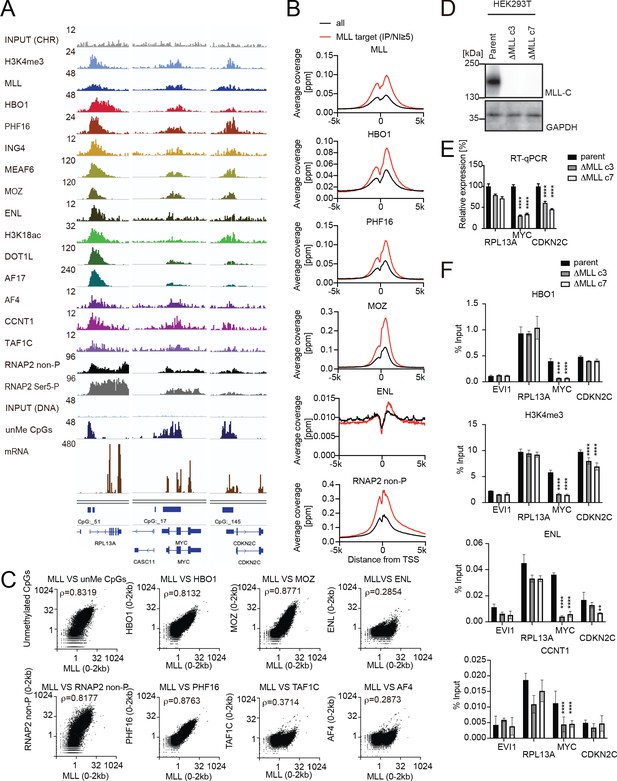
MLL recruits the HBO1 and AEP/SL1 complexes to promoters.
(A) Genomic localization of various transcriptional regulators/epigenetic marks in HEK293T cells. Chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq) analysis was performed on the chromatin of HEK293T cells for the indicated proteins/modifications. CIRA-seq data for unmethylated CpGs (unMe CpGs) and RNA-seq data are shown for comparison. (B) Average distribution of proteins near transcription start sites (TSSs) of HEK293T cells. Average ChIP signal distribution of indicated proteins at the MLL target genes (red) or all genes (black) is shown as in Figure 2E. (C) Relative occupation by multiple factors at all TSSs. ChIP-seq or CIRA-seq tags of indicated proteins at all genes were clustered into a 2 kb bin (0 to +2 kb from the TSS) and are presented as XY scatter plots. Spearman’s rank correlation coefficient (ρ) is shown. (D) Expression of MLL in MLL-deficient HEK293T cell lines. (E) Expression of MLL target genes in two independently generated MLL-deficient clones. Relative expression levels normalized to GAPDH are shown with error bars (mean ± SD of PCR triplicates) by RT-qPCR. Data are redundant with our previous report (Miyamoto et al., 2020). (F) Localization of MLL, the HBO1 complex, the AEP complex, and the SL1 complex in two independently generated MLL-deficient clones. ChIP-qPCR was performed for indicated genes using qPCR probes designed for the TSS of each gene. ChIP signals are expressed as the percent input with error bars (mean ± SD of biological triplicates). Statistical analysis was performed by ordinary two-way ANOVA comparing each sample with the parent cells. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
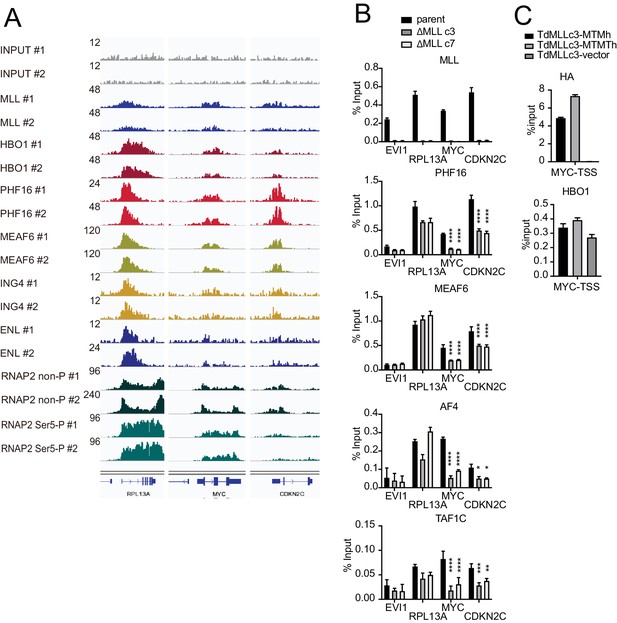
MLL colocalizes with the HBO1 and AF4/ENL/P-TEFb (AEP)/SL1 complexes at promoters containing H3K4me3 marks.
(A) Confirmation of chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq) data by repeated ChIP-seq analyses. Duplicated ChIP-seq analyses in HEK293T cells were performed on each target proteins individually. (B) Localization of MLL, the HBO1 complex, the AEP complex, and the SL1 complex in two independently generated MLL-deficient clones. ChIP-qPCR was performed for indicated genes using qPCR probes designed for the transcription start site (TSS) of each gene. ChIP signals are expressed as the percent input with error bars (mean ± SD of PCR triplicates). Statistical analysis was performed by ordinary two-way ANOVA comparing each sample with the parent cells. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. (C) Localization of MLL mutants and HBO1 in MLL-deficient cells. ChIP-qPCR analysis on the MYC promoter was performed on the chromatin of MLL-deficient HEK293T cells (ΔMLL c3) transiently expressing HA-tagged MTM or MTMT constructs.
