Cellular assays identify barriers impeding iron-sulfur enzyme activity in a non-native prokaryotic host
Figures
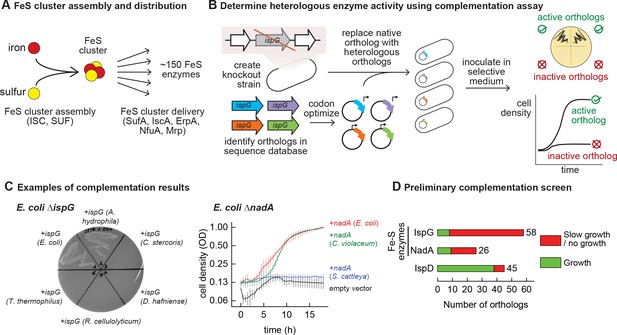
Mapping functional expression of heterologous iron-sulfur (Fe-S) enzymes using complementation experiments.
(A) In prokaryotes, two pathways that assemble and deliver Fe-S clusters have been identified (ISC, SUF), as have several Fe-S delivery proteins (SufA, IscA, ErpA, NfuA in Escherichia coli). (B) The complementation assays used to determine activity of heterologous orthologs expressed by E. coli. Orthologs of conditionally essential enzymes are identified in sequence databases, codon optimized, and cloned into a low-copy plasmid. Expression is controlled by an inducible promoter (PTet). The plasmids are then transformed into E. coli strains lacking the corresponding E. coli ortholog. Each strain is inoculated in selective media and expression of the heterologous ortholog is induced. Growth in selective media indicates that the heterologous ortholog retains sufficient activity to support growth of E. coli. (C) Exemplary results of complementation assay obtained using either solid or liquid selective medium. Error bars represent standard error of duplicate cultures inoculated from separate colonies. (D) Results of preliminary complementation tests comparing activity of Fe-S enzymes with a non-Fe-S enzyme. Orthologs tested for the preliminary screen are listed in Supplementary file 1.
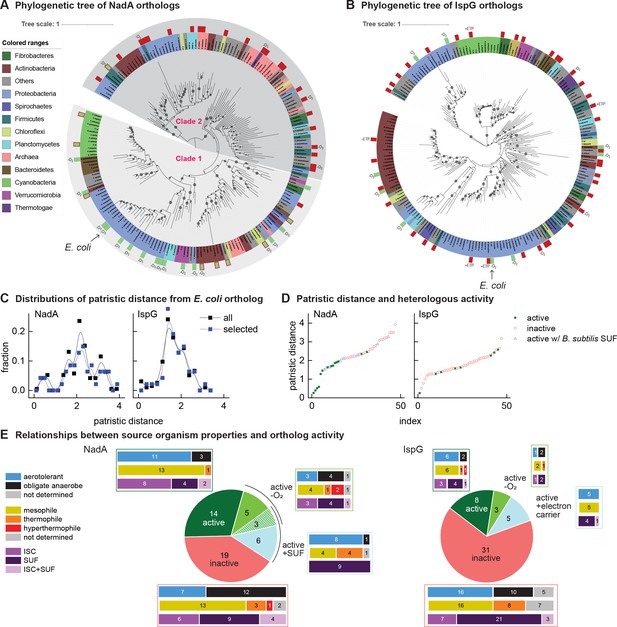
Phylogenetic diagrams contrasting the compatibility of iron-sulfur (Fe-S) enzymes NadA and IspG across a selection of orthologs representative of known prokaryotic sequence diversity.
(A, B) Phylogeny of NadA orthologs (A) and IspG orthologs (B). In each diagram, the position of Escherichia coli ortholog is highlighted by an arrow. Orthologs selected for our complementation assay are indicated by color rectangles (green, successful complementation; green surrounded by red, complementation by coexpression of Bacillus subtilis SUF; red, no complementation). Orthologs complementing growth in anaerobic conditions are indicated by –O2. IspG orthologs recovered by expression of compatible electron transfer protein indicated by +ETP. NadA Clades 1 and 2 are indicated. (C) Distributions of patristic distances from the corresponding E. coli ortholog of all sequences (black curves) and the sequences of orthologs selected for testing (blue curves). (D) Complementation and reactivation results correlated with patristic distance from the E. coli ortholog. (E) Summary of complementation and recovery results for heterologous orthologs. The bar plots adjoining each wedge indicate the characteristics of native hosts (aerotolerance, optimal growth temperature, Fe-S cluster biosynthesis system). SUF and ISC indicate that homologs to SufBD or IscU, respectively, are identified in native genome. Results and properties of individual orthologs are given in Tables 1 and 2.
