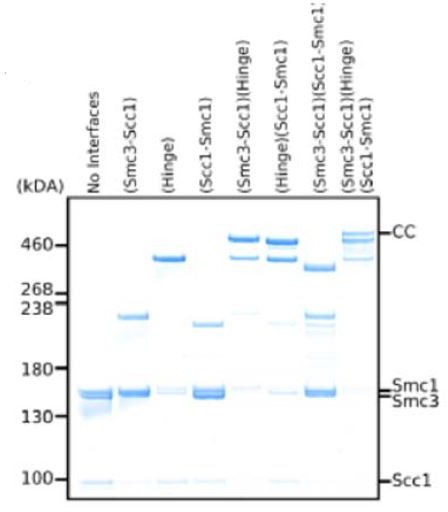
Folding of cohesin’s coiled coil is important for Scc2/4-induced association with chromosomes
Figures
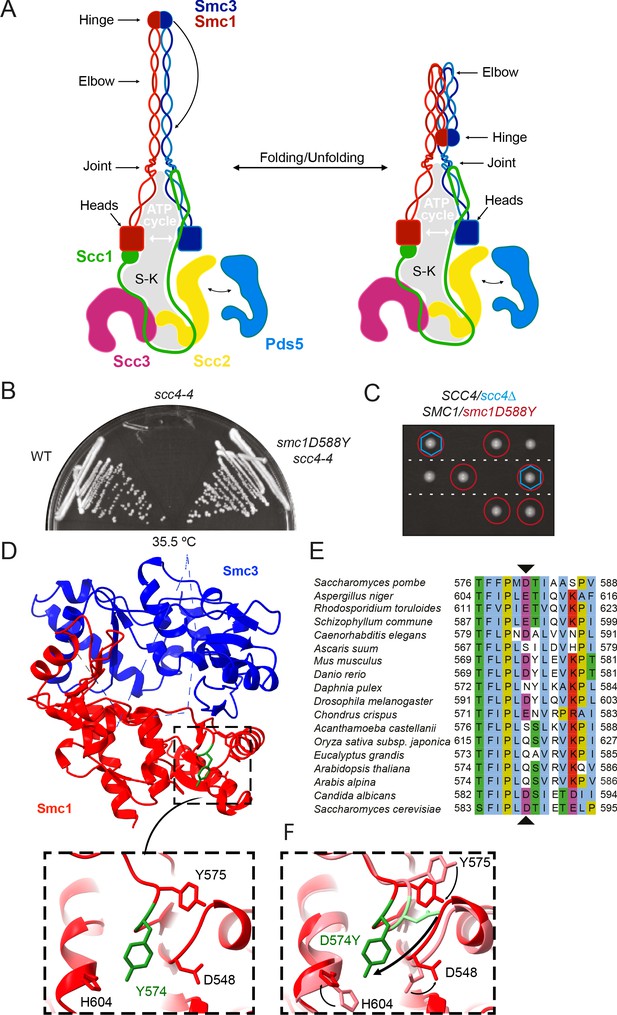
A mutation in the hinge domain of Smc1 restores viability in the absence of Scc4.
(A) Schematic representation of Saccharomycescerevisiae cohesin complex and its folding cycle. (B) Comparison of growth of wild-type (WT), scc4-4, and scc4-4 smc1D588Y strains at 35.5°C (K699, K8326, K19813). (C) Tetrad dissection of diploid strains containing SCC4/scc4Δ SMC1/smc1D588Y grown at 30°C. Spores expressing smc1D588Y are circled in red, and spores that lack Scc4 are indicated with blue hexagons. (D) Structure of the mouse Smc3-Smc1D574Y hinge domain (PDB: 7DG5). (E) Multiple sequence alignment indicating conservation of Smc1D588. (F) Structural superposition of the WT hinge and the D574Y mutant hinge. Tyr574 swings out relative to the position of D574 with a concomitant local conformational change of the mutated loop.
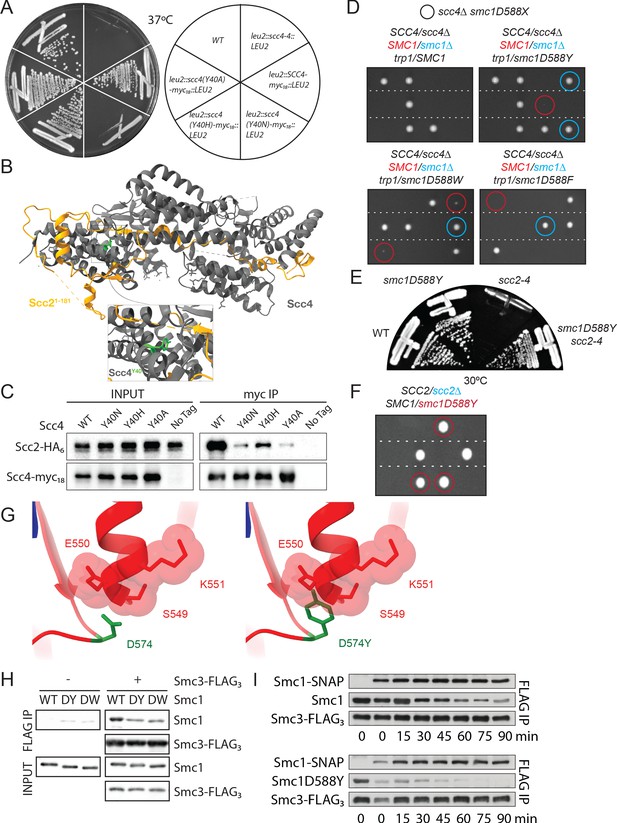
A mutation in the hinge domain of Smc1 restores viability in the absence of Scc4.