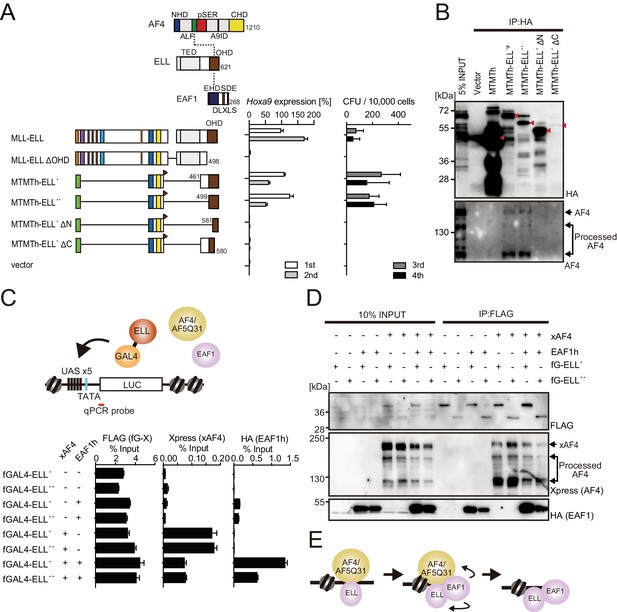
MLL-ELL transforms through the common binding platform for AF4 and EAF1.
(A) Requirement of the occludin homology domain (OHD) in MLL-ELL-mediated transformation. Various MLL-ELL constructs carrying mutations in the ELL portion were examined for the transformation of myeloid progenitors, as in Figure 1A. NHD: N-terminal homology domain; ALF: AF4/LAF4/FMR2 homology domain; pSER: poly-serine; A9ID: AF9 interaction domain; CHD: C-terminal homology domain; EHD: EAF family homology domain; DLXLS: DLXLS motif; SDE: SDE motif. (B) OHD-dependent association with AF4. Immunoprecipitation (IP)-western blotting of the chromatin fraction of virus-packaging cells, transiently expressing various HA-tagged MTMT-ELL fusion constructs, was performed. Endogenous AF4 proteins co-purified with MTMTh-ELL proteins were visualized by an anti-AF4 antibody. (C) Sequential recruitment of AF4 and EAF1 by ELL. HEK293TL cells (Okuda et al., 2015), which harbor GAL4-responsive reporter, were transfected with various combinations of FLAG-tagged GAL4 fusion proteins, xAF4, and HA-tagged EAF1 (EAF1h), and were subjected to chromatin immunoprecipitation (ChIP)-qPCR analysis. A qPCR probe near the GAL4-responsive elements (UAS) was used. The ChIP signals were expressed as the percent input with error bars (mean ± SD of PCR triplicates). TATA: TATA box; LUC: luciferase. (D) Effects of overexpression of AF4 or EAF1 on ELL complex formation. IP-western blotting of the chromatin fraction of HEK293TL cells, transiently expressing various combinations of FLAG-tagged GAL4-ELL proteins, xAF4, and EAF1h, was performed. (E) A model of the sequential association between ELL, AF4 family proteins, and EAF family proteins.
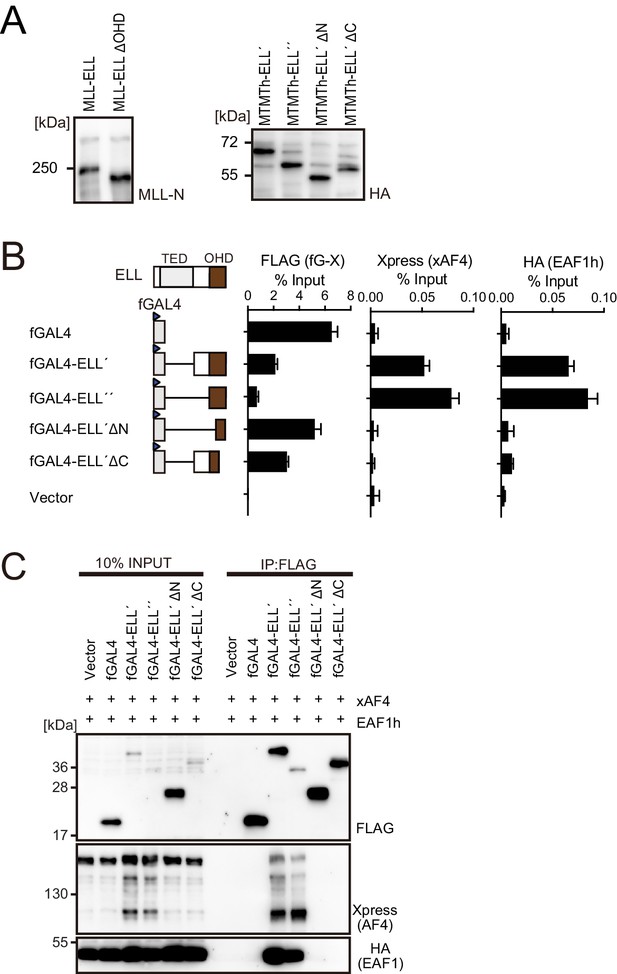
Functions of the ELL portion.
