A mechano-osmotic feedback couples cell volume to the rate of cell deformation
Figures
Cell volume in spreading cells.
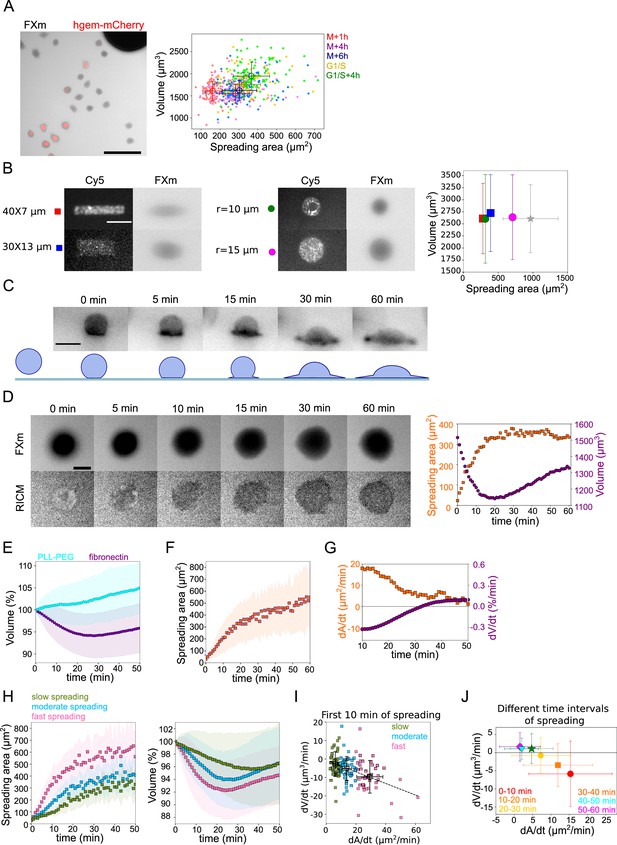
(A) Left: Composite of FXm in GFP channel and fluorescent image in mCherry channel of HeLa hgem-mCherry cells. Scale bar: 100 µm. Right: Relation between volume and spreading area of HeLa hgem-mCherry cells at the different cell cycle stages, N=3: M+1h (n=131) correlation coefficient R=0.11, M+4h (n=131) R=0.23, M+6h (n=131) R=0.26, G1/S (n=99) R=0.20, G1/S+4h (n=92) R=0.22. Error bars represent standard deviation. (B) Left: Typical images of micropatterns and typical images of cells plated on micropatterns. Scale bar: 10 µm. Right: Average volume of HeLa Kyoto cells plated on the patterns (measurements are done 4 hr after cell plating) of different shape and size in comparison with non-patterned cells. Blue: rectangle 30×13 µm2 (n=131, N=2); red: rectangle 40×7 µm2 (n=214, N=2); purple: circle, r=15 µm (n=338, N=4); green: circle, r=10 µm (n=242, N=3); gray: non-patterned cells (n=286, N=3). Error bars represent standard deviation. There is nos statistical difference between patterned cells and non-patterned cells: rectangle 30×13 p=0.15, rectangle 40×7 µm2 p=0.96, r=15 µm p=0.63, r=10 µm p=0.97. (C) Top: Side view of a HeLa-Lifeact (black) cell spreading on fibronectin-coated glass. Scale bar: 20 µm. Bottom: Scheme of shape transition during cell spreading. (D) Left: FXm and RICM imaging of a HeLa Kyoto cell spreading on fibronectin-coated glass. Scale bar: 20 µm. Right: Volume (red) and spreading area (blue) of cell represented on the left panel. (E) Average normalized volume of control HeLa Kyoto cells (blue, n=127, N=3) spreading on fibronectin-coated glass, or plated on PLL-PEG-coated glass (cyan, n=493, N=5). Error bars represent standard deviation. (F) Average spreading area of control HeLa Kyoto cells (n=125, N=3), spreading on fibronectin-coated glass. Error bars represent standard deviation. (G) Linear derivatives dA/dt (blue) and dV/dt (red) for average spreading area and volume represented on (F) and (E) for sliding window 10 min. (H) Average normalized volume (left) and spreading area (right) of control HeLa cells divided in three categories based on their initial spreading speed, N=3: slow (n=42), moderate (n=43), fast (n=42). Error bars represent standard deviation. (I) Volume flux (dV/dt) of single control HeLa Kyoto cells (n=194, N=3) plotted versus their spreading speed (dA/dt) at the first 10 min of spreading. The data are fitted with linear regression y=−0.31x−0.71, R2=0.19. Error bars represent standard deviation. Color code indicate three groups of cells represented on (H). (J) Median volume flux (dV/dt) of HeLa Kyoto cells plotted versus median spreading speed (dA/dt) at the different time intervals of spreading (n=194, N=3). Error bars represent standard deviation.
-
Figure 1—source data 1
Data tables related to quantifications in Figure 1.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig1-data1-v2.zip
Cell volume in spreading cells.
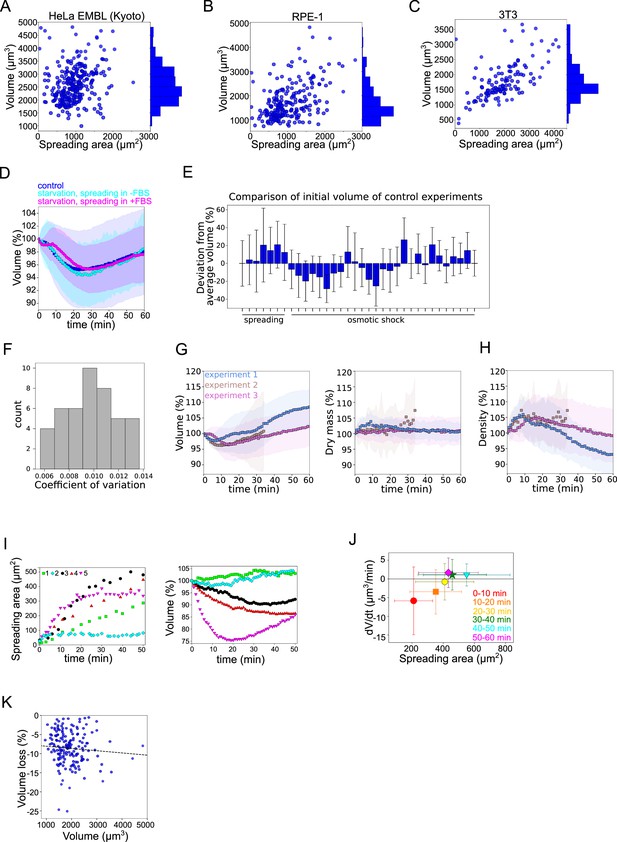
(A) Relation between volume and spreading area of HeLa Kyoto cells 4 hr after plating on the fibronectin-coated glass (n=286, N=3). Correlation coefficient R=0.23. (B) Relation between volume and spreading area of RPE-1 cells 4 hr after plating on the fibronectin-coated glass (n=222, N=3). Correlation coefficient R=0.47. (C) Relation between volume and spreading area of 3T3 cells 4 hr after plating on the fibronectin-coated glass (n=117, N=3). Correlation coefficient R=0.61. (D) Average normalized volume of control HeLa Kyoto cells (blue, n=99, N=1), cells incubated overnight in the serum-free medium (cyan, n=61, N=1) or cells incubated overnight in the serum-free medium and resuspended in FBS containing medium prior experiments (magenta, n=71, N=1) spreading on fibronectin-coated glass. Error bars represent standard deviation. (E) Comparison of the average initial volumes of HeLa Kyoto cells for individual experiments with the average initial volume value of these experiments. (F) Distribution of coefficient of variation of cell volume for n=44 cells recorded for 9 s with the time frame 30 ms. (G) Average relative cell volume (left) and dry mass (right) of HeLa Kyoto cells in response to hypoosmotic shock for three individual experiments (n=25, n=17, and n=50). Error bars represent standard deviation. (H) Average normalized density of cells represented on Figure 1—figure supplement 1G. (I) Left: Spreading area of individual HeLa Kyoto cells during spreading on fibronectin-coated glass. Right: Normalized volume of individual HeLa Kyoto cells spreading on fibronectin-coated glass. (J) Volume flux (dV/dt) of single control HeLa Kyoto cells (n=195, N=3) plotted versus their spreading area at the 50–60 min of spreading. Error bars represent standard deviation. Color bar indicates kernel density. (K) Relative volume loss while spreading plotted versus absolute initial cell volume (n=170, N=3). The data are fitted with linear regression y=−0.0006x−7.48, R2=0.0046.