(A) Comparison of growth of endogenous SCC4 with ectopically expressed SCC4, scc4Y40A, scc4Y40H, scc4Y40N, and scc4-4 at 37°C (K7564, K8504, K20350, K20351, K20352, K20353). (B) Crystal structure of the Scc21-181/Scc4 complex (PDB: 4XDN; Hinshaw et al., 2015). Scc2 is shown in orange, Scc4 in grey, and Scc4Y40 in green. (C) Co-immunoprecipitation (co-IP) of wild-type (WT) or mutant Scc4-myc18 from cells expressing Scc2-HA6 (K20110, K20111, K20112, K20113, K7564) (D) Tetrad dissection performed on heterozygous smc1∆/SMC1 scc4∆/SCC4 strains with SMC1, smc1(D588Y), smc1(D588W), or smc1(D588F) integrated at the trp1 locus (K21973, K21974, K22012, K21990). Spores in which scc4∆-related lethality is suppressed by ectopically expressed smc1 mutants are circled in blue (smc1∆) and red (SMC1). Following growth on YPD for 2 days at 30°C, 15.7% of 108 spores contained the markers for smc1(D588Y) and scc4∆ (12.0% smc1∆, 3.7% SMC1), 27.8% of 72 spores contained the markers for smc1(D588W) and scc4∆ (18.0% smc1∆, 9.8% SMC1), and 5.6% of 72 spores contained the markers for smc1(D588F) and scc4∆ (4.2% smc1∆, 1.4% SMC1). (E) Comparison of growth of WT, smc1D588Y, scc2-4, and scc2-4 smc1(D588Y) strains at 30°C (K699, K21416, K5828, K21995). (F) Tetrad dissection of a heterozygous SCC2/scc2∆ strain in a background heterozygous for suppressor mutation smc1(D588Y). Spores bearing the marker for smc1(D588Y) are encircled in red. No spores bearing the marker for scc2∆ were detected. (G) Graphical D574Y substitution in the structure of the WT cohesin hinge. Y574 adopting the same conformation of D574 crashes with a neighbouring loop (right, D574Y). (H) Smc1 WT, D588Y (DY) or D588W (DW), and Smc3-FLAG monomeric hinge proteins were mixed in equimolar ratio prior to co-IP with anti-FLAG beads. The amount of protein bound to beads was determined by western blot using anti-HIS antibody to detect SMC proteins. Non-specific binding of Smc1 to anti-FLAG beads is shown in left-hand panels. (I) WT Smc1-SNAP competitor was added to Smc1 (WT or D588Y) and Smc3-FLAG preformed heterodimeric hinges, samples were added to BSA-blocked anti-FLAG beads every 15 min for 90 min. Protein bound to beads was detected as in (E).
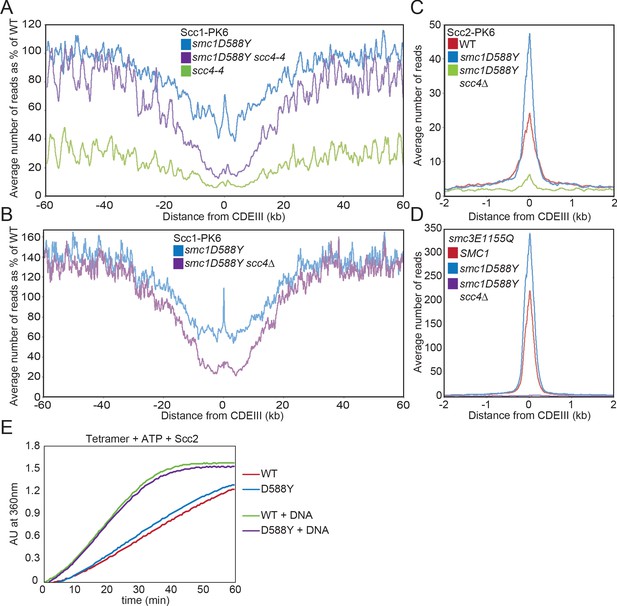
smc1D588Y restores cohesin occupancy on chromosome arms in the absence of Scc4.
(A) Average calibrated ChIP-seq profiles of Scc1-PK6 in smc1D588Y, scc4-4, and smc1D588Y scc4-4 cells 60 kb either side of CDEIII plotted as a percentage of the average number of reads obtained for wild-type (WT) cells. Cells were pheromone arrested in G1 at 25°C before release at 37°C into medium containing nocodazole. Samples were taken 75 min after release (K22005, K22009, K21999, K22001). (B) Average calibrated ChIP-seq profiles of Scc1-PK6 in smc1D588Y, and smc1D588Y scc4Δ cells 60 kb either side of CDEIII plotted as a percentage of the average number of reads obtained for WT cells. Cells were pheromone arrested in G1 at 25°C before release at 25°C into medium containing nocodazole. Samples were taken 60 min after release (K22005, K22009, K19624). (C) Average calibrated ChIP-seq profiles of Scc2-PK6 2 kb either side of CDEIII in cycling WT, smc1D588Y, and smc1D588Y scc4Δ cells at 25°C (K21388, K24680, K24678). (D) Average calibrated ChIP-seq profiles of ectopically expressed Smc3E1155Q-PK6 2 kb either side of CDEIII in cycling WT, smc1D588Y, and smc1D588Y scc4Δ cells at 25°C (K24562, K24689, K24564). (E) ATPase activity of WT or mutant tetramers on addition of ATP and Scc2 in the presence and absence of DNA.