(A) Virus-packaging cells transiently expressing each MLL fusion construct were analyzed by western blotting with indicated antibodies. (B) Recruitment of exogenously expressed AF4 or EAF1 by ELL. HEK293TL cells (Okuda et al., 2015), which harbor GAL4-responsive reporter, were transfected with FLAG-tagged GAL4 fusion proteins, xAF4, and HA-tagged EAF1 (EAF1h), and were subjected to chromatin immunoprecipitation (ChIP)-qPCR analysis. A qPCR probe near the GAL4-responsive elements (UAS) was used. The ChIP signals were expressed as the percent input with error bars (mean ± SD of PCR triplicates). TATA: TATA box; LUC: luciferase. (C) Association of ELL with exogenously expressed AF4 or EAF1 on chromatin. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293TL cells transiently expressing various FLAG-tagged GAL4-ELL proteins along with xAF4 and EAF1h was performed.
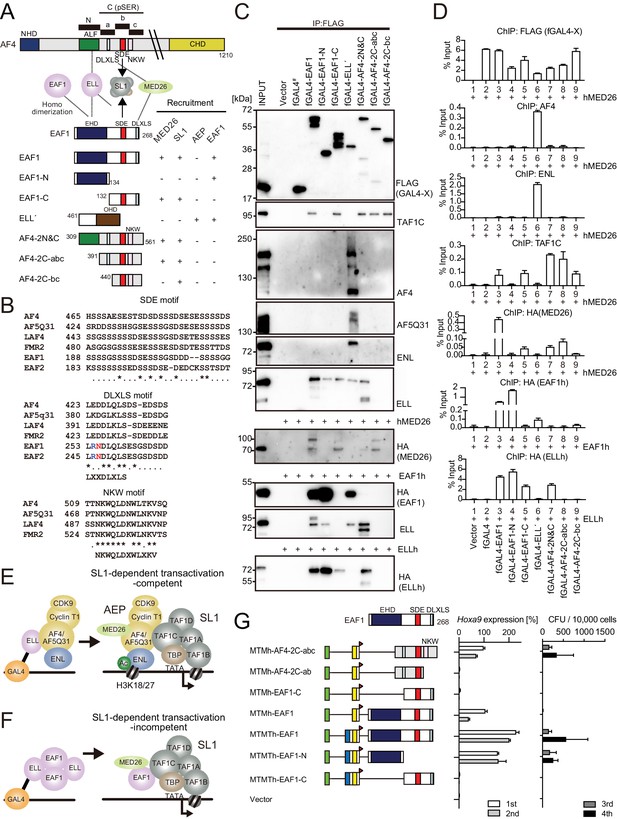
AF4 and EAF1 form distinct SL1/MED26-containing complexes.
(A) A schema of the structures of AF4, ELL, and EAF1. ALF is responsible for interaction with ELL, pSER is responsible for interaction with SL1, and DLXLS is responsible for interaction with MED26. EAF family homology domain (EHD) is responsible for homodimerization and interaction with ELL. NKW:NKW motif responsible for transcriptional activation. (B) Alignment of the amino acid sequences of the SDE, DLXLS, and NKW motifs of human AF4 family proteins and EAF family proteins. Conserved residues are indicated by asterisks. (C) Association of EAF1, ELL, and AF4 domains with various associating factors on chromatin. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293TL cells transiently expressing various FLAG-tagged GAL4 fusion proteins, with or without indicated HA-tagged constructs, was performed as in Figure 4D. Endogenous proteins were detected by specific antibodies for each protein, while exogenous proteins were detected by antibodies for FLAG or HA tag. (D) Recruitment of various transcriptional regulatory proteins by EAF1, ELL, and AF4 domains. Chromatin immunoprecipitation (ChIP)-qPCR analysis of HEK293TL cells transiently expressing various combinations of FLAG-tagged GAL4 fusion proteins along with indicated HA-tagged constructs was performed as in Figure 4C. (E) Putative complex recruitment mediated by ELL-AF4 interaction. (F) Putative complex recruitment mediated by ELL-EAF1 interaction. (G) Structural requirement of MTM-AF4 pSER fusion and MTMT-EAF1 fusion. Various MTMh/MTMTh constructs fused with EAF1 domains were examined for the transformation of myeloid progenitors as in Figure 1A.
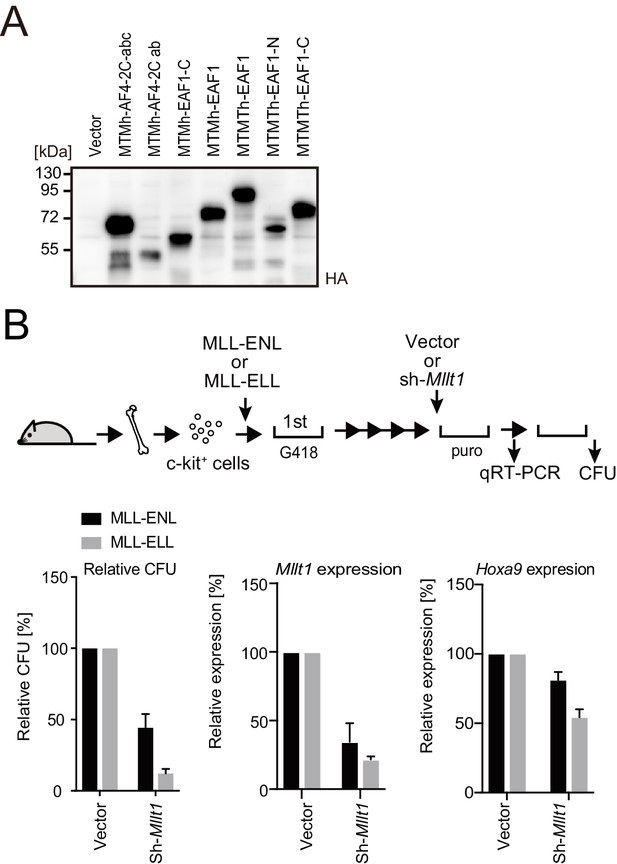
Roles for AF4/ENL/P-TEFb (AEP) in MLL-ELL-mediated leukemic transformation, related to Figure 5.
