METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance
Figures
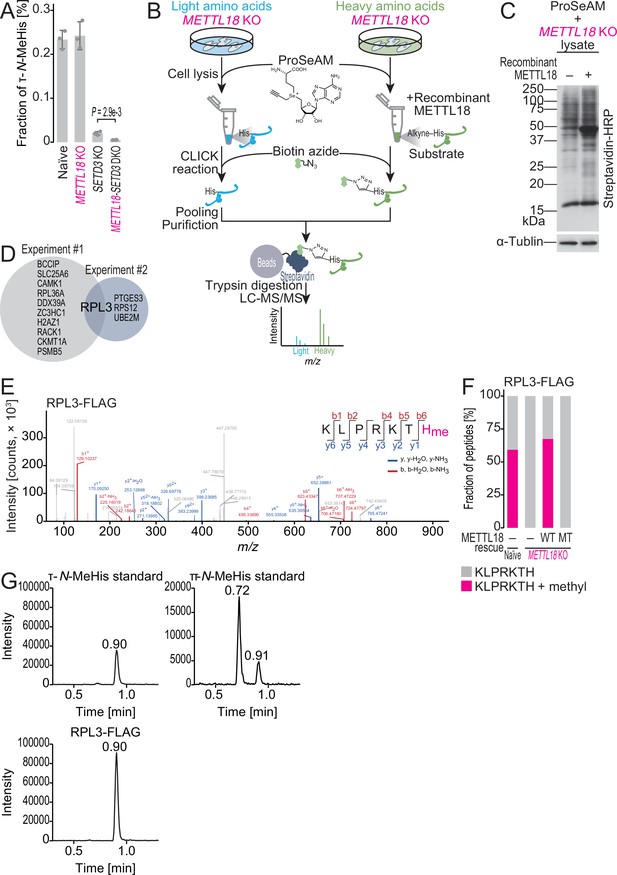
ProSeAM-SILAC identifies RPL3 as a substrate of METTL18.
(A) Multiple reaction monitoring (MRM)-based identification of τ-N-methylated histidine in bulk proteins from the indicated cell lines. Data from three replicates (points) and the mean (bar) with SD (error bar) are shown. Significance was determined by Student’s t-test (unpaired, two-sided). (B) Schematic representation of the ProSeAM-SILAC approach. (C) ProSeAM-labeled proteins in cell lysate with recombinant His-METTL18 protein. Biotinylated proteins were detected by streptavidin-HRP. Western blot for α-tubulin was used as a loading control. (D) Venn diagram of proteins identified in two independent ProSeAM-SILAC experiments. The reproducibly detected protein was RPL3. (E) Methylated histidine residue in ectopically expressed RPL3-FLAG was searched by liquid chromatography mass spectrometry (LC-MS/MS). (F) Quantification of methylated and unmethylated peptides (KLPRKTH) from the indicated cells. RPL3-FLAG was ectopically expressed and immunopurified for LC-MS/MS. WT, wild type; MT, Asp193Lys-Gly195Arg-Gly197Arg mutant. (G) MRM-based identification of τ-N-methylhistidine in peptides from RPL3. The τ-N-methylhistidine standard, π-N-methylhistidine standard, and RPL3-FLAG peptide (KLPRKTH) results are shown. MeHis, methylhistidine.
-
Figure 1—source data 1
Full and unedited blots corresponding to Figure 1C.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-data1-v1.tif
-
Figure 1—source data 2
Primary data for graphs in Figure 1.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-data2-v1.xlsx
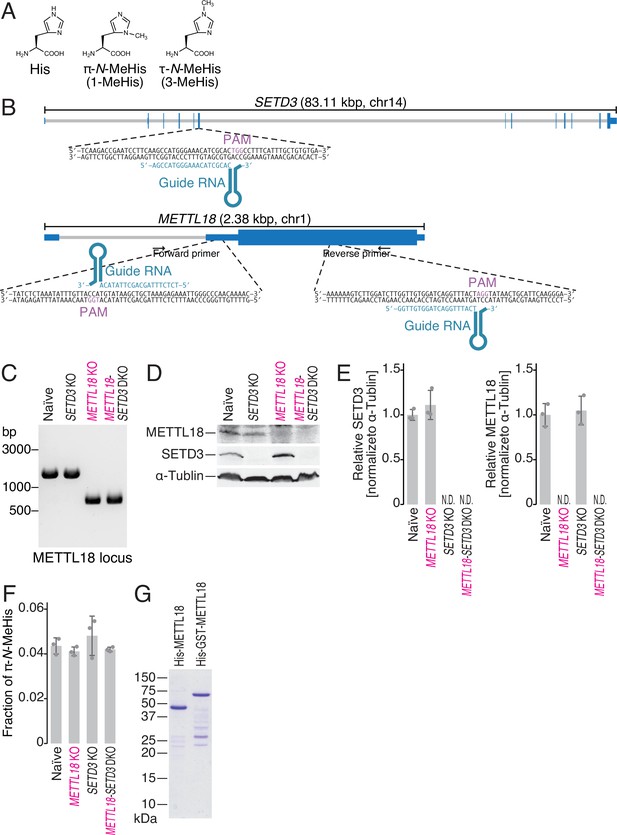
Generation of SETD3 and METTL18 knockout (KO) cells.