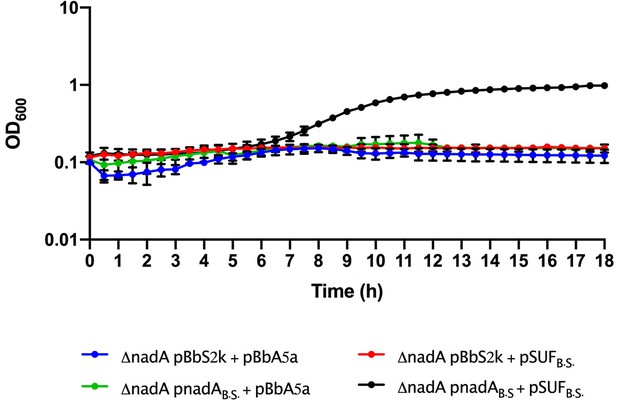
Growth curves of Escherichia coli ΔnadA grown in M9, Kan, Amp, aTc, and isopropyl β-D-1-thiogalactopyranoside complemented with pBbS2k and pBbA5a plasmids carrying different genes: empty vectors (blue line), pBbS2k and Bacillus subtilis SUF (red line), B. subtilis nadA and pBbA5a (green line), and B. subtilis nadA and B. subtilis SUF (black line).
B.S., B. subtilis. The average of two biological replicates is reported with SD.
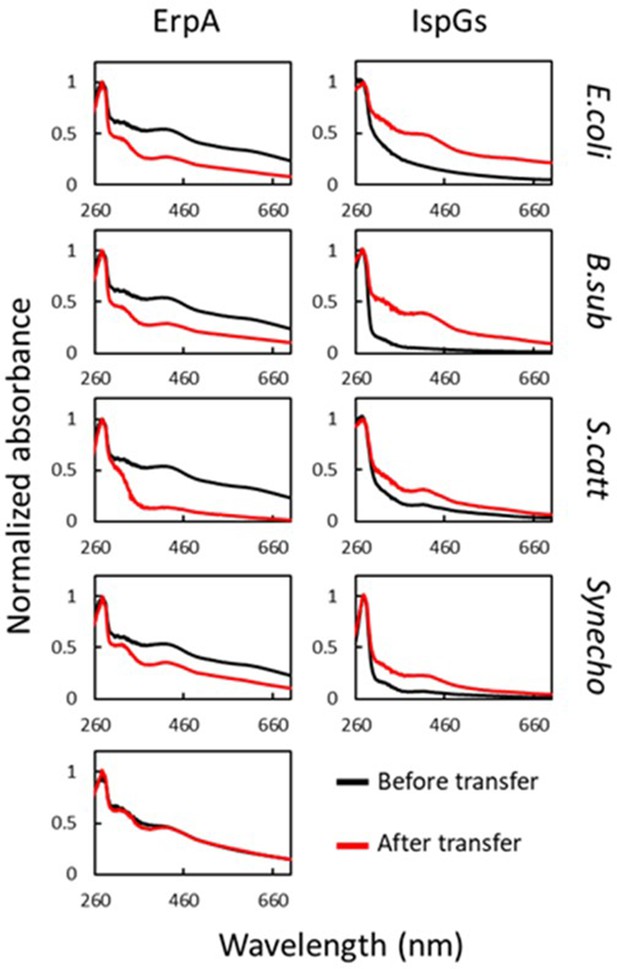
Escherichia coli ErpA mediates iron-sulfur (Fe-S) cluster transfer into heterologous IspGs.
UV-visible spectra of (Fe-S)-ErpA and apo-IspGs were recorded before ErpA/IspG coincubation (black traces). One equivalent of apo-IspG was incubated with 2.2 equivalents of (Fe-S)-ErpA. After 1 hr incubation, proteins were separated on an affinity column (IspGs in the flow-through [FT], ErpA in the eluate). Spectra were recorded for both (red traces). The bottom left panel corresponds to spectra of E. coli ErpA incubated without IspG. IspG orthologs used: E. coli, E. coli; B. sub, B. subtilis; S. catt: Streptomyces cattleya; Synecho, Synechocystis sp. CACIAM05.
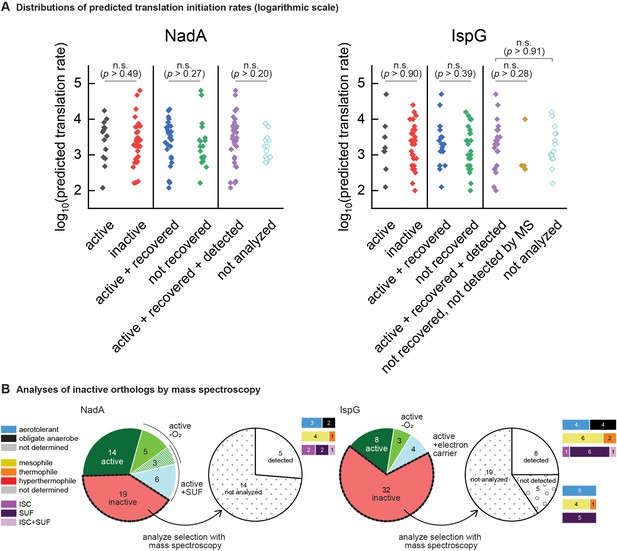
Confirming expression of heterologous orthologs.