(A) Protein expression from various MLL fusion constructs. Virus-packaging cells transiently expressing each MLL fusion construct were analyzed by western blotting with indicated antibodies. (B) Effects of Mllt1 knockdown on MLL-ELL- and MLL-ENL-ICs. MLL-ELL- and MLL-ENL-ICs were transduced with shRNA for endogenous Mllt1 and analyzed for gene expression normalized to Gapdh by qRT-PCR (mean ± SD of PCR triplicates) and relative colony-forming units (mean ± SD of ≥3 biological replicates).
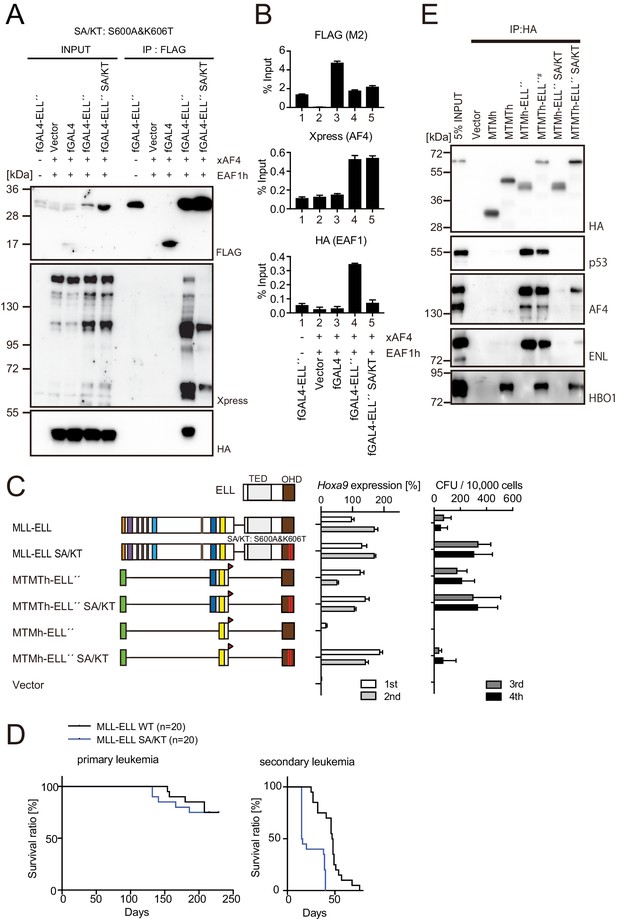
MLL-ELL transforms hematopoietic progenitors via association with AF4/ENL/P-TEFb (AEP), but not with EAF1 or p53.
(A) Mutations of ELL selectively abrogate interaction with EAF1. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293TL cells transiently expressing FLAG-tagged GAL4-ELL proteins with or without S600A/K606T substitutions (SA/KT) along with xAF4 and EAF1h was performed as in Figure 4D. (B) Recruitment of exogenously expressed AF4 or EAF1 by ELL mutant proteins. Chromatin immunoprecipitation (ChIP)-qPCR analysis of HEK293TL cells transiently expressing FLAG-tagged GAL4-ELL proteins with or without the SA/KT mutation along with xAF4 and EAF1h was performed as in Figure 4C. (C) Effects of the SA/KT mutation on MLL-ELL-mediated leukemic transformation ex vivo. Various MLL-ELL constructs with or without the SA/KT mutation were examined for the transformation of myeloid progenitors as in Figure 1A. (D) Effects of the SA/KT mutation on MLL-ELL-mediated leukemic transformation in vivo. MLL-ELL or its SA/KT mutant was transduced to c-Kit-positive hematopoietic progenitors and transplanted into syngeneic mice (n = 20). Primary leukemia cells were harvested from the bone marrow and transplanted into secondary recipient mice (n = 20). (E) Enhanced interaction of ELL with AEP mediated by the trithorax homology domain 2 (THD2) domain. IP-western blotting of the chromatin fraction of virus-packaging cells, transiently expressing various HA-tagged MTMT-ELL fusion constructs (with or without the SA/KT mutation), was performed. Endogenous proteins co-purified with MLL-ELL proteins were visualized by specific antibodies.