-
Figure 1—figure supplement 1—source data 1
Data tables related to quantifications in Figure 1 Supplement 1.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig1-figsupp1-data1-v2.zip
Side view of a HeLa-LifeAct cell spreading on fibronectin-coated glass.
Top: bright field, bottom: LifeAct. Scale bar: 10 µm. 20× PA.
Spreading of HeLa EMBL cells on fibronectin-coated glass.
Left: FXm, right: RICM. 20× PA.
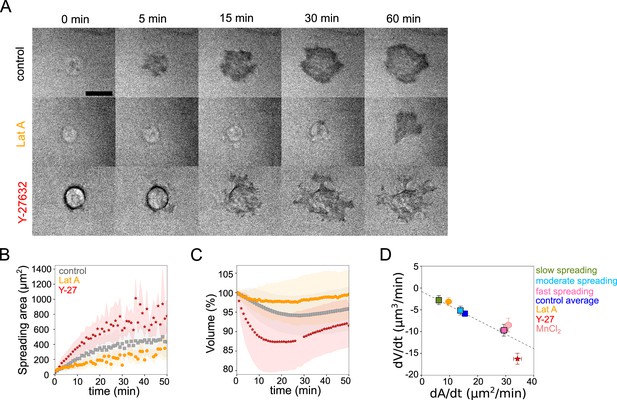
Cell volume depends on spreading speed.
(A) RICM imaging of control HeLa Kyoto cell or cell treated with 100 nM Latrunculin A or with 100 µM Y-27632 spreading on fibronectin-coated glass. Scale bar: 20 µm. (B) Average spreading area of control HeLa Kyoto cells (gray, n=125, N=3), cells treated with 100 nM Latrunculin A (orange, n=30, N=2) or 100 µM Y-27632 (red, n=98, N=3). Error bars represent standard error. (C) Average normalized volume of control HeLa Kyoto cells (gray, n=125, N=3), cells treated with 100 nM Latrunculin A (orange, n=30, N=2) or 100 µM Y-27632 (red, n=98, N=3). Error bars represent standard error. (D) Median volume flux (dV/dt) of control (blue, n=194, N=3), 100 µM Y-27632 (red, n=121, N=4), 100 nM Latrunculin A (orange, n=41, N=3) or 1 mM MnCl2 (N=3, n=57) treated HeLa Kyoto cells plotted versus their spreading speed (dA/dt) at the first 10 min of spreading. Average dV/dt(dA/dt) for three groups of control cells from Figure 1I are shown on the graph. The dashed line is a linear regression for control cells from panel 1I. Error bars represent standard error.
-
Figure 2—source data 1
Data tables related to quantifications in Figure 2.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig2-data1-v2.zip
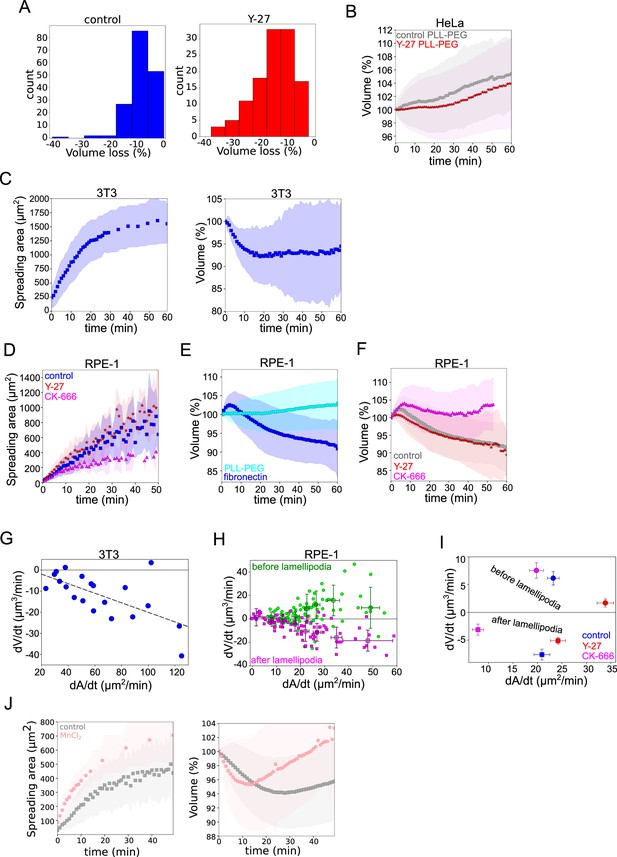
Cell volume depends on spreading speed, additional cell lines.
(A) Distributions of maximum volume loss of individual cells induced by spreading for control (n=170, N=3) and Y-27 (n=120, N=4) treated cells. (B) Average normalized volume of control HeLa Kyoto cells (gray, n=493, N=5) or treated with 100 µM Y-27632 (red, n=292, N=3) plated on PLL-PEG-coated glass. Error bars represent standard deviation. (C) Left: Average spreading area of 3T3-ATCC cells spreading on fibronectin-coated glass (n=20, N=3). Error bars represent standard deviation. Right: Average normalized volume of 3T3-ATCC cells spreading on fibronectin-coated glass (n=20, N=3). Error bars represent standard deviation. (D) Average normalized spreading area of control RPE-1 cells (gray, n=90, N=3), treated with 100 µM Y-27632 (red, n=85, N=3) or 100 µM CK-666 treated (magenta, n=48, N=3) spreading on fibronectin-coated glass. Error bars represent standard deviation. (E) Average normalized volume of control RPE-1 cells (blue, n=90, N=3) or control cells plated on PLL-PEG-coated glass (cyan, n=168, N=3). Error bars represent standard deviation. (F) Average normalized volume of control RPE-1 cells (gray, n=90, N=3), 100 µM Y-27632 treated (red, n=85, N=3) or 100 µM CK-666 treated (magenta, n=48, N=3) spreading on fibronectin-coated glass. Error bars represent standard deviation. (G) Volume flux (dV/dt) of 3T3-ATCC cells plotted versus their spreading speed (dA/dt) at the first 10 min of spreading (n=20, N=3). The data are fitted with linear regression y=−0.23x+2.51, R2=0.42. (H) Volume flux (dV/dt) of RPE-1 cells plotted versus their spreading speed (dA/dt) before (green) and after (magenta) lamellipodia formation (n=90, N=3). Error bars represent standard error. (I) Median volume flux (dV/dt) of control (blue, n=89, N=3), 100 µM Y-27632 (red, n=85, N=3) or 100 µM CK-666 (magenta, n=38, N=2) treated RPE-1 cells plotted versus their spreading speed (dA/dt) before and after lamellipodia formation. Error bars represent standard error. (J) Average spreading area (left) and average normalized volume (right) of control HeLa cells and treated with 1 mM MnCl2 (N=3, n=57).