(A) Translation rates predicted by the RBS calculator for NadA and IspG orthologs from the phylogenetically based set. Predicted rates are sorted into distributions corresponding to the results of complementation assays (‘active’ vs ‘inactive’), recovery assays (‘active + recovered’ vs ‘not recovered’), and mass spectroscopy detection assays (‘active + recovered + detected’ vs ‘not detected by MS’ and ‘not analyzed’). Points indicate individual value obtained for each ortholog tested and all values are depicted. Statistical comparisons as depicted between groups revealed no significant differences (Kruskal-Wallis ANOVA test, degree of freedom = 1). n.s., not significant (p<0.05). Test applied using Origin 2019 software. (B) Pie charts depicting outcomes of mass spectroscopy detection experiments analyzing subsets of the inactive orthologs. The bar plots adjoining each wedge indicate the characteristics of native hosts (aerotolerance, optimal temperature, iron-sulfur cluster biosynthesis system).
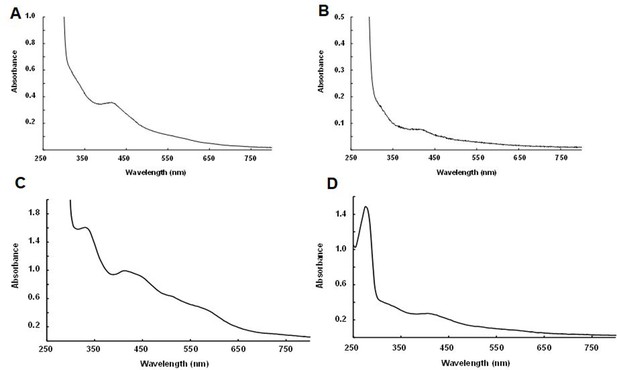
Biochemical detection of iron-sulfur (Fe-S) clusters within purified NadA and IspG proteins.
UV-visible spectra together with iron and sulfur content of orthologs expressed within aerobic E. coli cultures followed by anaerobic purification. Spectra from (A) Dictyoglomus thermophilum NadA (33 mg/ml, Fe and S content: 0.1 ± 0.02 Fe/monomer and 0.1 ± 0.02 S/monomer); (B) Desulfurispirillum indicum NadA (2 mg/ml, 0.15 ± 0.01 Fe/monomer and 0.13 ± 0.02 S/monomer); (C) Thermotoga maritima IspG (20 mg/ml, 0.4 ± 0.02 Fe/monomer and 0.38 ± 0.03 S/monomer); (D) Symbiobacterium thermophilum IspG (2.2 mg/ml, 0.95 ± 0.02 Fe/monomer 0.85 ± 0.1 S/monomer).
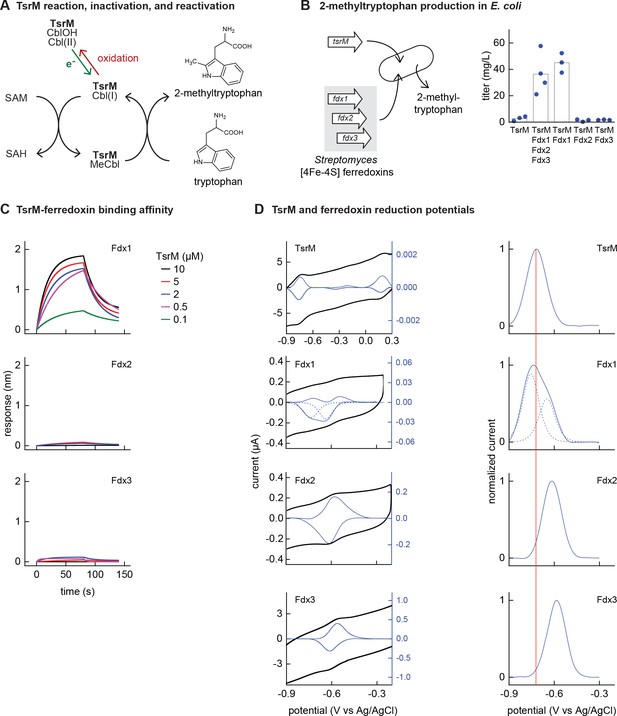
Identifying the barrier to optimal activity of an iron-sulfur (Fe-S)-dependent rSAM enzyme.