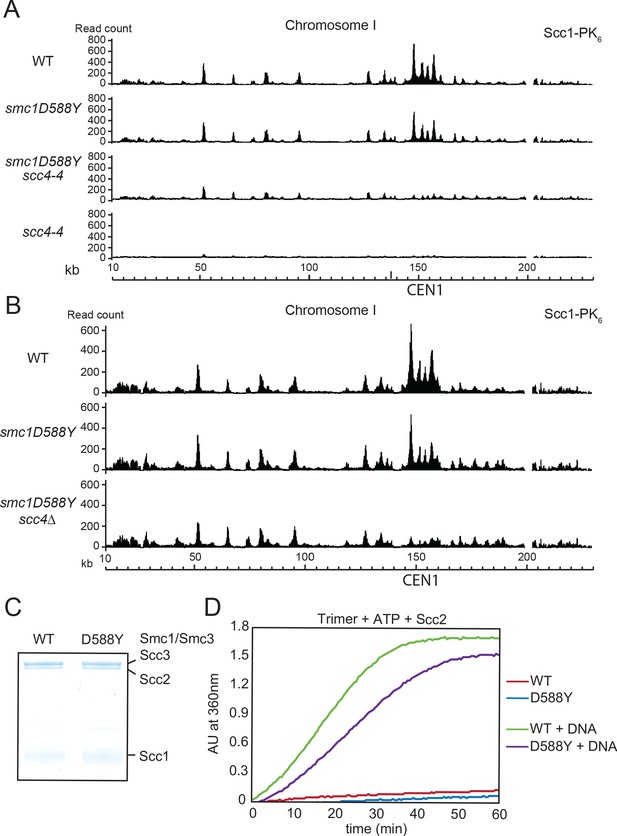
smc1D588Y restores cohesin occupancy on chromosome arms in the absence of Scc4.
(A) Calibrated ChIP-seq profiles of Scc1-PK6 in wild-type (WT), smc1D588Y, scc4-4, and smc1D588Y scc4-4 cells across chromosome I. Cells were pheromone arrested in G1 at 25°C before release at 37°C into medium containing nocodazole. Samples were taken 75 min after release (K22005, K22009, K21999, K22001). (B) Calibrated ChIP-seq profiles of Scc1-PK6 in WT, smc1D588Y, and smc1D588Y scc4Δ cells across chromosome I. Cells were pheromone arrested in G1 before release into medium containing nocodazole at 25°C. Samples were taken 60 min after release (K22005, K22009, K19624). (C) A fraction of the ATPase reaction stained with Coomassie after SDS-PAGE to confirm protein levels. (D) ATPase activity of WT or mutant trimers on addition of ATP and Scc2 in the presence or absence of DNA.
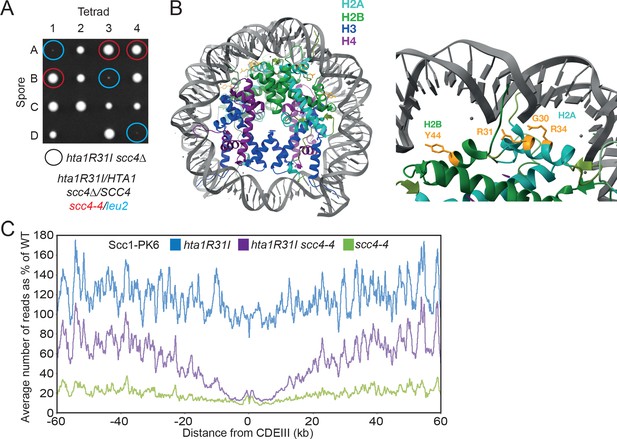
Mutations in SCC2 and histone genes also suppress scc4Δ lethality.
(A) Tetrad dissection of diploid strains containing SCC4/scc4Δ leu2/scc4-4 ΗTΑ1/hta1R31I. Spores in which scc4Δ is rescued by hta1R31I are circled in blue. (B) Structure of the yeast nucleosome (PDB: 1ID3; White et al., 2001). H2A is shown in blue and H2B in green. Suppressor mutations are shown in yellow. (C) Average calibrated ChIP-seq profiles of Scc1-PK6 in hta1R31I, scc4-4, and hta1R31I scc4-4 cells 60 kb either side of CDEIII plotted as a percentage of the average number of reads obtained for wild-type (W)T cells. Cells were pheromone arrested in G1 at 25°C before release at 35.5°C into medium containing nocodazole. Samples were taken 60 min after release (K22005, K24574, K24568, K22001).
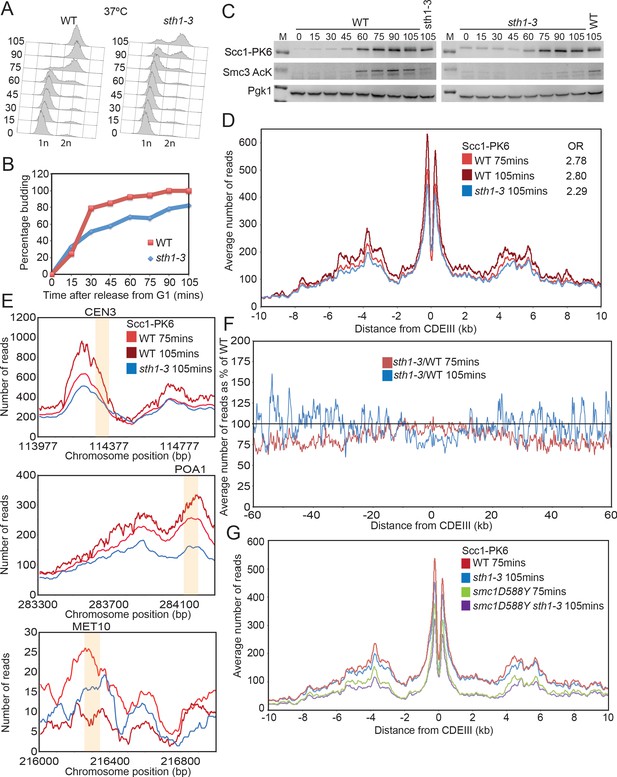
Scc4 helps overcome inhibition of loading by nucleosomes.