-
Figure 2—figure supplement 1—source data 1
Data tables related to quantifications in Figure 2 Supplement 1.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig2-figsupp1-data1-v2.zip
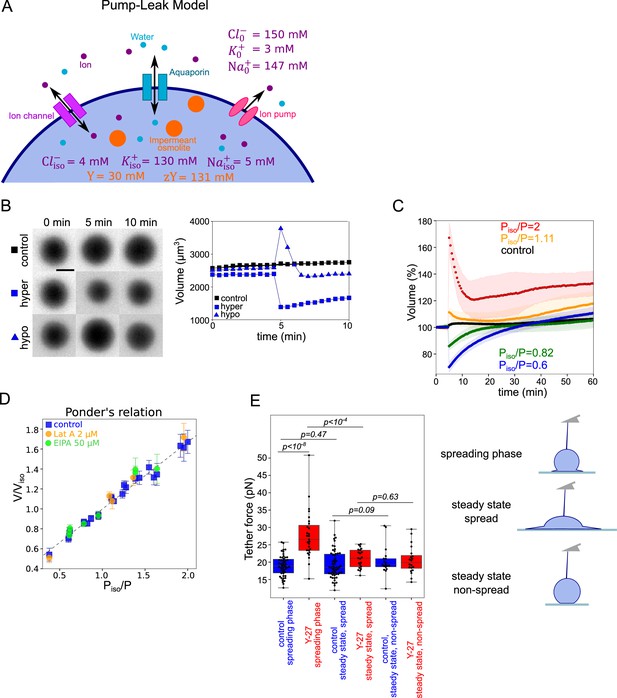
Verifying the Pump and Leak Model and its mecano-sensitive extension.
(A) Schematic of “pump-leak” model (PLM). In brief, the plasma membrane let ions and water pass, with specific channels which increase their permeation coefficient (the ‘leak’). Ions can also be pumped out of the cell (the ‘pump’). Outside the cell, the concentration of ions is about 300 mM, while it is only of about 150 mM inside the cell. The quantity of ion species also differs, with more anions outside the cell (because proteins are on average negatively charged), more sodium outside the cell, and more potassium inside. To achieve the equilibrium, sodium and chloride need to be constantly pumped outside of the cell. The cell also contains nonpermeant osmolytes: charged osmolytes (‘zY’ at the scheme) and neutral osmolytes (‘Y’ at the scheme), for example, proteins, amino-acids, and sugars (which cannot pass the membrane at the same rate as water or ions). Considering these equilibria and the resulting osmotic balance, the PLM predicts the volume of the cell (see the model in Appendix 1 and Cadart et al., 2019). Numbers at the panels are taken from the model in Appendix 1. (B) Left: FXm images of HeLa Kyoto cells exposed to media exchange of same osmolarity, hypertonic and hypotonic. Right: Volume of the cells represented on the left panel. (C) Average normalized HeLa Kyoto cells volume response to osmotic shocks of different magnitudes acquired every 30 s. Number of cells in the experiments: control Piso/P=1 (n=51, N=1), Piso/P=1.11 (n=30, N=1), Piso/P=2 (n=17, N=1), Piso/P=0.82 (n=33, N=1), Piso/P=0.6 (n=67, N=1). (D) Ponder’s relation for control HeLa Kyoto cells (blue), treated with 2 µM Lat A (orange), or 50 µM EIPA (green). Each point represents average value of single experiment. Average number of cells in each experiment n~58. Error bars represent standard deviation. Dashed line represents linear regression fit for control cells y=0.67x+0.33, R2=0.98. Coefficient 0.67 refers to the ratio of osmotically active volume to total volume named ‘R’ in the Appendix 1. (E) Tether force measurements of control HeLa Kyoto cells (blue) and treated with 100 µM Y-27632 (red) at the ‘spreading phase’ (measurements are performed within 30–90 min after cell plating), ‘steady state, spread’ (measurements are performed within 4–5 hr after plating) or ‘steady state, non-spread’ (measurements are performed within 4–5 hr after plating on 20 µm diameter micropatterns). For ‘control, spreading phase’ n=50, N=6; for ‘Y-27, spreading phase’ n=27, N=3; for ‘control, steady state, spread’ n=55, N=9; for ‘Y-27, steady state’ n=21, N=3, for ‘control, steady state, non-spread’ n=18, N=3, for ‘Y-27, steady state, non-spread’ n=20, N=3. Error bars represent standard deviation. The results of statistical tests are shown at the graph.
-
Figure 3—source data 1
Data tables related to quantifications in Figure 3.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig3-data1-v2.zip
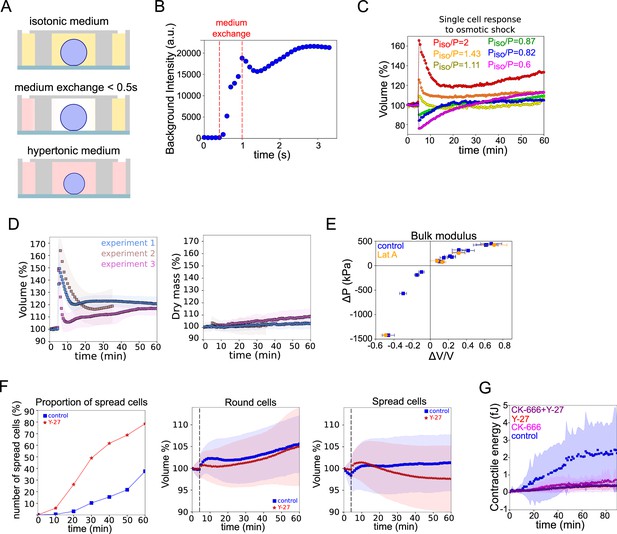
Osmotic response of HeLa cells and effect of contractility on cell volume.
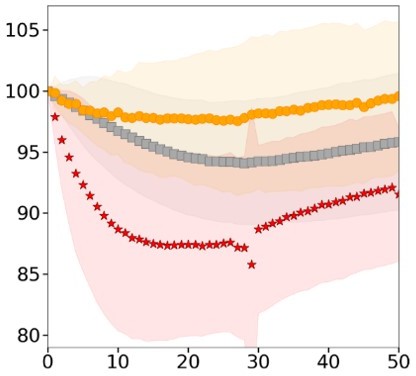
(A) Schematic of osmotic shock experiments in FXm chamber. (B) Timescale of medium exchange inside the volume measurement chamber. Full replace of medium (between two dashed lines) inside the chamber occurred in ~0.5 s. N=1. (C) Examples of single HeLa Kyoto cells volume changes in response to osmotic shocks of different magnitudes acquired every 30 s over 60 min. (D) Average relative cell volume (left) and dry mass (right) of HeLa Kyoto cells in response to hypoosmotic shock for three individual experiments (n=63, n=20, n=28). Error bars represent standard deviation. (E) Relative changes in osmotic pressure induced by osmotic shock plotted versus deformation for control (blue) and 2 µM Lat A treated (orange) HeLa Kyoto cells. Based on the same data as in Figure 3E. Error bars represent standard deviation. (F) Proportion of HeLa cells plated on PLL that spread during the experiments (left); average relative volume of round (middle) and spread (right) control cells (N=3, n=77) and treated with Y-27 during the experiment (N=4, N=297). Dashed line indicates the time point of the medium exchange to the control medium or to the medium supplemented with Y-27. (G) Contractile energy of control (n=9, N=2), treated with Y-27632 (n=16, N=2), CK-666 (n=13, N=2), or CK-666+Y-27 (n=13, N=2) HeLa EMBL cells during spreading on fibronectin-coated glass.