(A) The enzyme TsrM synthesizes 2-methyltryptophan as part of the synthesis pathway of the antibiotic thiostrepton. A one-electron reduction (in green) converts the bound cobalamin to cob(I)alamin, which is converted to MeCbl via reaction with SAM. TsrM can be inactivated during catalysis by adventitious oxidation of the bound cob(I)alamin to cob(II)alamin (in red). Recovering the catalytically active cob(I)alamin form requires a one-electron reduction (green). (B) Titers of 2-methyltryptophan in a TsrM-expressing E. coli strain increase substantially upon coexpression of a specific Streptomyces cattleya ferredoxin (Fdx1) whereas coexpression of alternative S. cattleya [4Fe-4S] ferredoxins, Fdx2 and Fdx3, does not increase titers. (C, D) Identifying the molecular basis enabling Fdx1 to recover TsrM activity. (C) Biolayer interferometry of TsrM with Fdx1, Fdx2, and Fdx3 indicates that TsrM interacts measurably with Fdx1 alone. (D) Cyclic voltammetry of TsrM and the three ferredoxins indicates that the reducing potentials of one of the (Fe-S) clusters of Fdx1 are sufficiently low potential to reduce TsrM metal cofactors (cob(II)alamin to cob(I)alamin and [4Fe-4S]+2 to [4Fe-4S]+1) and thus restore activity after adventitious oxidation.
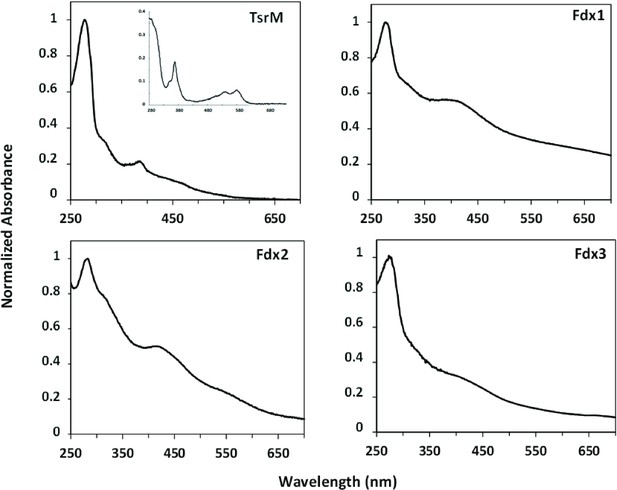
TsrM, Fdx1, Fdx2, and Fdx3 with their metallic cofactors.
Protein concentration was normalized with Abs280 = 1. Consistent with previous observations, as-purified TsrM (0.9 Cbl and 3.5 Fe ± 0.3 per monomer) shows features consistent with the presence both of iron-sulfur [Fe-S] cluster and of cobalamin, including the broad absorption between 350 and 600 nm, indicative of a Fe-S cluster, and the distinct feature at 390 nm, suggestive of the presence of cob(I)alamin. Inset: TsrM treated with potassium cyanide yields the dicyanocobalamin adduct of cobalamin, which has a strong feature at 367 nm (ε367 = 30,800 M−1 cm–1). Concerning Streptomyces cattleya ferredoxins (Fdx1, Fdx2, Fdx3), their iron content was consistent with their annotations as dicluster and single-cluster ferredoxins (7.4 Fe ± 0.5/monomer, 2.9 Fe ± 0.1/monomer, and 2.8 Fe ± 0.1/monomer for Fdx1, Fdx2, and Fdx3, respectively).
Tables
Compiled results of complementation and protein expression experiments for each NadA ortholog.
Complementation of Escherichia coli ΔnadA and recovery of NadA activity with either SUF coexpression (pNadA + pBsSUF for Bacillus subtilis SUF, or pNadA + pEcSUF for E. coli SUF) or anaerobic growth (pNadA −O2). Minus (−), negative complementation; plus (+), positive complementation; N.T., not tested. Protein expression, predicted translation initiation rate from the RBS calculator (log10 scale) and outcome of SDS-PAGE and MS detection experiments. SDS-PAGE column, (+) band observed on an SDS-PAGE gel. MS column, (+) confirmed detection by mass spectroscopy; N.T., not tested; detection N.T., negative complementation and detection by MS not tested. The properties of native species for each ortholog are indicated: O2 tolerance: (+) aerotolerant, (−) obligate anaerobe, (?) unknown; iron-sulfur (Fe-S) synthesis system describes homologs to IscU or SufB identified in native genome. SufBD, SufB homolog detected; IscU, IscU homolog detected; SufBC, IscU, both systems detected.