(A) Cell cycle progression as measured by FACS of wild-type (WT) and sth1-3 cells arrested in G1 with pheromone prior to release into nocodazole containing medium at 37°C (K23997, K22005). (B) Fraction of cells with buds of cells treated as described in (A). (C) Western blot to measure the levels of Scc1-PK6 and acetylation of Smc3 of cells treated as described in (A). (D) Average calibrated ChIP-seq profile of Scc1-PK6 10 kb either side of CDEIII at 75 min and 105 min after release described in (A). The occupancy ratios (OR) were derived as described in Hu et al., 2015. (E) ChIP-seq profiles of Scc1-PK6 as in (D) at individual loci. Sequences measured in Lopez-Serra et al., 2014 are shaded in orange. (F) Average calibrated ChIP-seq profile of Scc1-PK6 in sth1-3 cells at 105 min after release 60 kb either side of CDEIII plotted as a percentage of the average number of reads obtained for WT cells at either 75 or 105 min after release. (G) Average calibrated ChIP-seq profiles of Scc1-PK6 10 kb either side of CDEIII of cells expressing SMC1 or smc1D588Y in the presence of STH1 or sth1-3. Cells were pheromone arrested in G1 at 25°C prior to release into nocodazole containing medium at 37°C. Samples were taken at 75 min and 105 min post release, and samples at similar cell cycle stages were compared (K22005, K22009, K23997, K24031).
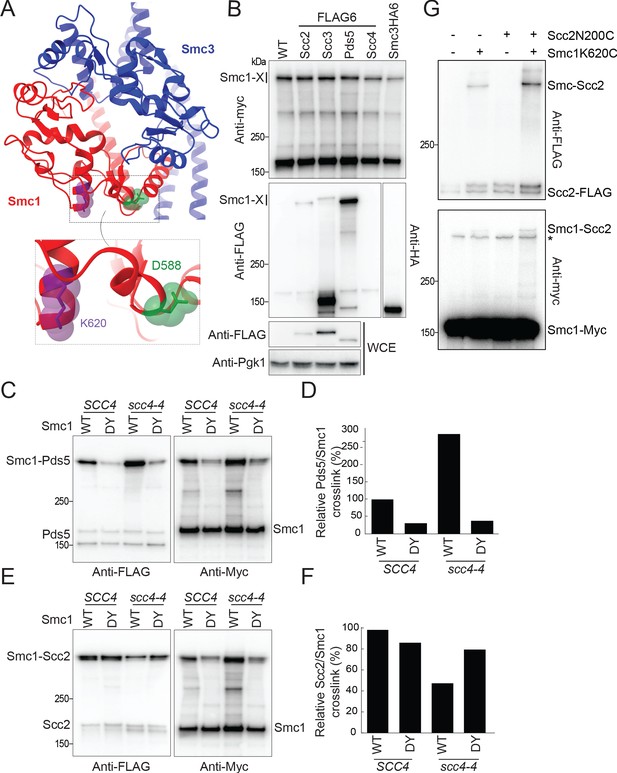
Scc4 regulates an interaction between the hinge domain and HAWKs.
(A) Modelled structure of the yeast cohesin hinge domain based on bacterial SMC hinge from Thermotoga maritima (PDB: 1GXL; Haering et al., 2002). (B) Identification of proteins that crosslink to Smc1 hinge. Strains expressing various cohesin regulators tagged with either FLAG6 or HA6 in combination with Smc1K620BPA-myc were treated with UV prior to immunoprecipitation with PK-tagged Scc1 and the products analysed by western blotting (B1969, B1976, B1983, B2020, B2072, B2079). (C) Effect of Scc4 and Smc1D588Y on crosslinking between Pds5 and Smc1 hinge. Cells expressing Smc1K620BPA in the presence or absence of scc4-4 and Smc1D588Y were exponentially grown at 25°C and shifted to 35.5°C for 1 hr. Cells were irradiated with UV, and the cohesin complex was isolated by immunoprecipitation of PK-tagged Scc1. The Myc-tagged Smc1K620BPA was examined by western blot (B2072, B2212, B2214, B2215). (D) Quantification of the crosslinks in (C) as a percentage of the wild-type (WT) Smc1 crosslinking efficiency. (E) Effect of Scc4 and Smc1D588Y on crosslinking between Scc2 and Smc1 hinge. Strains were treated as described in (C) (B1969, B2213, B2216, B2217). (F) Quantification of the crosslinks in (E) as a percentage of the WT Smc1 crosslinking efficiency. The experiments shown in (C–F) were performed twice with the same result. (G) In vivo cysteine crosslinking of Smc1 hinge with Scc2 protein. Yeast cells expressing Smc1K620C and Scc2N200C were incubated with bismaleimidoethane (BMOE) (B3082, B3107, B3114, and B3116). The crosslinked Smc1/Scc2 was isolated by immunoprecipitation of PK-tagged Scc1 and examined by western blot. * Unspecific crosslink band.