-
Figure 3—figure supplement 1—source data 1
Data tables related to quantifications in Figure 3 Supplement 1.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig3-figsupp1-data1-v2.zip
FXm imaging of HeLa EMBL cells attached on PLL-coated glass exposed to osmotic shock 20× LD.
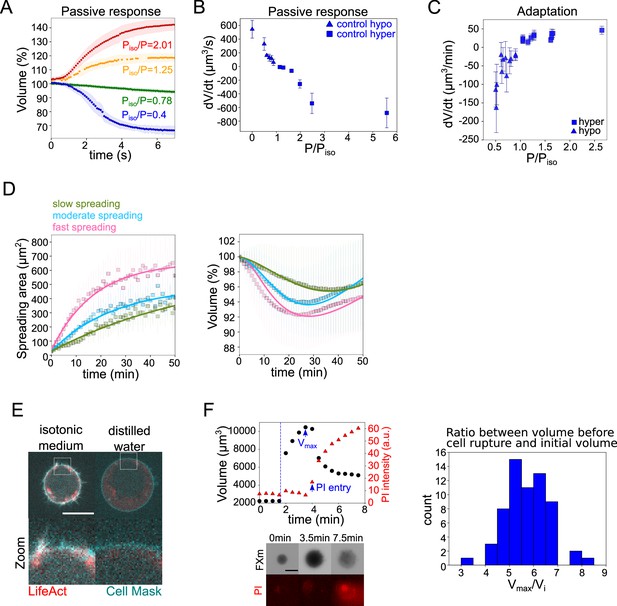
The mecano-sensitive PLM explains the volume loss in spreading cells.
(A) Average normalized volume of HeLa Kyoto cells during initial response to osmotic shocks of different magnitudes measured with high time resolution, 100 ms. Number of cells in the experiments: control Piso/P=1.25 (n=13, N=1), Piso/P=2.01 (n=19, N=1), Piso/P=0.78 (n=15, N=1), Piso/P=0.4 (n=17, N=1). Error bars represent standard deviation. (B) Average volume flux in HeLa Kyoto cells during initial response to osmotic shocks of different magnitudes. Each point represents average value of single experiment, average number of cells in each experiment n~12. Error bars represent standard deviation. (C) Average volume flux in HeLa Kyoto cells during regulatory volume adaptation. Each point represents average value of single experiment, average number of cells in each experiment n~48. Error bars represent standard deviation. (D) Fits of the spreading data from the model using best fit parameters on average normalized volume (right) and spreading area (left) of control cells divided into three categories represented in Figure 1H. (E) Z-plane of HeLa LifeAct-mcherry (red) cell before and after addition of distilled water, cell membrane is stained with CellMask Green (cyan). Scale bar 10 µm. (F) Left top: Volume (black) and propidium iodide (PI) intensity of single HeLa Kyoto cell exposed to distilled water. Dashed line indicates the time of distilled water addition. Reaching of maximum cell volume is followed by cell membrane rupture, volume decrease, and PI entry into the cell. Left bottom: Corresponding FXm images and PI staining. Right: Distribution of ratio between maximum volume cells reach before bursting induced by exposure of distilled water and their initial volume (n=63, N=3).
-
Figure 4—source data 1
Data tables related to quantifications in Figure 4.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig4-data1-v2.zip
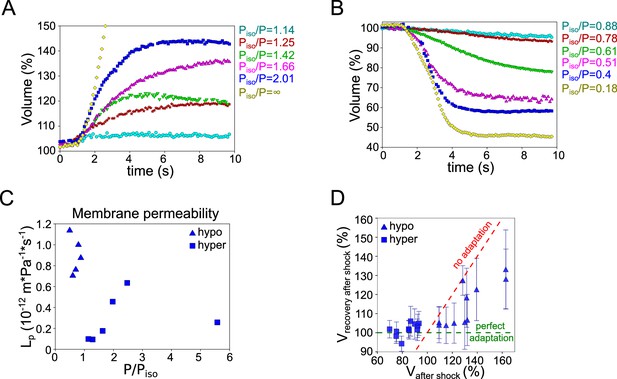
Experimental estimation of parameters for the PLM.
(A) Volume of single HeLa Kyoto cells during initial response to hypoosmotic shocks of different magnitudes acquired every 100 ms over ~7 s. (B) Volume of single HeLa Kyoto cells during initial response to hyperosmotic shocks of different magnitudes acquired every 100 ms over ~7 s. (C) Estimated hydraulic conductivity for osmotic shock of different magnitudes. Calculations are based on the data presented in Figure 4B. (D) Relative volume changes in HeLa Kyoto cells followed 30 min by passive volume response to osmotic shock plotted versus the values reached during passive response (presented in Figure 3D). Green dashed line corresponds to the ‘perfect adaptation,’ red dashed lines correspond to the absence of adaptation. Average number of cells in each condition n~48, for each condition N=1. Error bars represent standard deviation.
-
Figure 4—figure supplement 1—source data 1
Data tables related to quantifications in Figure 4 Supplement 1.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig4-figsupp1-data1-v2.zip
FXm imaging of HeLa EMBL cells attached on PLL-coated glass exposed to osmotic shock recorded with high frame rate.
20× LD.
3D-shape reconstruction by FXm of HeLa EMBL cells spread for 20 min at fibronectin-coated glass.
Control, Y-27632, CK-666, CK-666+Y-27632. 63×.
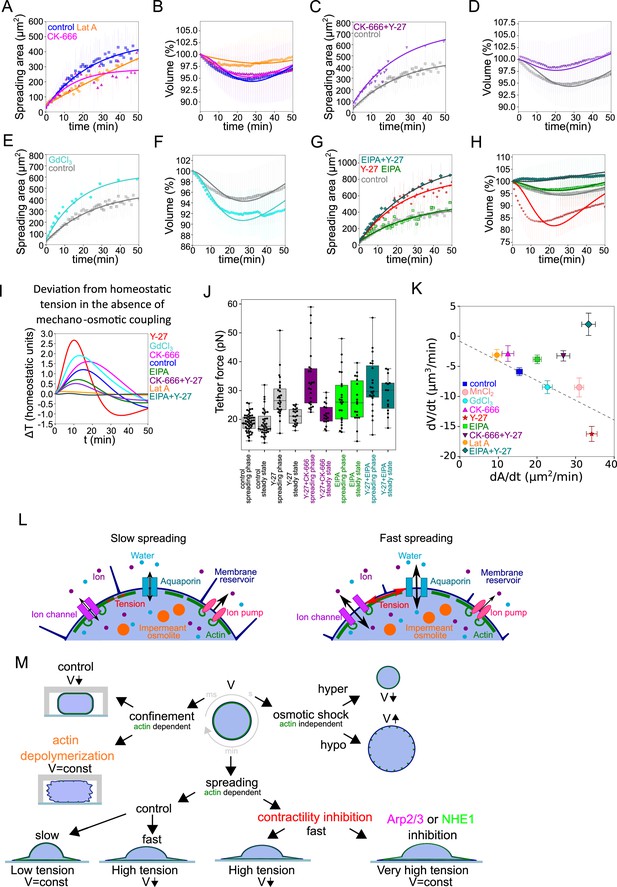
The mecano-sensitive PLM depends on branched actin and modulation of ion fluxes and constitutes a surface tension homeostasis mecanism.