| Phylum | Species/strain | Complementation/recovery | Protein expression | Native host characteristics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pNadA | pNadA + pBsSUF | pNadA + pEcSUF | pNadA −O2 | Complemented or recovered | Predicted translationinitiation (log10) | SDS-PAGE | MS | Complemented/recovered/detected | O2 tolerance | Temperature | Fe-S synthesis system | ||
| Actinobacteria | Mycolicibacterium smegmatis MC2 155 | − | + | + | + | + | 3.3 | − | + | + | + | Mesophilic | SufBD |
| Rubrobacter xylanophilus DSM 9941 | + | N.T. | N.T. | + | + | 2.7 | − | N.T. | + | + | Thermophilic | SufBD | |
| Collinsella stercoris DSM 13279 | − | − | − | − | − | 3.4 | − | N.T. | Detection N.T. | − | Mesophilic | SufBD | |
| Acidimicrobium ferrooxidans DSM 10331 | − | + | − | − | + | 2.2 | − | N.T. | + | + | Thermophilic | SufBD | |
| Streptomyces cattleya NRRL 8057 = DSM 46488 | − | − | − | − | − | 4.8 | − | + | + | + | Mesophilic | SufBD | |
| Bacteroidetes | Salinivirga cyanobacteriivorans | + | N.T. | N.T. | + | + | N.T. | + | N.T. | + | − | Mesophilic | SufBD |
| Candidatus Fermentibacteria | Candidatus Fermentibacter daniensis | − | − | − | − | − | 3.9 | − | N.T. | Detection N.T. | − | Mesophilic | IscU |
| Candidatus Melainabacteria | Candidatus Gastranaerophilales bacterium HUM_7 | − | − | − | + | + | 3.8 | − | N.T. | + | − | ? | IscU |
| Chloroflexi | Sphaerobacter thermophilus DSM 20745 | − | + | − | + | + | 4.1 | − | N.T. | + | + | Thermophilic | SufBD |
| Thermogemmatispora tikiterensis | − | − | − | − | − | 2.8 | − | N.T. | Detection N.T. | + | Thermophilic | SufBD | |
| Chrysiogenetes | Desulfurispirillum indicum S5 | − | − | − | + | + | 3.6 | + | N.T. | + | − | Mesophilic | IscU |
| Cyanobacteria | Synechocystis sp. PCC 6803 | − | + | − | − | + | 3.3 | − | N.T. | + | + | Mesophilic | SufBD |
| Crocosphaera watsonii WH 8501 | − | + | − | − | + | 2.8 | − | N.T. | + | + | ? | SufBD | |
| Deinococcus-Thermus | Thermus thermophilus HB27 | − | + | − | − | + | 3.5 | − | N.T. | + | + | Thermophilic | SufBD |
| Dictyoglomi | Dictyoglomus thermophilum H-6–12 | − | − | − | + | + | 3.6 | + | N.T. | + | − | Hyperthermophilic | SufBD, IscU |
| Euryarchaeota | Methanopyrus kandleri AV19 | − | − | − | − | − | 2.9 | − | N.T. | Detection N.T. | − | Hyperthermophilic | SufBD |
| Haloferax volcanii DS2 | − | − | − | − | − | 3.1 | − | N.T. | Detection N.T. | + | Mesophilic | SufBD | |
| Haloterrigena turkmenica DSM 5511 | − | − | − | − | − | 3.8 | − | N.T. | Detection N.T. | + | Mesophilic | SufBD | |
| Candidatus Methanomethylophilus alvus Mx1201 | − | − | − | − | − | 3.0 | − | N.T. | Detection N.T. | − | ? | SufBD | |
| Fibrobacteres | Chitinivibrio alkaliphilus ACht1 | − | − | − | − | − | 3.2 | − | N.T. | Detection N.T. | − | Mesophilic | IscU |
| Firmicutes | Bacillus subtilis subsp. subtilis str. 168 | − | + | + | + | + | 3.8 | − | N.T. | + | + | Mesophilic | SufBD |
| Desulfitobacterium hafniense DCB-2 | − | − | − | + | + | 3.5 | − | + | + | − | Mesophilic | IscU | |
| Ruminiclostridium cellulolyticum H10 | − | − | − | − | − | 2.9 | − | N.T. | Detection N.T. | − | Mesophilic | IscU | |
| Heliobacterium modesticaldum Ice1 | − | − | − | − | − | 3.4 | − | + | + | − | Thermophilic | IscU | |
| Nitrospinae | Nitrospina gracilis | + | N.T. | N.T. | + | + | 3.2 | + | N.T. | + | + | Mesophilic | SufBD |
| Planctomycetes | Gemmata obscuriglobus | + | N.T. | N.T. | + | + | 2.1 | − | N.T. | + | + | Mesophilic | IscU |
| Pirellula staleyi DSM 6068 | − | − | − | − | − | 2.7 | − | + | + | + | Mesophilic | SufBD | |
| Proteobacteria | Aeromonas hydrophila | + | N.T. | N.T. | + | + | 3.8 | − | N.T. | + | + | Mesophilic | IscU |