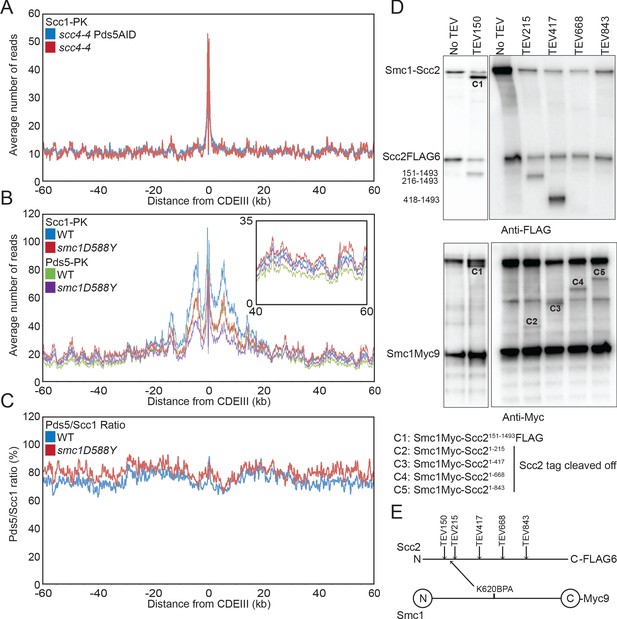
Scc4 regulates an interaction between the hinge domain and HAWKs.
(A) Average calibrated ChIP-seq profiles of Scc1-PK6 60 kb either side of CDEIII of cells expressing scc4-4 in the presence or absence of Pds5-AID. Cells were pheromone arrested in G1 at 25°C before release at 37°C into medium containing nocodazole and auxin. Samples were taken 75 min after release (K22001, K27751). (B) Average calibrated ChIP-seq profiles of Scc1-PK6 and Pds5-PK6 60 kb either side of CDEIII of cycling cells expressing wild-type (WT) or Smc1D588Y. Inset shows magnification of the region 40–60 kb away from CDEIII (K19012, K25378, K22005, K22009). (C) Data shown in (B) plotted as a ratio of Pds5:Scc1 for WT and Smc1D588Y. (D) Determination of the Scc2 region crosslinked by Smc1K620BPA. Yeast strains expressing Smc1K620BPA and indicated alleles of TEV-cleavable Scc2 were subjected to UV irradiation (B1969, B2143, B2144, B2145, B2149, and B2298). The crosslink products were co-immunoprecipitated with Scc1-PK and treated with TEV proteinase. The cleaved Scc2/Smc1 crosslinked products were analysed by western blot. (E) Schematic of TEV cleavage sites introduced into Scc2 with respect to the crosslink to Smc1K620BPA in (D).
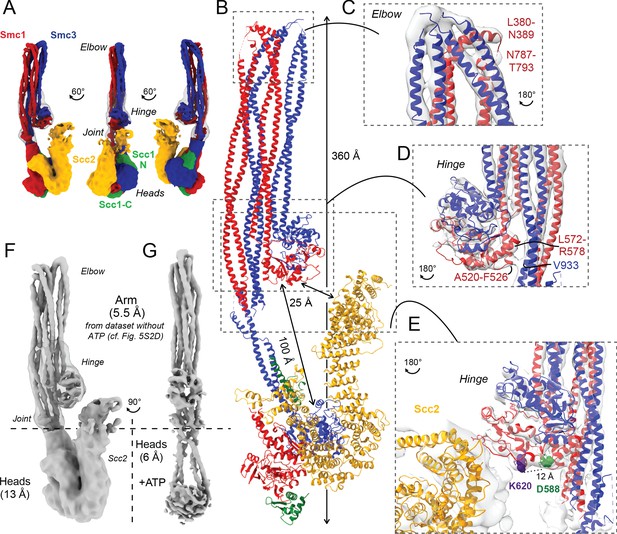
Folded cohesin allows interaction of hinge with Scc2 N-terminus.
(A) Views of cryo-EM reconstruction of Scc2-bound cohesin coloured by subunit. (B) Full pseudo-atomic model of folded cohesin trimer bound to Scc2. (C) Close-up of breaks in the coiled coils of Smc3 and Smc1 that constitute the elbow region of cohesin (PDB: 7OGT; EMD-12887). (D) Close-up of the interaction between the hinge and Smc3 that stabilises the folded state. (E) Close-up of Scc2 N-terminus in proximity of hinge residues K620 and D588Y. (F, G) Comparison of cryo-EM densities between Scc2-bound and ATP-free cohesin seen in (F) (EMD-12880) and ATP-bound cohesin seen in (G) (EMD-12889), demonstrating that head engagement is not sufficient for coiled coil unzipping.
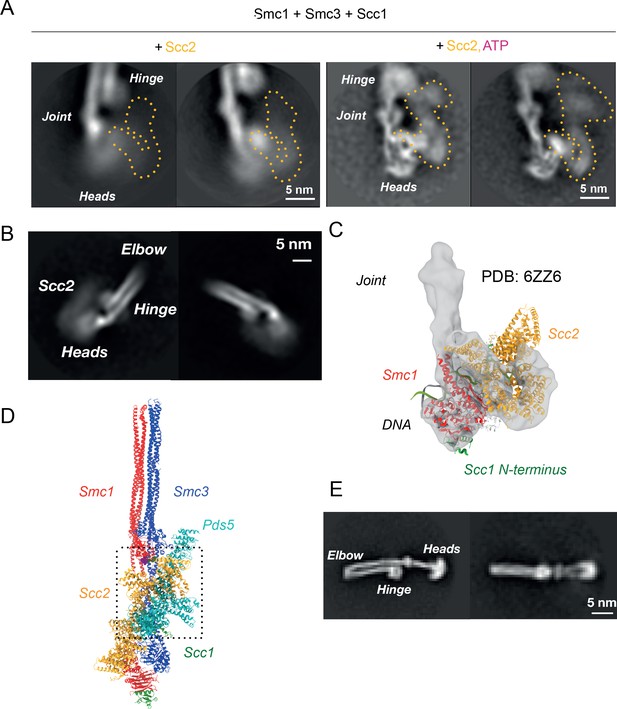
Folded cohesin allows interaction of hinge with Scc2 N-terminus.