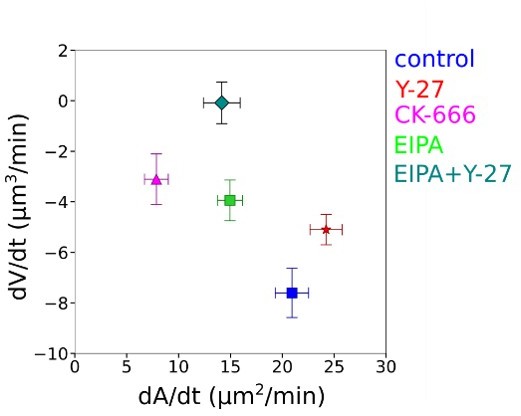
(A) Two parameter fits for the spreading kinetics using the exponential saturation anzatz (see text) on average area of control cells (blue, n=73, N=1), 100 nM Latrunculin A (orange, n=30, N=2) or 100 µM CK-666 (magenta, n=37, N=2) treated. Error bars represent standard deviation. (B) Fits from the model using best fit parameters on average normalized volume of control cells (blue, n=73, N=1), 100 nM Latrunculin A (orange, n=30, N=2) or 100 µM CK-666 (magenta, n=37, N=2) treated. Error bars represent standard deviation. (C) Two parameter fits for the spreading kinetics using the exponential saturation anzatz (see text) on average area of control cells (gray, n=73, N=1) or combination of 100 µM CK-666 and 100 µM Y-27632 (violet, n=24, N=1) treated. Error bars represent standard deviation. (D) Fits from the model using best fit parameters on average normalized volume of control cells (gray, n=73, N=1) or combination of 100 µM CK-666 and 100 µM Y-27632 (violet, n=24, N=1) treated. Error bars represent standard deviation. (E) Two parameter fits for the spreading kinetics using the exponential saturation anzatz (see text) on average area of control cells (gray, n=73, N=1) or 100 µM GdCl3 (cyan, n=30, N=2) treated. Error bars represent standard deviation. (F) Fits from the model using best fit parameters on average normalized volume of control cells (gray, n=73, N=1) or 100 µM GdCl3 (cyan, n=30, N=2) treated. Error bars represent standard deviation. (G) Two parameter fits for the spreading kinetics using the exponential saturation anzatz (see text) on average area of control cells (gray, n=73, N=1), 100 µM Y-27632 (red, n=21, N=1), 50 µM EIPA (green, n=73, N=1), or combination of 50 µM EIPA and 100 µM Y-27632 (dark cyan, n=30, N=2) treated. Error bars represent standard deviation. (H) Fits from the model using best fit parameters on average normalized volume of control cells (gray, n=73, N=1), 100 µM Y-27632 (red, n=21, N=1), 50 µM EIPA (green, n=73, N=1), or combination of 50 µM EIPA and 100 µM Y-27632 (dark cyan, n=30, N=2) treated. Error bars represent standard deviation. (I) Predicted by model, plots for difference between tension without mechano-osmotic coupling (for α = 0 and = 100) and tension with mechano-osmotic coupling (for fitted and = 100). (J) Tether force measurements of control HeLa Kyoto cells (gray, for ‘spreading phase’ n=50, N=6; for ‘steady state’ n=55, N=9), treated with Y-27632 (gray, for ‘spreading phase’ n=27, N=3; for ‘steady state’ n=21, N=3), CK-666+Y-27 (purple, for ‘spreading phase’ n=25, N=3; for ‘steady state’ n=19, N=3), EIPA (green, for ‘spreading phase’ n=23, N=3; for ‘steady state’ n=18, N=3), EIPA+Y-27 (dark cyan, for ‘spreading phase’ n=23, N=3; for ‘steady state’ n=15, N=3) during the first 30–90 min after plating or 4–5 hr after plating. Error bars represent standard deviation. The results of statistical tests are shown at the graph. (K) Volume flux (dV/dt) of single control HeLa Kyoto cells (n=194, N=3), treated with Lat A (n=41, N=3), CK-666 (n=54, N=3), Y-27 (n=121, N=4), EIPA (n=117, N=3), GdCl3 (n=53, N=3), CK-666+Y-27 (n=74, N=3), EIPA+Y-27 (n=50, N=3), MnCl2 (N=3, n=57) plotted versus their spreading speed (dA/dt) at the first 10 min of spreading. Error bars represent standard error. (L) Scheme of mechanosensistive “pump-leak” model. (M) Scheme representing cell volume regulation in response to deformations.
-
Figure 5—source data 1
Data tables related to quantifications in Figure 5.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig5-data1-v2.zip
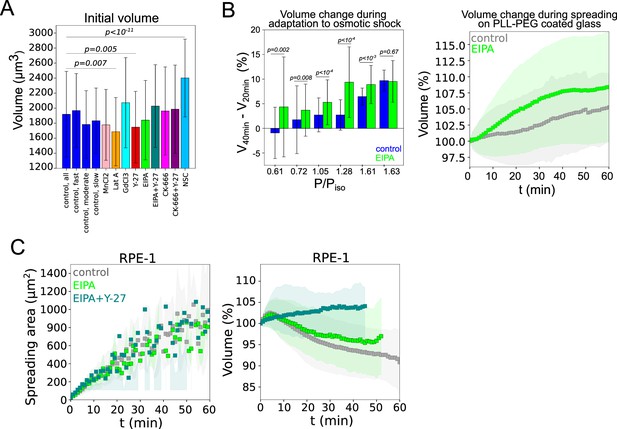
Effect of drug treatments on initial cell volume, adaptation to osmotic shocks and volume loss in RPE1 cells.
(A) Average absolute initial volume of three groups of control HeLa Kyoto (slow, moderate, or fast spreading on the panel 1H) cells and cells after 30 min incubation with drugs prior the experiments. All control cells (n=194, N=3), slow (n=42, N=3), moderate (n=43, N=3), and fast (n=42, N=3), treated with Lat A (n=41, N=3), CK-666 (n=54, N=3), Y-27 (n=121, N=4), EIPA (n=117, N=3), GdCl3 (n=53, N=3), CK-666+Y-27 (n=74, N=3), EIPA+Y-27 (n=50, N=3), and MnCl2 (N=3, n=57). Error bars represent standard error. There is statistically significant difference for initial volume for Lat A, Y-27, and NSC. For the rest of conditions p values are more than 0.05. (B) Left: Average relative volume change between 40 and 20 min of osmotic shock experiments (corresponds to the adaptation to osmotic shock) for control HeLa cells and cell treated with 50 µM EIPA for the different magnitude of the shock. Error bars represent standard deviation. The results of statistical tests are shown at the graph. Right: average relative volume of control (N=5, n=493) and 50 µM EIPA (N=1, n=111) treated HeLa cells plated on PLL-PEG. Error bars represent standard deviation. (C) Average spreading area (left) and average normalized volume (right) of control RPE-1 cells (N=3, n=90), treated with EIPA (N=3, n=72) or EIPA+Y-27 (N=3, n=55). Error bars represent standard deviation.
-
Figure 5—figure supplement 1—source data 1
Data tables related to quantifications in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig5-figsupp1-data1-v2.zip
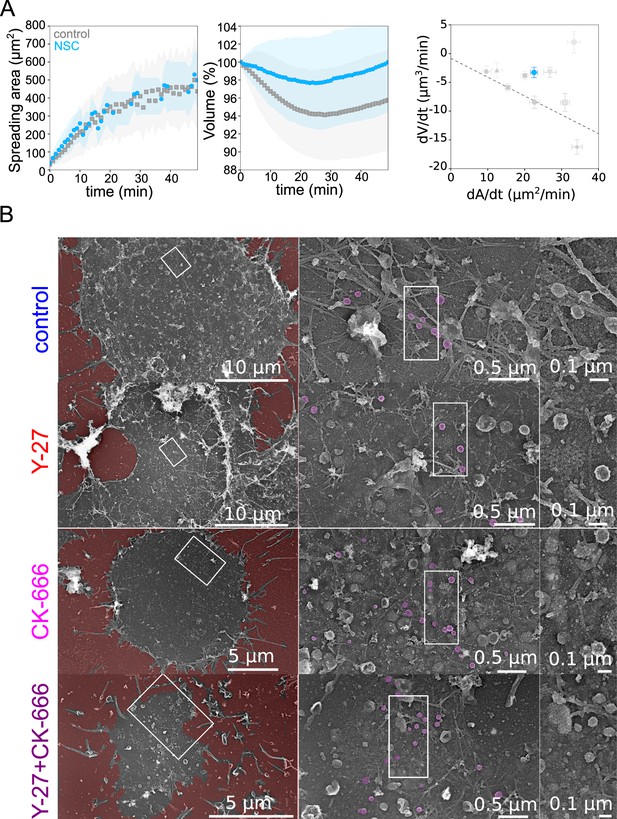
Role of membrane to actin attachment on volume loss and state of the plasma membrane in spreading cells.