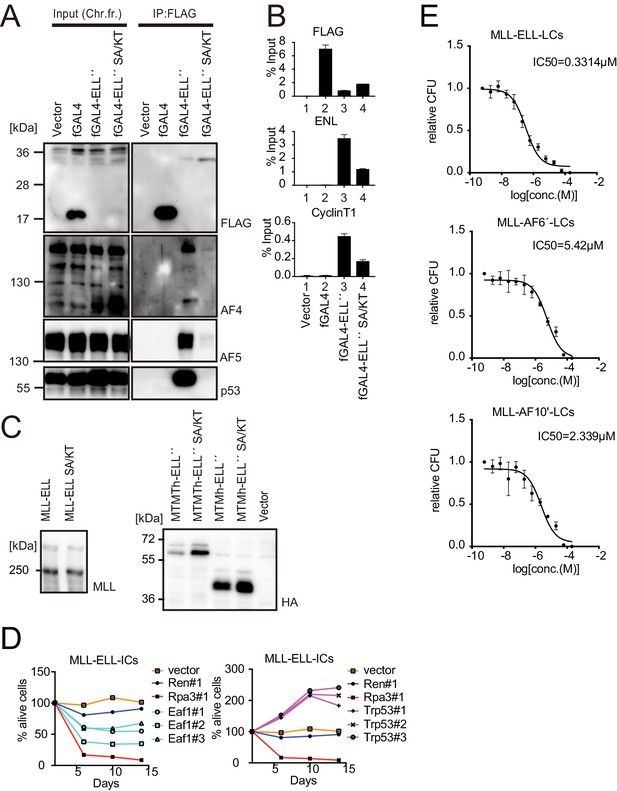
Roles for EAF1 and p53 in MLL-ELL-mediated transformation 6.
(A) Mutations in ELL selectively abrogated the interaction with p53. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293TL cells transiently expressing FLAG-tagged GAL4-ELL proteins with or without S600A/K606T substitutions (SA/KT) was performed as in Figure 4D. Endogenous AF4 family proteins and p53 co-purified with GAL4-ELL proteins were detected by anti-AF4-, AF5Q31-, and p53-antibodies. (B) Recruitment of endogenous AF4/ENL/P-TEFb complex (AEP) proteins by ELL mutant proteins. Chromatin immunoprecipitation (ChIP)-qPCR analysis of HEK293TL cells transiently expressing FLAG-tagged GAL4-ELL proteins with or without the SA/KT mutation was performed as in Figure 4C. ChIP signals of endogenous ENL and CyclinT1 were detected using the anti-ENL and CyclinT1 antibodies. (C) Protein expression from various MLL fusion constructs. Virus-packaging cells, transiently expressing each MLL fusion construct, were analyzed by western blotting with the indicated antibodies. (D) Requirement of EAF1 and p53 for MLL-ELL-immortalized cells. sgRNA competition assays for Eaf1 and Trp53 were performed as in Figure 2F. (E) Sensitivity to pan MYST HAT inhibitor. Leukemia cells (LCs) for various MLL fusions were cultured in the presence of WM1119, a pan MYST HAT inhibitor, and their relative colony-forming units (mean ± SD of thee biological replicates) were monitored.
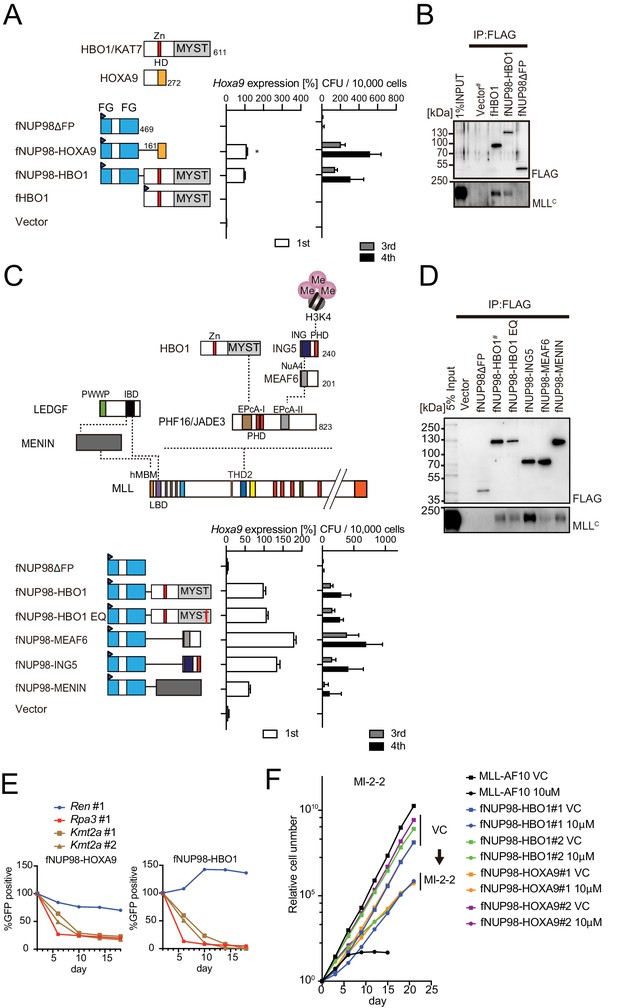
Nucleoporin-98 (NUP98)-HBO1 fusion transforms myeloid progenitors through interaction with MLL.