(A) 2D classes of Scc2-bound ATPase heads in the absence (left) and presence (right) of ATP demonstrating the stabilising effect of head engagement. (B) 2D classes showing flexibility within Scc2 and between the heads and the joint. (C) Fitting of atomic map from Collier et al., 2020 (6ZZ6) in cryo-EM map made by focused classification. The map originates from the same data as that of Figure 5A and has been processed to remove the floppy C-terminal head domain of Scc2. (D) Overlay of Pds5- and Scc2-bound pseudo-atomic model of cohesin tetramer. The binding of the respective HAWKS is mutually exclusive. (E) 2D classes of engaged cohesin in the absence of any HAWKs that demonstrate that folding through the elbow is constitutive.
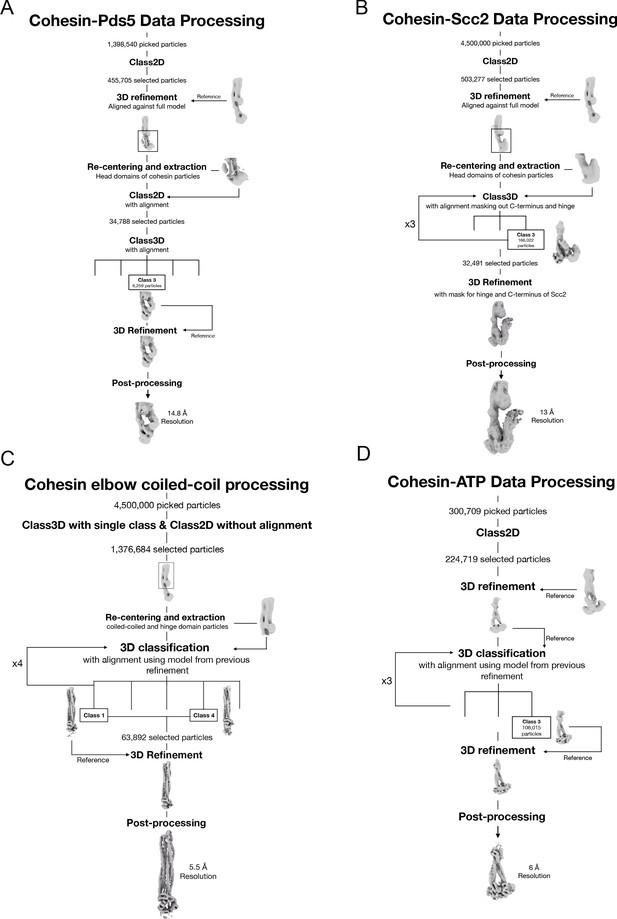
Data processing and reconstruction schematics of all cryo-EM maps.
Processing workflow to obtain the maps of the folded elbow structure (A), the Scc2-bound cohesin complex (B), the Pds5-bound cohesin complex (C), and the engaged ATPase heads (D).
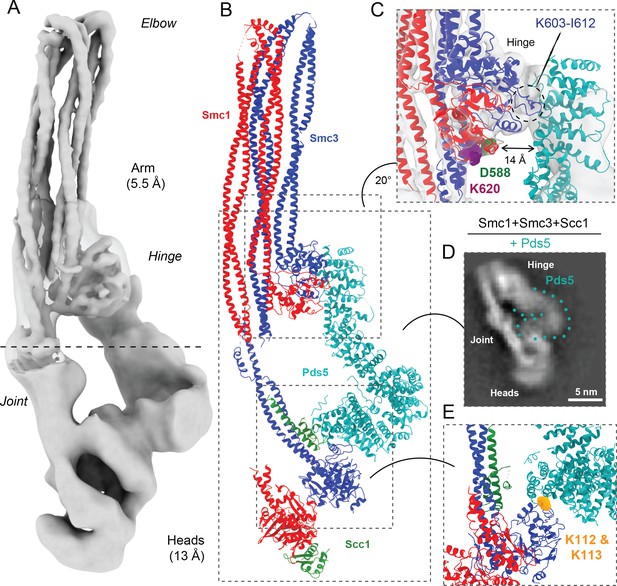
Pds5 binds to Smc3 head while contacting the hinge.
(A) Composite map of cryo-EM reconstructions of Pds5-bound cohesin (EMD-12888). (B) Full pseudo-atomic model of folded cohesin trimer bound to Pds5 coloured by subunit. (C) Close-up of interaction between hinge and Pds5 showing proximity of N-terminus of the HAWK to hinge residues D588 and K620. (D) 2D classes of Pds5-bound ATPase heads. (E) Close-up of Pds5 binding to K112- and K113-proximal region of the Smc3 head.