(A) Left: Average spreading area of control HeLa Kyoto cells (gray, n=125, N=1) or 20 µM NSC (light blue, n=101, N=3). Middle: Average normalized volume of control HeLa Kyoto cells (gray, n=125, N=1), or 20 µM NSC (light blue, n=101, N=3). Error bars represent standard deviations. Right: Volume flux (dV/dt) plotted versus their spreading speed (dA/dt) of single control HeLa Kyoto cells and treated with various drugs and represented in Figure 5K and treated with 20 µM NSC (light blue, n=101, N=3). Error bars represent standard error. (B) Platinum replica electron microscopy survey views of the cytoplasmic surface in control, Y-27632, CK-666, or CK-666+Y-27632-treated unroofed Hela cells spread on glass coverslips for 30 min. Extracellular substrate is pseudo-colored in red. For each panel, high magnification views corresponding to the boxed regions are shown on the right.
-
Figure 6—source data 1
Data tables related to quantifications in Figure 6.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig6-data1-v2.zip
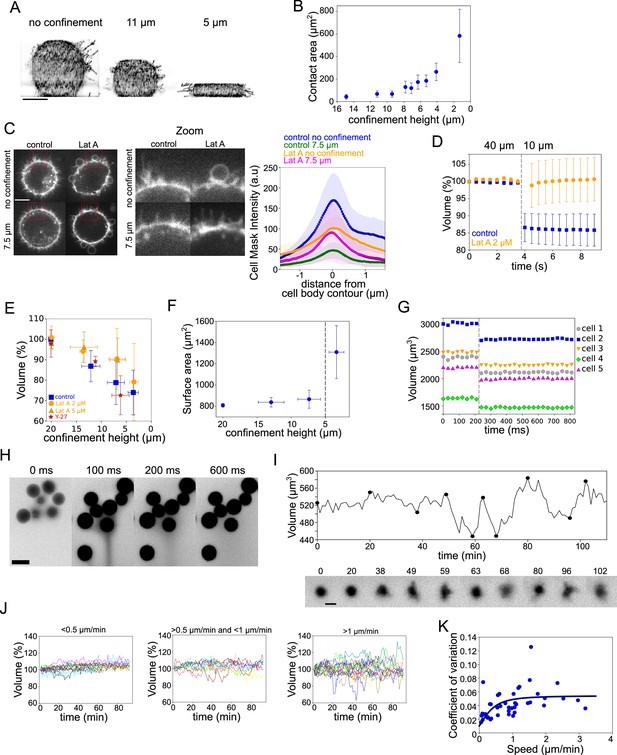
Volume modulation during ultra-fast cell flattening and during cell migration through collagen matrices.
(A) 3D-membrane reconstruction of HeLa expressing MyrPalm-GFP (black) cells cell shape under different confinement heights, side view. Scale bar: 10 µm. (B) Contact area with bottom glass substrate of Hela Kyoto cells under different confinement heights. Average number of cells in each experiment n~79, for each condition N=1. Error bars represent standard deviation. (C) Left: Z-plane of control and 2 µM Lat A treated HeLa cells under 20 µm and 7.6 µm confinement heights. Cell membrane is stained with CellMask Far Red (white). Scale bar: 10 µm. Right: Average CellMask intensity plotted versus distance from cell body contour on the middle Z-plane of HeLa-MYH9-GFP-LifeAct-mcherry cells. Number of cells in each condition n=10, N=1 for each condition. Error bars represent standard deviation. (D) Average normalized volume of control (blue, n=48, N=1) and 2 µM Lat A treated (orange, n=32, N=1) HeLa Kyoto cells during dynamic confinement experiment. Dashed line indicates the moment of confinement. Error bars represent standard deviation. (E) Average normalized volume of HeLa Kyoto cells (blue) and cells treated with Lat A 2 µM (orange) or 5 µM (yellow) or 100 µM Y-27 (red) under different confinement heights. Each data point represents an average of N~10 experiments; each experiment contains n~160 individual cells. Error bars represent standard deviation. There is statistically significant difference between control and Lat A for the heights ~12 µm and ~7 µm (p=0.02 and p<10–8). There is no statistically significant difference between control and Y-27 for the heights ~12 µm and ~7 µm (p=0.4 and p=0.3). (F) Projected surface (computed from volume represented in panel Figure 5G) of HeLa Kyoto cells under different confinement heights. Dashed line indicates the confinement height that corresponds to blebs appearance. Error bars represent standard deviation. (G) Volume of single HeLa Kyoto cells during dynamic confinement experiment. Dashed line indicates the moment of confinement. (H) FXm images of HeLa Kyoto cells during dynamic confinement experiment taken with high NA objective. (I) Top: Volume of single DC migrating in collagen. Bottom: Corresponding FXm images. (J) Volume of single DCs migrating in collagen with the different speeds, N=1. Left: <0.5 µm/min (n=14), middle: >0.5 µm/min (n=10) and <1 µm/min, right: >1 µm/min (n=19). (K) Coefficient of variation of volume flux dV/dt computed for 10 min intervals during single DCs migration in collagen plotted versus their average speed (n=43, N=1). The line is the fit for coefficient of variation of DCs volume using best-fit parameters (see Appendix 1).
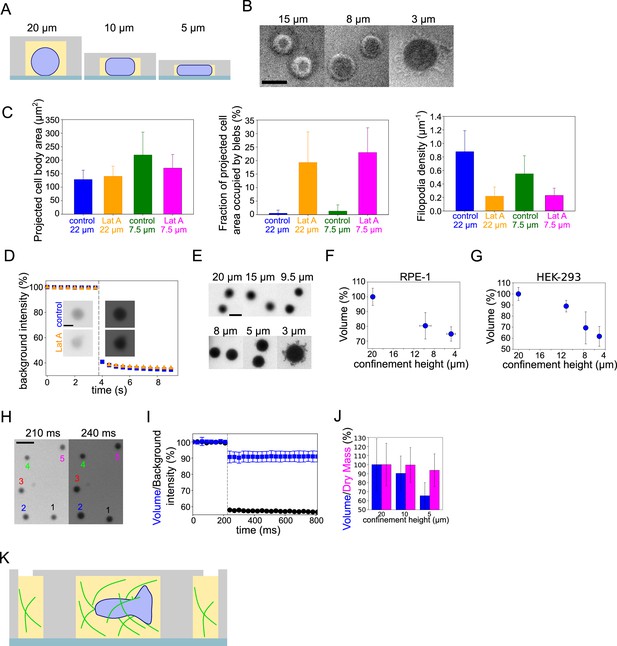
Characterisation of cell protrusions and dry mass in ultra-fast HeLa cell flattening and volume modulation in ultra-fast flattening of RPE1 and HEK-293 cells.