(A) Structural requirement of NUP98-HBO1 fusion. Various NUP98 fusion constructs were examined for the transformation of myeloid progenitors (along with an HBO1 construct) as in Figure 1A. Relative Hoxa9 expression in first round colonies was analyzed. Asterisk indicates that the qPCR probe detected a human HOXA9 coding sequence included in the NUP98-HOXA9 construct in addition to endogenous murine Hoxa9. Zn: zinc finger; MYST: MYST HAT domain; HD: homeodomain; FG: phenylalanine-glycine repeat. (B) Association with MLL by NUP98-HBO1. The chromatin fraction of virus-packaging cells transiently expressing the indicated transgenes was subjected to immunoprecipitation (IP)-western blotting. Endogenous MLLC fragment was detected by a specific anti-MLL antibody. (C) Structural requirement of various NUP98 fusions for leukemic transformation. Indicated NUP98 fusion constructs were examined for the transformation of myeloid progenitors as in Figure 1A. ING: inhibitor of growth domain of ING4 and ING5; NuA4: histone acetyltransferase subunit NuA4; EPcA-I/II: enhancer of polycomb A domains I and II. (D) Association with MLL by various NUP98 fusions. IP-western blotting was performed as in (B). (E) Requirement of MLL for NUP98 fusion-immortalized cells. sgRNA competition assays for Kmt2a were performed on NUP98-HOXA9- and NUP98-HBO1-immortalized cells as in Figure 2F. (F) Effects of pharmacologic inhibition of MENIN-MLL interaction on NUP98 fusion-immortalized cells. NUP98-HBO1- and NUP98-HOXA9-immortalized cells were cultured in the presence of 10 μM of MI-2–2 MENIN-MLL interaction inhibitor, and their proliferation was monitored every 3 days. MLL-AF10-immortalized cells were also analyzed as a positive control. VC: vehicle control.
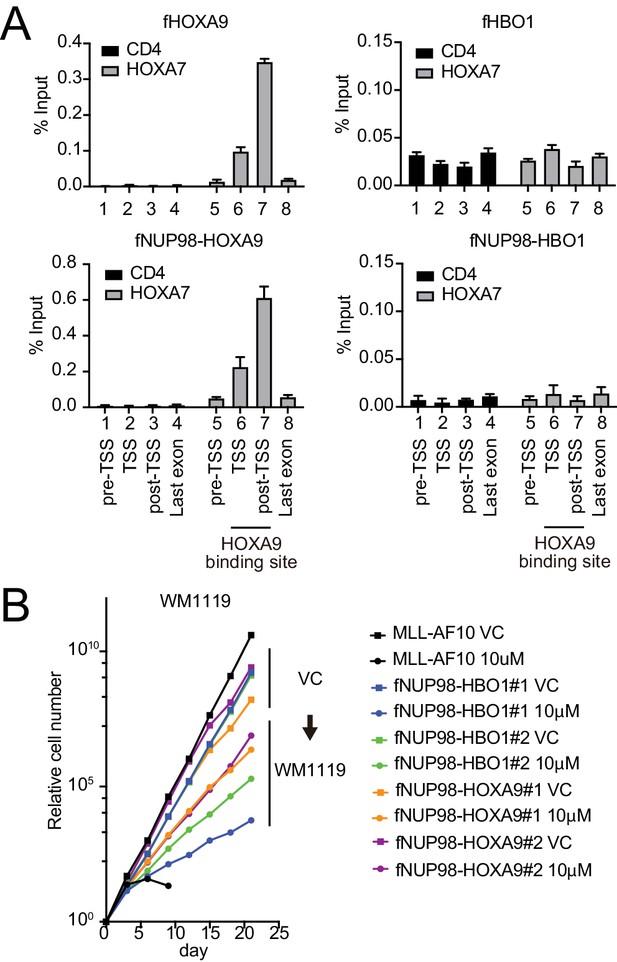
Functional differences between nucleoporin-98 (NUP98)-HOXA9 and NUP98-HBO1.
(A) Targeting ability to the HOXA9-binding site. Virus-packaging cells transiently expressing indicated transgenes were subjected to chromatin immunoprecipitation (ChIP)-qPCR analysis. The percent input values of each probe (mean ± SD of PCR triplicates) are shown. A HOXA9-binding site was previously identified at the HOXA7 locus (Shima et al., 2017). The CD4 locus was analyzed as a negative control. (B) Effects of pharmacologic inhibition of MYST HATs on NUP98 fusion-immortalized cells. NUP98-HBO1- and NUP98-HOXA9-ICs were cultured in the presence of 10 μM of WM1119 MYST HAT inhibitor and their proliferation was monitored every 3 days. MLL-AF10-immortalized cells were also analyzed as the positive control. VC: vehicle control.