| Desulfovibrio vulgaris str. Hildenborough | − | − | − | − | − | 3.2 | − | N.T. | Detection N.T. | − | Mesophilic | SufBD, IscU | |
| Allochromatium vinosum | + | N.T. | N.T. | + | + | 2.9 | + | N.T. | + | − | Mesophilic | SufBD, IscU (NifU) | |
| Arcobacter butzleri | − | − | − | − | − | 2.2 | − | + | + | + | Mesophilic | SufBD, IscU | |
| Novosphingobium stygium | − | + | + | − | + | 2.3 | − | N.T. | + | + | Mesophilic | SufBD | |
| Herminiimonas arsenicoxydans | + | N.T. | N.T. | + | + | 3.5 | − | N.T. | + | + | Mesophilic | IscU | |
| Chromobacterium violaceum ATCC 12472 | + | N.T. | N.T. | + | + | 3.4 | − | N.T. | + | + | Mesophilic | IscU | |
| Bdellovibrio bacteriovorus HD100 | + | N.T. | N.T. | + | + | 3.9 | − | N.T. | + | + | Mesophilic | SufBD | |
| Methylobacillus flagellatus KT | − | − | − | − | − | 4.7 | − | + | + | − | Mesophilic | IscU | |
| Anaeromyxobacter dehalogenans 2 CP-C | + | N.T. | N.T. | + | + | 3.7 | − | N.T. | + | − | Mesophilic | SufBD, IscU | |
| Candidatus Pelagibacter ubique HTCC1002 | − | − | − | − | − | 3.4 | − | N.T. | Detection N.T. | + | ? | SufBD | |
| Syntrophobacter fumaroxidans MPOB | − | − | − | − | − | 3.5 | − | N.T. | Detection N.T. | − | Mesophilic | SufBD, IscU | |
| Cellvibrio japonicus Ueda107 | + | N.T. | N.T. | + | + | 2.9 | − | N.T. | + | + | Mesophilic | SufBD | |
| Escherichia coli str. K-12 substr. MG1655 | + | N.T. | N.T. | + | + | 3.8 | + | + | + | + | Mesophilic | SufBD, IscU | |
| Desulfarculus baarsii DSM 2075 | − | − | − | − | − | 2.8 | − | N.T. | Detection N.T. | − | Mesophilic | IscU | |
| Salinisphaera sp. LB1 | + | N.T. | N.T. | + | + | 4.0 | − | N.T. | + | + | Mesophilic | SufBD | |
| Spirochaetes | Leptospira interrogans serovar Lai str. 56,601 | + | N.T. | N.T. | + | + | 4.2 | + | N.T. | + | + | Mesophilic | SufBD |
| Spirochaeta thermophila DSM 6578 | − | + | − | − | + | 3.2 | − | N.T. | + | − | Thermophilic | SufBD | |
| Synergistetes | Thermanaerovibrio acidaminovorans DSM 6589 | − | − | − | − | − | N.T. | − | N.T. | Detection N.T. | − | Thermophilic | SufBD, IscU |
| Thermotogae | Thermotoga maritima MSB8 | − | − | − | + | + | 4.3 | − | N.T. | + | − | Hyperthermophilic | SufBD |
| Verrucomicrobia | Coraliomargarita akajimensis DSM 45221 | + | N.T. | N.T. | + | + | 3.6 | + | N.T. | + | + | Mesophilic | SufBD |
Compiled results of complementation and protein expression experiments for each IspG ortholog.
Complementation of Escherichia coli ΔispG and recovery of IspG activity with either coexpression of electron transfer proteins (pIspG + Fld or Fd) or anaerobic growth (pIspG −O2). Electron transfer proteins found to activate each ortholog are listed in Supplementary files 5 and 6. Minus (−), negative complementation; plus (+), positive complementation; N.T., not tested. Protein expression, predicted translation initiation rate from the RBS calculator (log10 scale) and outcome of SDS-PAGE and mass spectroscopy detection experiments. SDS-PAGE column, (+) band observed on an SDS-PAGE gel. MS column, (+) confirmed detection by mass spectroscopy; (−), detection attempted by mass spectroscopy but no signal matching expected peptides; N.T., not tested; detection N.T., negative complementation and detection by MS not tested. The properties of native species for each ortholog are indicated: O2 tolerance: (+) aerotolerant, (−) obligate anaerobe, (?) unknown; iron-sulfur (Fe-S) synthesis system describes homologs to IscU or SufB identified in native genome. SufBD, SufB homolog detected; IscU, IscU homolog detected; SufBC, IscU, both systems detected.