(A) Schematic of FXM experiments under confinement. (B) RICM images of cell contact area under different confinement heights. Scale bar: 10 µm. (C) Analysis of control and 2 µM Latrunculin A treated cell morphological changes under confinement. Left: Average projected cell body area of cells. Middle: Average fraction of projected cell area occupied by blebs. Right: Filopodia density. For each panel analysis is performed on the middle Z-plane of HeLa-MYH9-GFP-LifeAct-mcherry cells. Number of cells in each condition (n=10, N=1). Error bars represent standard deviation. (D) Corresponding to Figure 5F background intensity values and FXm images of single cells before and after confinement. Scale bar: 10 µm. Error bars represent standard deviation. (E) FXm images of Hela Kyoto cells under different confinement heights. Scale bar: 10 µm. (F) Average normalized volume of RPE-1 cells under different confinement heights. Each data point represents an average of N=3 experiments, each experiment contains n~140 individual cells. Error bars represent standard deviation. (G) Average normalized volume of HEK-293 cells under different confinement heights. Each data point represents an average of N=5 experiments, each experiment contains n~118 individual cells. Error bars represent standard deviation. (H) Corresponding to Figure 5I FXm images. (I) Corresponding to Figure 5I background intensity values (black) and average normalized volume (blue, n=19). Dashed line indicates the moment of confinement. (J) Average cell volume (blue) and dry mass (magenta) of HeLa Kyoto cells under different confinement heights. Number of single cells analyzed for each height, N=2: 20 µm (n=192), 10 µm (n=174), and 5 µm (n=113). (K) Schematic of volume measurements of DCs migrating in collagen in FXm chamber.
-
Figure 7—figure supplement 1—source data 1
Data tables related to quantifications in Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/72381/elife-72381-fig7-figsupp1-data1-v2.zip
3D-membrane reconstruction of HeLa expressing MyrPalm-GFP (black) cells cell shape under different confinement heights 63×.
Z-planes of control and 2 µM Lat A treated HeLa-MYH9-GFP-LifeAct-mcherry, cells under 20 µm and 7.6 µm confinement heights.
Cell membrane is stained with CellMask Far Red (white). 63×.
FXm imaging of HeLa EMBL cells during dynamic confinement recorded with 20× LD.
FXm imaging of HeLa EMBL cells during dynamic confinement recorded with 20× PA.
FXm imaging of DCs migrating in collagen gel with 20× LD.
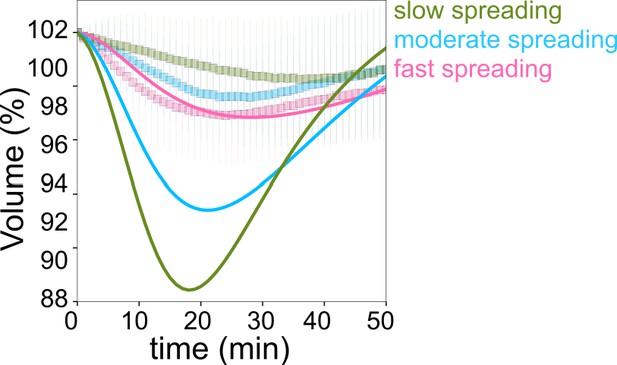
Model Figure 1.
Fits for volume taking the contact area of the fast-spreading cells for the three sets of parameters. Model predictions for the cell volume, based on fits from data, for slow (green), moderate (cyan), and fast (pink) spreading.
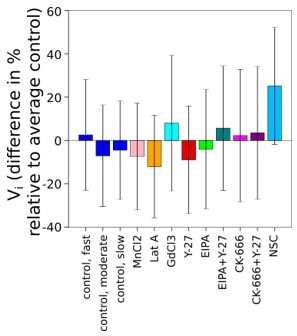
Graph showing difference in % of average initial volume of 3 groups of control cells and cells treated with drugs comparing with average initial volume of control cells.
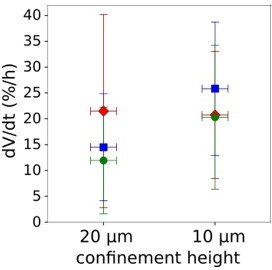
Graph showing the rate of volume increase after initial volume loss induced by confinement.
Each point represents the average rate of individual experiment. Average amount of cells in each experiment n=73.

Graph showing average relative volume of control and Y-27 treated cells under different confinement heights.
Each point is average relative volume of N=14-30 (average amount of cells n=165) experiments and N=4-5 (average amount of cells n=166) for Y-27 treated cells. The results of t-test for each height: h=20 µm p=0.13, h=11 µm p=0.41, h=5 µm p=0.29.
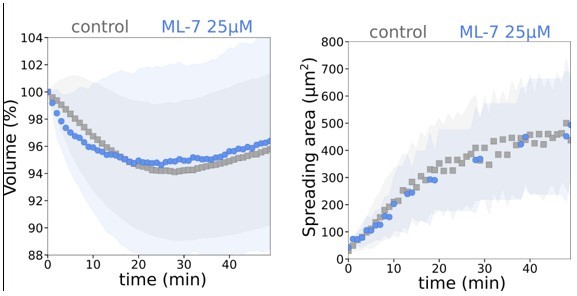
Graph showing average relative volume and average spreading area of control and treated with 25 µM ML-7 cells (N=3, n=25).
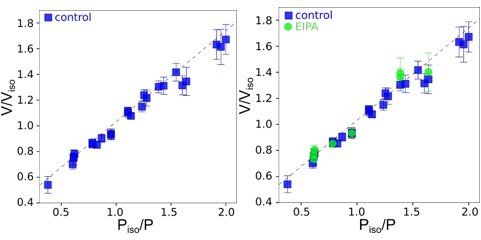
Graphs showing Ponder’s relation of control cells (the line indicated linear regression for control cells); only control cells, linear regression y=0.
67+0.33, R2=0.98 (left); only EIPA treated cells, linear regression y=0.71+0.32, R2=0.96 (right).
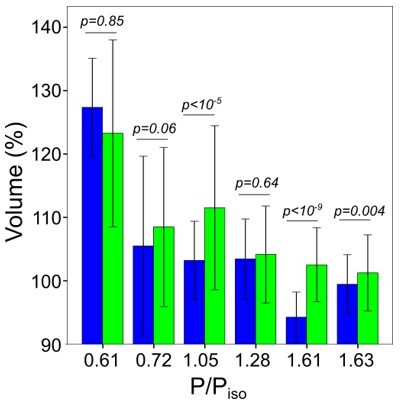
Graph showing the effect on RVI and RVD.
We plotted average volume at 30 min after osmotic shock for control and EIPA treated cells (where P/Piso=0.61 and 0.72 are hypoosmotic shock). EIPA treatment, in a majority of cases, increased the cell volume gained during adaptation..