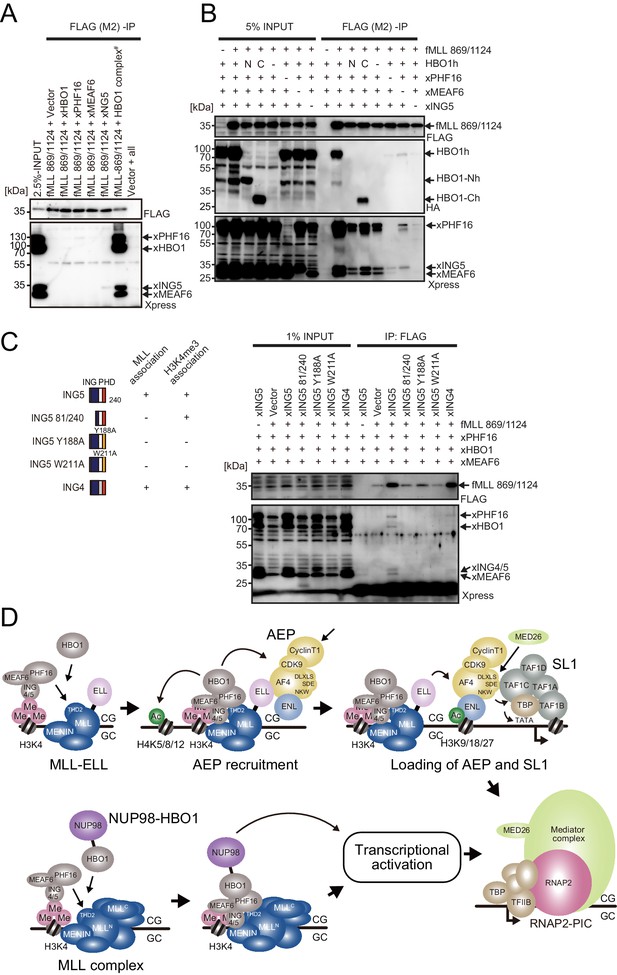
MLL-HBO1 complex association is mediated by multiple contacts and in a chromatin-bound context containing H3K4 methylation.
(A) Association of the MLL bait protein with the HBO1 complex. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293T cells transiently expressing FLAG-tagged MLL 869/1124 construct and Xpress-tagged HBO1 complex components was performed. (B) Influence of each HBO1 complex component for MLL-HBO1 complex association. (C) Domain mapping of ING5 responsible for MLL-HBO1 complex association. (D) Models of MLL-ELL- and nucleoporin-98 (NUP98)-HBO1-mediated gene activation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Other | MNase | Sigma-Aldrich | Cat#:N3755-200U | |
| Other | Protein-G Magnetic beads | Thermo Fisher Scientific | Cat#:1004D | |
| Other | FLAG M2 antibody-conjugated beads | Sigma-Aldrich | Cat#:M8823 RRID:AB_2637089 | |
| Other | c-Kit magnetic beads | Miltenyi Biotec | Cat#:130-091-224 RRID:AB_2753213 | |
| Other | MI-2–2 | Calbiochem | Cat#:444825 | |
| Other | WM1119 | Enamine | Cat#:EN300-1719156 | |
| Antibody | MLL(N) | Cell Signaling Technology | Cat#:14689 RRID:AB_2688009 | WB(1000:1) |
| ChIP(1 μL/400 μL) | ||||
| Antibody | MLL(N) | Generated in-house Yokoyama et al., 2002 | rpN1 | ChIP(1 μg/400 μL) |
| Antibody | MOZ | Active motif | Cat#:39868 | ChIP(1 μg/400 μL) |
| Antibody | AF4 | Santa Cruz Biotechnology | Cat#:sc-49350 RRID:AB_2226113 | ChIP(1 μg/400 μL) |
| Antibody | TAF1C | Bethyl Laboratories | Cat#:A303-698A RRID:AB_11203194 | ChIP(1 μg/400 μL) |
| Antibody | AF17 | Bethyl Laboratories | Cat#:A302-198A RRID:AB_1659777 | ChIP(1 μg/400 μL) |
| Antibody | DOT1L | Bethyl Laboratories | Cat#:A300-953A RRID:AB_805775 | ChIP(1 μg/400 μL) |
| Antibody | CyclinT1 | Santa Cruz Biotechnology | Cat#:sc-8127 RRID:AB_2073892 | ChIP(1 μg/400 μL) |
| Antibody | ENL | Cell Signaling Technology | Cat#:14893S | WB(1000:1) |
| ChIP(5 μL/400 μL) | ||||
| Antibody | Histone H3K4me3 | Active motif | Cat#:39159 RRID:AB_2615077 | ChIP(1 μg/400 μL) |
| Antibody | Histone H3K18ac | Abcam | Cat#:ab1191 RRID:AB_298692 | ChIP(1 μg/400 μL) |
| Antibody | RNAP2 non-P | Abcam | 8WG16/Cat#:ab817 RRID:AB_306327 | ChIP(1 μg/400 μL) |
| Antibody | RNAP2 Ser5-P | Millipore | Cat#:05–623 AB_309852 | ChIP(1 μg/400 μL) |
| Antibody | FLAG | Sigma-Aldrich | Cat#:F3165 RRID:AB_259529 | ChIP(1 μg/400 μL) |
| Antibody | FLAG | Sigma-Aldrich | Cat#:F7425 RRID:AB_439687 | WB(1000:1) |
| Antibody | HA | Roche | 3F10/Cat#:11867423001 RRID:AB_390918 | WB(1000:1) |
| ChIP(0.2 μg/400 μL) | ||||
| Antibody | Xpress | Santa Cruz Biotechnology | Cat#:sc-7270 RRID:AB_675763 | WB(1000:1) |
| ChIP(1 μg/400 μL) | ||||
| Antibody | ELL | Cell Signaling Technology | Cat#:14468S RRID:AB_2798489 | WB(1000:1) |
| Antibody | HBO1 | Abcam | Cat#:70183 RRID:AB_1269226 | WB(1000:1) |
| ChIP(1 μg/400 μL) | ||||
| Antibody | PHF16 | Abcam | Cat#:129495 RRID:AB_11157084 | WB(1000:1) |
| ChIP(1 μg/400 μL) | ||||
| Antibody | MEAF6 | STJ | Cat#:116836 | WB(1000:1) |
| ChIP(1 μg/400 μL) | ||||
| Antibody | ING4 | Abcam | Cat#:108621 RRID:AB_10860023 | WB(1000:1) |
| ChIP(1 μg/400 μL) | ||||
| Antibody | MLL(C) | Cell Signaling Technology | Cat#:14197S RRID:AB_2688010 | WB(1000:1) |
| Antibody | AF4 | Bethyl Laboratories | Cat#:A302-344A RRID:AB_1850255 | WB(1000:1) |
| Antibody | AF5Q31 | Bethyl Laboratories | Cat#:A302-538A RRID:AB_1998985 | WB(1000:1) |