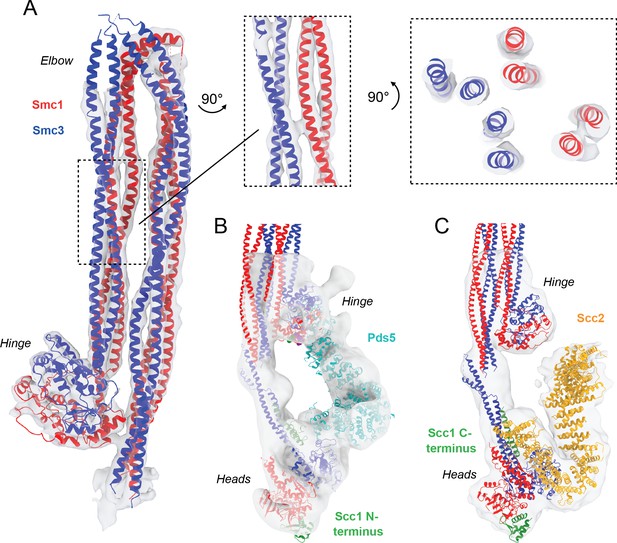
Detailed view of fitted atomic structures in cryo-EM maps.
(A) Coiled coil elbow and hinge pseudo-atomic model fitted into its corresponding cryo-EM density with views of coiled coils from the side and through an intersection. (B) Fitting of Pds5 (PDB: 5F0N; Lee et al., 2016), Smc1 (PDB: 1W1W Haering et al., 2004), and Smc3 (PDB: 4UX3; Gligoris et al., 2014) into the Pds5-bound cryo-EM map. (C) Fitting of Scc2 (PDB: 5T8V; Kikuchi et al., 2016), Smc1 (PDB: 1W1W; Haering et al., 2004), and Smc3 (PDB: 4UX3; Gligoris et al., 2014) into the Scc2-bound cryo-EM map.
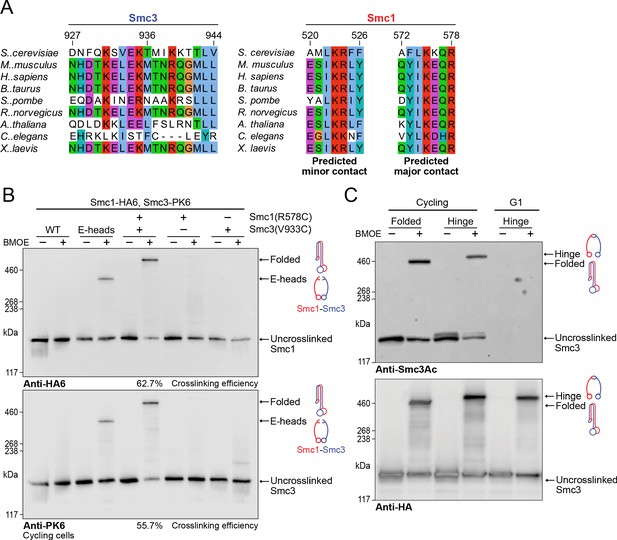
Folding of cohesin’s coiled coils occurs in vivo and is a feature of sister chromatid cohesion.
(A) Sequence conservation analysis for the Smc3 coiled coil and Smc1 hinge helices shown in Figure 4D shows that the residues are highly conserved. (B) Whole-cell extract western blot analysis for the crosslink between Smc1R578C-HA6 and Smc3V933C-PK6 with single cysteine controls probing for hemagglutinin (HA) (top) and PK (bottom). A band shift is observed at the same molecular weight for both blots, confirming the identity of the crosslinked species. Crosslinking of the engaged heads (Chapard et al., 2019) was used as a positive control (K28401, K27359, K28585, K28546, K28583). (C) Western blot analysis of crosslinking measuring the folded state (Smc1R578C-HA6 and Smc3V933C-PK6) and Smc1-Smc3 hinge dimerisation (Haering et al., 2008) probing for acetylated Smc3 (top) and HA (bottom) in logarithmic or pheromone arrested cells (K26081, K28586).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Spodoptera frugiperda) | Sf9 insect cells | Thermo Fisher | Cat# 11496015 | N/A |
| Genetic reagent (Saccharomyces cerevisiae) | NCBITaxon:4932 | This paper | Yeast strains | Supplementary file 4 |
| Biological sample | α-factor peptide | CRUK Peptide Synthesis Service | N/A | N/A |
| Antibody | Mouse monoclonal Anti-V5 | Bio-Rad | Cat# MCA1360 | (1:1000) |
| Antibody | Anti-HA High Affinity (3F10) (Rat) | Roche | Cat# 11867423001 | (1:1000) |
| Antibody | Anti-His (mouse) | GenScript | Cat# A00186 | (1:1000) |
| Antibody | Anti-c-Myc A-14 (9E10) (rabbit) | Santa Cruz Biotech | Cat# sc-789 | (1:1000) |
| Antibody | Anti-Myc 4A6 (mouse) | Millipore | Cat# 05-724 | (1:1000) |
| Antibody | Anti-FLAG (rabbit) | Sigma | Cat# F7425 | (1:1000) |
| Recombinant DNA reagent | pACEbac1 2xStrepII-Scc2151-1493 | Collier et al., 2020 | N/A | N/A |
| Recombinant DNA reagent | pACEbac1 Smc1-8xHis-Smc3/pIDC Scc1-2xStrepII (trimer) | Petela et al., 2018 | N/A | N/A |
| Recombinant DNA reagent | pIDS Pds5-Flag | Petela et al., 2018 | N/A | N/A |
| Commercial assay or kit | Talon Superflow Metal Affinity Resin | Takara Bio. | Cat# 635669 | N/A |
| Commercial assay or kit | NuPAGE 3–8% Tris-Acetate Protein gels | Thermo Fisher | Cat# EA0378BOX | N/A |
| Commercial assay or kit | Trans-Blot Turbo Midi0.2 µm Nitrocellulose Transfer Packs | Bio-Rad | Cat# 1704159 | N/A |
| Commercial assay or kit | Protein G Dynabeads | Thermo Fisher | Cat# 300385 | N/A |
| Commercial assay or kit | ChIP DNA Clean and Concentrator kit | Zymo Research | Cat# D5205 | N/A |
| Commercial assay or kit | NEBNext Fast DNA Library Prep Set for Ion Torrent | NEB | Cat# Z648094 | N/A |
| Commercial assay or kit | Ion Xpress Barcode Adaptors | Thermo Fisher | Cat# 4471250 | N/A |
| Commercial assay or kit | E-Gel SizeSelect II 2% Agarose gels | Thermo Fisher | Cat# G661012 | N/A |
| Commercial assay or kit | KAPA Ion Torrent DNA standards | Roche | Cat# 07960395001 | N/A |
| Commercial assay or kit | EnzChek phosphate assay kit | Thermo Fisher | Cat# E6646 | N/A |