| Phylum | Species/strain | Complementation/recovery | Protein expression | Native host characteristics | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pIspG | pIspG + Fld or fd | pIspG −O2 | Complemented or recovered | Predicted translation initiation rate (log10) | SDS-PAGE | MS | Complemented/recovered/detected | O2 tolerance | Temperature | Fe-S scaffold | ||
| Acidobacteria | Candidatus Koribacter versatilis Ellin345 | − | N.T. | − | − | 3.6 | − | N.T. | Detection N.T. | + | Mesophilic | IscU |
| Actinobacteria | Thermoleophilum album | − | N.T. | − | − | 2.9 | − | N.T. | Detection N.T. | + | Thermophilic | SufBD |
| Streptomyces coelicolor A3 | − | − | − | − | 2.6 | − | − | − | + | Mesophilic | SufBD | |
| Rubrobacter xylanophilus DSM 9941 | + | N.T. | + | + | 2.6 | − | N.T. | + | + | Thermophilic | SufBD | |
| Cutibacterium acnes KPA171202 | + | N.T. | + | + | 4.7 | − | N.T. | + | − | Mesophilic | SufBD | |
| Collinsella stercoris DSM 13279 | − | − | + | + | 3.3 | − | − | + | − | Mesophilic | SufBD | |
| Acidimicrobium ferrooxidans DSM 10331 | + | N.T. | + | + | 3.5 | − | N.T. | + | + | Mesophilic | SufBD | |
| Streptomyces cattleya NRRL 8057 = DSM 46488 | − | + | − | + | 2.7 | + | N.T. | + | + | Mesophilic | SufBD | |
| Egibacter sp. | − | N.T. | − | − | 3.4 | − | N.T. | Detection N.T. | ? | ? | SufBD | |
| Aquificae | Aquifex aeolicus VF5 | + | N.T. | + | + | 3.8 | − | N.T. | + | + | Hyperthermophilic | IscU |
| Thermovibrio ammonificans HB-1 | − | N.T. | − | − | 2.9 | − | N.T. | Detection N.T. | − | Thermophilic | SufBD | |
| Bacteroidetes | Pedobacter heparinus DSM 2366 | − | N.T. | − | − | N.T. | − | N.T. | Detection N.T. | + | Mesophilic | SufBD, IscU |
| Salinivirga cyanobacteriivorans | − | N.T. | − | − | N.T. | − | N.T. | Detection N.T. | − | Mesophilic | SufBD | |
| Candidatus Fermentibacteria | Candidatus Fermentibacter daniensis | − | N.T. | − | − | 3.0 | − | N.T. | Detection N.T. | − | Mesophilic | IscU |
| Candidatus Melainabacteria | Candidatus Caenarcanum bioreactoricola | − | N.T. | − | − | 4.1 | − | N.T. | Detection N.T. | ? | ? | IscU |
| Candidatus Gastranaerophilales bacterium HUM_7 | − | N.T. | − | − | 4.2 | − | N.T. | Detection N.T. | − | ? | IscU | |
| Candidatus Sumerlaeota | Candidatus Sumerlaea chitinovorans | − | N.T. | − | − | 2.9 | − | N.T. | Detection N.T. | ? | ? | SufBD |
| Chlamydiae | Chlamydia caviae GPIC | − | − | − | − | 2.7 | − | − | − | + | Mesophilic | SufBD |
| Chloroflexi | Sphaerobacter thermophilus DSM 20745 | − | N.T. | − | − | N.T. | − | N.T. | Detection N.T. | + | Thermophilic | SufBD |
| Cyanobacteria | Gloeobacter violaceus PCC 7421 | − | − | − | − | 3.6 | − | N.T. | Detection N.T. | + | ? | SufBD |
| Synechocystis sp. PCC 6803 | − | + | − | + | 3.5 | − | N.T. | + | + | Mesophilic | SufBD | |
| Deinococcus-Thermus | Thermus thermophilus HB27 | − | − | − | − | 2.7 | − | − | − | + | Thermophilic | SufBD |
| Fibrobacteres | Fibrobacter succinogenes subsp. succinogenes S85 | − | N.T. | − | − | 2.6 | − | N.T. | Detection N.T. | − | Mesophilic | SufBD |
| Firmicutes | Bacillus subtilis subsp. subtilis str. 168 | − | + | − | + | 2.7 | − | N.T. | + | + | Mesophilic | SufBD |
| Desulfitobacterium hafniense DCB-2 | − | − | − | − | 3.4 | − | + | + | − | Mesophilic | SufBD | |
| Symbiobacterium thermophilum IAM 14863 | − | − | − | − | 2.0 | − | + | + | + | Thermophilic | SufBD | |
| Ruminiclostridium cellulolyticum H10 | − | − | − | − | 3.4 | − | + | + | − | Mesophilic | IscU | |
| Lentisphaerae | Victivallales bacterium CCUG 44730 | − | N.T. | − | − | N.T. | − | N.T. | Detection N.T. | ? | ? | IscU |
| Nitrospinae | Nitrospina gracilis | − | − | − | − | 4.0 | − | − | − | + | Mesophilic | SufBD |
| Planctomycetes | Blastopirellula marina DSM 3645 | +/− | + | + | + | 3.1 | − | N.T. | + | + | Mesophilic | SufBD |
| Isosphaera pallida ATCC 43644 | − | − | − | − | 2.5 | − | + | + | + | Mesophilic | SufBD | |
| Phycisphaera mikurensis NBRC 102666 | − | N.T. | − | − | 3.8 | − | N.T. | Detection N.T. | + | Mesophilic | SufBD | |
| Proteobacteria | Aeromonas hydrophila | + | N.T. | + | + | 2.1 | − | N.T. | + | + | Mesophilic | IscU |
| Allochromatium vinosum | − | − | − | − | 3.7 | − | + | + | − | Mesophilic | SufBD, IscU (NifU) | |
| Azoarcus sp. BH72 | + | N.T. | + | + | 3.1 | + | N.T. | + | − | Mesophilic | IscU | |
| Helicobacter pylori J99 | − | − | + | + | 3.3 | + | + | + | + | Mesophilic | IscU | |