| Antibody | P53 | Santa Cruz Biotechnology | Cat#:sc-126 RRID:AB_628082 | WB(1000:1) |
| Sequence-based reagent | GAPDH | Thermo Fisher Scientific | Hs02786624_g1 | |
| Sequence-based reagent | HOXA9 | Thermo Fisher Scientific | Hs00365956_m1 | |
| Sequence-based reagent | MYC | Thermo Fisher Scientific | Hs01570247_m1 | |
| Sequence-based reagent | RPL13A | Thermo Fisher Scientific | Hs04194366_g1 | |
| Sequence-based reagent | CDKN2C | Thermo Fisher Scientific | Hs00176227_m1 | |
| Sequence-based reagent | Gapdh | Thermo Fisher Scientific | Mm99999915_g1 | |
| Sequence-based reagent | Hoxa9 | Thermo Fisher Scientific | Mm00439364_m1 | |
| Sequence-based reagent | Mllt1 | Thermo Fisher Scientific | Mm00452080_m1 | |
| Sequence-based reagent | sgRNAs | This study | See Supplementary file 1 | |
| Sequence-based reagent | Custom made qPCR primers | Thermo Fisher Scientific | See Supplementary file 1 | |
| Recombinant DNA reagent | pMSCV-neo | Clontech | Cat#:634401 | |
| Recombinant DNA reagent | pLKO5.EFS.GFP | Addgene (gift from Benjamin Ebert) Heckl et al., 2014 | Addgene #57822 RRID:Addgene_57822 | |
| Recombinant DNA reagent | pKLV2-Cas9.bsd | Addgene (gift from Kosuke Yusa) Tzelepis et al., 2016 | Addgene #68343 RRID:Addgene_68343 | |
| Recombinant DNA reagent | pMDLg/pRRE | Addgene (gift from Didier Trono) Dull et al., 1998 | Addgene #12251 RRID:Addgene_12251 | |
| Recombinant DNA reagent | pRSV-rev | Addgene (gift from Didier Trono) Dull et al., 1998 | Addgene #12253 RRID:Addgene_12253 | |
| Recombinant DNA reagent | pMD2.G | Addgene (gift from Didier Trono) Dull et al., 1998 | Addgene #12259 RRID:Addgene_12259 | |
| Recombinant DNA reagent | pLKO.1-puro | Addgene (gift from Bob Weinberg) Stewart et al., 2003 | Addgene #8453 RRID:Addgene_84530 | |
| Recombinant DNA reagent | pLKO.1-sh-Mllt1-puro | GE Healthcare | TRCN0000084405 | |
| Software, algorithm | GraphPad Prism7 | GraphPad Software Inc | RRID:SCR_002798 | |
| Software, algorithm | Integrative Genomics Viewer | Thorvaldsdóttir et al., 2013 | RRID:SCR_011793 | |
| Software, algorithm | Cutadapt | Martin, 2011 | RRID:SCR_011841 | |
| Software, algorithm | BWA | Li and Durbin, 2009 | RRID:SCR_010910 | |
| Cell line (Homo sapiens) | Human: HEK293T | Gift from Michael Cleary | Authenticated by JCRB Cell Bank in 2019 | |
| Cell line (Homo sapiens) | Human: HB1119 | Gift from Michael Cleary Tkachuk et al., 1992 | The cell line was verified by the expression of MLL-ENL | |
| Cell line (Homo sapiens) | Human: REH | Gift from Michael Cleary Yokoyama et al., 2004 | RRID:CVCL_1650 | Authenticated by JCRB Cell Bank in 2021 |
| Cell line (Homo sapiens) | Human: PLAT-E | Gift from Toshio Kitamura Morita et al., 2000 | RRID:CVCL_B488 | |
| Cell line (Homo sapiens) | Human: HEK293TN | System Bioscience | Cat#:LV900A-1 | |
| RRID:CVCL_UL49 | ||||
| Cell line (Homo sapiens) | Human: HEK293T dMLLc3 | Generated in-house Miyamoto et al., 2020 | ||
| Cell line (Homo sapiens) | Human: HEK293T dMLLc7 | Generated in-house Miyamoto et al., 2020 | ||
| Cell line (Homo sapiens) | Human: HEK293TL | Generated in-house Okuda et al., 2015 | ||
| Strain, strain background (Mus musculus) | Mouse: C57BL/6J | CLEA Japan |
Additional files
-
Source data 1
Gel images.
- https://cdn.elifesciences.org/articles/65872/elife-65872-data1-v1.pdf
-
Supplementary file 1
Sequence data.
- https://cdn.elifesciences.org/articles/65872/elife-65872-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65872/elife-65872-transrepform-v1.docx