| Commercial assay or kit | StrepTrap HP | Fisher Scientific | Cat# 11540654 | N/A |
| Commercial assay or kit | Superose 6 Increase10/300 GL | VWR | Cat# 29-0915-96 | N/A |
| Chemical compound | Nocodazole | Sigma | Cat# M1404 | N/A |
| Chemical compound | Bismaleimidoethane (BMOE) | Thermo Fisher | Cat# 22323 | 5 mM |
| Chemical compound | Complete EDTA-free protease inhibitor cocktail | Roche | Cat# 4693132001 | (1:50 mL) |
| Chemical compound | PMSF | Sigma | Cat# 03115836001 | 1 mM |
| Chemical compound | Immobilon Western ECL | Millipore | Cat# WBLKS0500 | N/A |
| Chemical compound | RNase A | Roche | Cat# 10109169001 | N/A |
| Chemical compound | Proteinase K | Roche | Cat# 03115836001 | N/A |
| Chemical compound | BPA | Bachem | Cat# 4017646.0005 | N/A |
| Chemical compound | TCEP | Thermo Fisher | Cat# 20490 | N/A |
| Chemical compound | Desthiobiotin | Fisher Scientific | Cat# 12753064 | N/A |
| Software, algorithm | FastQC | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | N/A |
| Software, algorithm | Fastx_trimmer | Hannon Lab | http://hannonlab.cshl.edu/fastx_toolkit/index.html | N/A |
| Software, algorithm | FilterFastq.py | Petela et al., 2018 | https://github.com/naomipetela/nasmythlab-ngs | N/A |
| Software, algorithm | Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | N/A |
| Software, algorithm | Samtools | Samtools | http://www.htslib.org | N/A |
| Software, algorithm | IGB browser | Nicol et al., 2009 | https://www.bioviz.org | N/A |
| Software, algorithm | chr_position.py | Petela et al., 2018 | https://github.com/naomipetela/nasmythlab-ngs | N/A |
| Software, algorithm | filter.py | Petela et al., 2018 | https://github.com/naomipetela/nasmythlab-ngs | N/A |
| Software, algorithm | Bcftools call | Samtools | http://www.htslib.org | N/A |
| Software, algorithm | MutationFinder.py | Petela et al., 2018 | https://github.com/naomipetela/nasmythlab-ngs | N/A |
| Software, algorithm | yeastmine.py | Petela et al., 2018 | https://github.com/naomipetela/nasmythlab-ngs | N/A |
| Software, algorithm | RELION 3.1 | doi:10.1016/j.jsb.2012.09.006 | N/A | N/A |
| Software, algorithm | CtfFind4 | doi:10.1016/j.jsb.2015.08.008 | N/A | N/A |
| Software, algorithm | CrYOLO 1.5 | doi:10.1038/s42003-019-0437 | N/A | N/A |
| Software, algorithm | Chimera | https://www.cgl.ucsf.edu/chimera/ | N/A | N/A |
| Software, algorithm | ChimeraX 1.0 | https://www.cgl.ucsf.edu/chimera/ | N/A | N/A |
| Software, algorithm | COOT | doi:10.1107/S0907444910007493 | N/A | N/A |
| Software, algorithm | MAIN | doi:10.1107/S0907444913008408 | N/A | N/A |
| Software, algorithm | Phenix.real_ space_refinement | doi:10.1107/S2059798318006551 | N/A | N/A |
| Software, algorithm | PYMOL 2 | https://pymol.org/2/ | N/A | N/A |
| Software, algorithm | SWISS-MODEL | https://swissmodel.expasy.org | N/A | N/A |
| Other | Quantifoil R 2/2 grid: Cu/Rh 200 cryoEM grids | Quantifoil GmbH | N/A | N/A |
Additional files
-
Supplementary file 1
Table detailing the amino acid substitutions made at position 588 in Smc1, with their respective ability to complement smc1Δ and scc4Δ.
- https://cdn.elifesciences.org/articles/67268/elife-67268-supp1-v1.docx
-
Supplementary file 2
Data collection and refinement statistics for the Smc1D574Y-Smc1 mouse hinge structure.
- https://cdn.elifesciences.org/articles/67268/elife-67268-supp2-v1.docx
-
Supplementary file 3
Data and model building statistics for all cryo-EM structures.
- https://cdn.elifesciences.org/articles/67268/elife-67268-supp3-v1.docx
-
Supplementary file 4
List of yeast strains and genotypes.
- https://cdn.elifesciences.org/articles/67268/elife-67268-supp4-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67268/elife-67268-transrepform-v1.docx