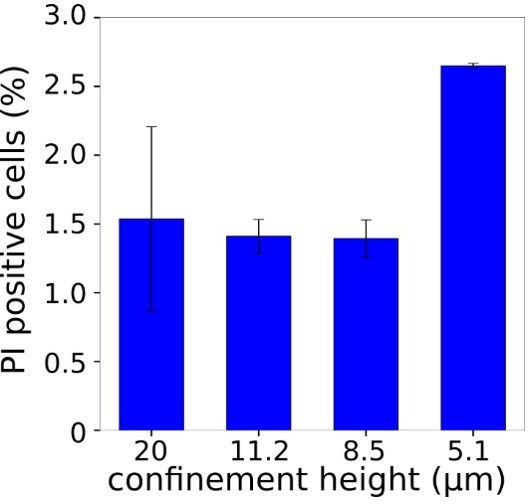
Graph showing percentage of cells with positive propidium iodide staining for different confinement heights (for each height N=2-3, average number of cells in each experiment n=196).</Author response image 8 title/legend>.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HeLa EMBL (Kyoto) | Gift from Valérie Doye | ||
| Cell line (H. sapiens) | RPE1 | ATCC | ||
| Cell line (H. sapiens) | 3T3 | ATCC – from Alba Diez-Munoz Lab, EMBL, Heidelberg, Germany | ||
| Cell line (H. sapiens) | HEK-293 | Gift from Liam Holt lab, NYU, NewYork | ||
| Chemical compound, drug | Fetal bovine serum | PAN-Biotech | P30-193306 | Use at 10% |
| Chemical compound, drug | Dextran, Alexa Fluor 647; 10,000 MW, Anionic, Fixable | Sigma-Aldrich | D22914 | Stock at 10 mg/ml in PBS |
| Chemical compound, drug | Fluorescein isothiocyanate-dextran; 10,000 MW | Sigma-Aldrich | FD10S | Stock at 50 mg/ml in PBS |
| Chemical compound, drug | Fibronectin | Sigma-Aldrich | F1141-1MG | 50 µg/ml in PBS |
| Chemical compound, drug | Poly-L-lysine | Sigma-Aldrich | P8920 | Use at 0.01% |
| Chemical compound, drug | PLL-PEG | SuSoS | 0.1 mg/ml solution in HEPES | |
| Software, algorithm | Software for FXm image analysis and volume calculation | Available upon request to the authors | RRID:SCR_001622 |
List of fitted parameter values for the control cells are grouped based on spreading speed when the tension depends on change in contact area.
| Condition/parameters | A0 (µm2) | |||
|---|---|---|---|---|
| Slow spreading | 620 | 63.3 | 98.5 | 0.21 |
| Moderate spreading | 488 | 25.6 | 64.2 | 0.16 |
| Fast spreading | 656 | 17.2 | 99.4 | 0.16 |
List of fitted parameter values for different drug treatment when the tension depends on change in contact area.
| Condition/parameters | A0 (µm2) | |||
|---|---|---|---|---|
| control | 479 | 25.9 | 53.5 | 0.16 |
| Y-27 | 833 | 24.4 | 16.4 | 0.67 |
| EIPA | 549 | 33.7 | 30.7 | 0.18 |
| Lat A | 1522 | 212 | 13.8 | 0.88 |
| GdCl3 | 628 | 18.3 | 171.2 | 0.17 |
| CK-666 | 285 | 14.9 | 0.09 | |
| CK-666+Y-27 | 738 | 25.6 | 36 | 0.1 |
| EIPA+Y-27 | 1035 | 29 | 0.02 |
List of fitted parameter values for the control cells are grouped based on spreading speed when the tension depends on change in total surface area.
| Condition/parameters | A0 (µm2) | |||
|---|---|---|---|---|
| Slow spreading | 620 | 63.3 | 5 | 2.5 |
| Moderate spreading | 488 | 25.6 | 13 | 0.9 |
| Fast spreading | 656 | 17.2 | 51 | 0.3 |
List of fitted parameter values for different drug treatment when the tension depends on change in total surface area.
| Condition/parameters | A0 (µm2) | |||
|---|---|---|---|---|
| Control | 479 | 25.9 | 12 | 1 |
| Y-27 | 833 | 24.4 | 8.5 | 1.3 |
| EIPA | 549 | 33.7 | 8.5 | 0.8 |
| Lat A | 1522 | 212 | 2 | 3.4 |
| GdCl3 | 628 | 18.3 | 34.5 | 0.4 |
| CK-666 | 285 | 14.9 | 29.5 | 0.9 |
| CK-666+Y-27 | 738 | 25.6 | 18 | 0.2 |
| EIPA+Y-27 | 1,035 | 29 | 4.5 | 0.1 |
List of parameter values.
| Parameter | Definition | Estimate |
|---|---|---|
| Concentration of chloride ions in the medium | 150 mM (Kay, 2017) | |
| Concentration of potassium ions in the medium | 3 mM (Kay, 2017) | |
| Concentration of sodium ions in the medium | 147 mM (Kay, 2017) | |
| Concentration of chloride ions in the cell | 4 mM (Equation (A17)) | |
| Concentration of potassium ions in the cell | 130 mM (Equation (A17)) | |
| Concentration of sodium ions in the cell | 5 mM (Equation (A17)) | |
| Concentration of trapped particles in the cell | 30 mM (osmotic balance) | |
| Concentration of trapped charges in the cell | 131 mM (electro-neutrality) | |
| Potential difference across the plasma membrane | –90 mV (Equation (A5)) | |
| Temperature (25°C) | 4.1*10–21 J | |
| Permeability of | 2*10–8 moles/(m2*s) (Kay, 2017) | |
| Permeability of | 6*10–7 moles/(m2*s) (Kay, 2017) | |
| Permeability of | 4*10–7 moles/(m2*s) (Kay, 2017) | |
| NaK Atpase pumping rate | 1.4*10–7 moles/(m2*s) | |
| Hydraulic conductivity | 10–(12–13) m/(Pa*s) (Equation (A9)) | |
| R | Ratio of osmotically active volume to total volume | 0.7 |
| Old panel | New panel | Statistics improvement |
|---|---|---|
| 1A | 1A | It was an error in the legend, actual N=3 |
| 1E | 1E | There were 2 more experiments in the attached datasets. During the revision 2 additional experiment were done. N=5 |
| 3C-D,4A-C | 3C-D,4A-C | The osmolarity of control medium and osmotic shock solution differ from experiment to experiment. Thus instead of averaging of experiments with similar osmolarity prior and after shock we plotted them as individual points representing the average of each individual experiment at the panels 3D, 4B and 4C. These panels also represent the total amount of osmotic shock experiment performed in this study. For the experiment with 30 s time lapse the total amount of experiments is: control N=28, Lat A N=4, EIPA N=8. For the experiment with 100 ms time lapse the total amount of experiments is: control N=11, N=5.Panels 3C and 4A are the representative examples. |
| 5D | 7B | The actual confinement height differs from experiment to experiment. Thus instead of averaging of experiments with similar heights we plotted them as individual points representing the average of each individual experiment. The total amount of experiments is N=9. |
| 5E | 7C | No improvements of statistics due to the limited revision time. |
| 5F, 5I | 7D, 7G | No improvements of statistics due to the limited revision time. |
| S1A | S1B | There were 2 more experiments in the attached datasets. We merged it with the experiment showed on the panel. N=3. |
| S1B | S1A | There were 2 more experiments in the attached datasets. We merged it with the experiment showed on the panel. N=3. |
| S1E | S1D | No improvements of statistics due to the limited revision time. |
| S1F | S1G | No improvements of statistics due to the limited revision time. |
| S2A | S2B | During the revision 2 additional experiment were done. N=3 |
| S2D | There were 2 experiments for CK-666. An additional experiment was performed. N=3. | |
| S2E | S2D-F | There were 2 more experiments for PLL-PEG in the attached datasets. We merged it with the experiment showed on the panel. N=3 for PLL-PEG.There were 2 experiments for CK-666. An additional experiment was performed. N=3. |
| S3B | We removed this panel. | |
| S3C | S3B | No improvements of statistics due to the limited revision time. |
| S3E | S3D | During the revision 2 additional experiment were done. N=3 |
| S5J | S7J | There was 1 more experiment in the attached datasets. We merged it with the experiment showed on the panel. N=2. |
| S6A | S3G | It was an error in the legend, actual N=2 |
| S6B-C | 6A | It was an error in the legend, actual N=2. But one more experiment was analyzed and added. N=3. |
| S6I | 7K | No improvements of statistics due to the limited revision time |