| Methylococcus capsulatus str. Bath | + | N.T. | + | + | 3.2 | − | N.T. | + | + | Mesophilic | SufBD, IscU | |
| Rhodobacter sphaeroides 2.4.1 | − | + | − | + | 3.4 | − | N.T. | + | + | Mesophilic | SufBD, IscU (NifU) | |
| Mariprofundus ferrooxydans | − | + | − | + | 3.7 | − | + | + | + | Mesophilic | SufBD | |
| Cellvibrio japonicus Ueda107 | − | − | − | − | 2.6 | − | − | − | + | Mesophilic | SufBD | |
| Escherichia coli str. K-12 substr. MG1655 | + | N.T. | + | + | 3.1 | + | + | + | + | Mesophilic | SufBD, IscU | |
| Anaplasma phagocytophilum str. CRT38 | − | − | − | − | 2.2 | − | N.T. | Detection N.T. | + | ? | IscU | |
| Spirochaetes | Spirochaeta thermophila DSM 6578 | − | − | − | − | 4.0 | − | + | + | − | Thermophilic | SufBD |
| Synergistetes | Thermanaerovibrio acidaminovorans DSM 6589 | − | N.T. | − | − | 3.1 | − | N.T. | Detection N.T. | − | Thermophilic | SufBD, IscU |
| Tenericutes | Spiroplasma citri | − | N.T. | − | − | 3.0 | − | N.T. | Detection N.T. | + | Mesophilic | SufBD |
| Thermodesulfobacteria | Thermodesulfatator indicus DSM 15286 | − | N.T. | − | − | 3.3 | − | N.T. | Detection N.T. | − | Thermophilic | SufBD |
| Thermotogae | Thermotoga maritima MSB8 | − | − | + | + | 4.4 | + | + | + | − | Hyperthermophilic | SufBD |
| Verrucomicrobia | Coraliomargarita akajimensis DSM 45221 | − | − | − | − | 2.6 | − | + | + | + | Mesophilic | SufBD |
Additional files
-
Supplementary file 1
Accession numbers and nucleotide sequences of heterologous NadA, IspG, and IspD orthologs used in the preliminary screen.
Complementation results indicated with minus (-): negative complementation; plus (+): positive complementation; (+/-): growth detected but substantially slower than with E. coli ortholog; N.A.: not analyzed.
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp1-v1.xlsx
-
Supplementary file 2
List of genomes used for the phylogenetic analysis.
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp2-v1.xlsx
-
Supplementary file 3
List of the NadA and IspG orthologs retrieved from the database depicted in Supplementary file 2.
Sequences that have been selected for the complementation test are indicated by a (+) in the adjacent column.
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp3-v1.xlsx
-
Supplementary file 4
Accession numbers and nucleotide sequences used to create a phylogenetically-representative selection of NadA and IspG orthologs.
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp4-v1.xlsx
-
Supplementary file 5
List of the Fe-S biogenesis proteins and electron carriers used to test re-activation of IspG orthologs using complementation, split into organisms from the phylogenetically-based set (top) and the preliminary survey set (bottom).
Re-activation of IspG orthologs was tested first by co-expressing all genes as a multi-cistronic operon: (+) indicating positive complementation; (-) indicating no complementation observed against any IspG ortholog tested. For cases in which individual electron carriers and Fe-S biogenesis proteins were sub-cloned for complementation testing, proteins activating at least one heterologous IspG ortholog (positive complementation) are indicated by (+), while proteins that did not activate IspG (no complementation) are indicated by (-).
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp5-v1.xlsx
-
Supplementary file 6
IspG reactivation using plasmids listed in Supplementary file 5, including cross-activation of IspG orthologs by non-native electron carrier proteins.
IspG orthologs belonging to the phylogenetically-based set and orthologs from the preliminary survey that were not included in the phylogenetic set are depicted separately. Minus (-): negative complementation; plus (+): positive complementation; (+/-): growth detected but substantially slower than with E. coli ortholog; N.T.: not tested. The rightmost column, provided to summarize reactivation experiments, indicates IspG orthologs that were successfully activated by at least one plasmid.
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp6-v1.xlsx
-
Supplementary file 7
Results of SDS-PAGE and shotgun proteomics of selected orthologs.
Orthologs detected by visible bands on the gel are indicated by a (+).
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp7-v1.xlsx
-
Supplementary file 8
List of the bacterial strains, plasmids, and oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/70936/elife-70936-supp8-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70936/elife-70936-transrepform1-v1.docx