(A) Chemical structure of histidine, π-N-methylhistidine, and τ-N-methylhistidine. (B) Schematic representation of guide RNAs (gRNAs) designed for CRISPR-Cas9-mediated gene KO. (C) Genomic PCR validated the partial DNA deletion in the METTL18 gene locus. (D, E) Western blot of the indicated proteins to confirm the KO of SETD3 and METTL18 (D) and the quantification (E). α-Tubulin was probed as a loading control and for normalization. (E) Data from three replicates (points) and the mean (bar) with SD (error bar) are shown. (F) Multiple reaction monitoring (MRM)-based identification of π-N-methylated histidine in bulk proteins from the indicated cell lines. Data from three replicates (points) and the mean (bar) with SD (error bar) are shown. MeHis, methylhistidine. (G) Coomassie brilliant blue (CBB) staining of recombinant METTL18 proteins used in this study.
-
Figure 1—figure supplement 1—source data 1
Full and unedited blots corresponding to Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Full and unedited blots corresponding to Figure 1—figure supplement 1D.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-figsupp1-data2-v1.zip
-
Figure 1—figure supplement 1—source data 3
Full and unedited gel images corresponding to Figure 1—figure supplement 1G.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-figsupp1-data3-v1.tif
-
Figure 1—figure supplement 1—source data 4
Primary data for graphs in Figure 1—figure supplement 1E and F.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-figsupp1-data4-v1.xlsx
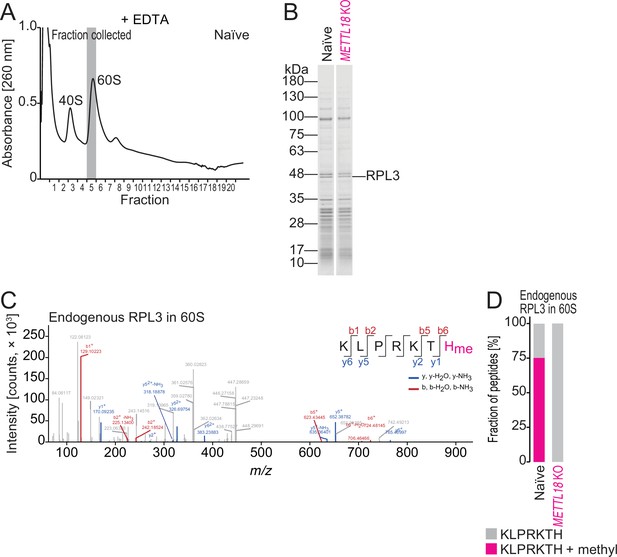
Characterization of methylhistidine in endogenous RPL3.
(A) Sucrose density gradient for ribosomal complexes. Lysate was prepared with a buffer containing EDTA to dissociate 80S into 40S and 60S. The 60S fraction used for liquid chromatography mass spectrometry (LC-MS/MS) analysis is highlighted in gray. (B) Coomassie brilliant blue (CBB) staining of proteins in the 60S fraction in naïve and METTL18 KO HEK293T cells. (C) Methylated histidine residue in endogenous RPL3 in 60S was searched by LC-MS/MS. (D) Quantification of methylated and unmethylated peptide (KLPRKTH) from endogenous RPL3 in 60S cells.
-
Figure 1—figure supplement 2—source data 1
Full and unedited gel images corresponding to Figure 1—figure supplement Figure 1—figure supplement 2B.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-figsupp2-data1-v1.tif
-
Figure 1—figure supplement 2—source data 2
Primary data for graphs in Figure 1—figure supplement 2D.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig1-figsupp2-data2-v1.xlsx
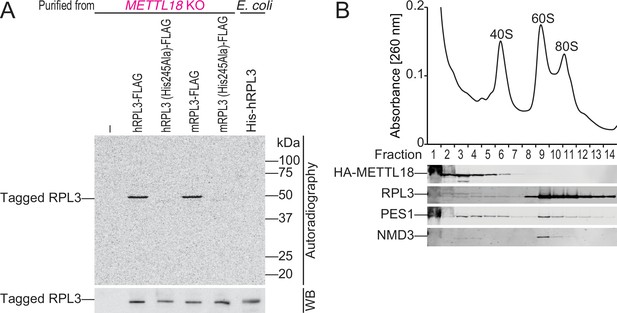
METTL18 associates with pre-60S.
(A) In vitro methylation assay with recombinant His-GST-METTL18 protein and 14C-labeled S-adenosyl-l-methionine (SAM). Immunopurified human or mouse RPL3 expressed in METTL18 knockout (KO) cells and recombinant human RPL3 expressed in bacteria were used as substrates. (B) Western blot for the indicated proteins in ribosomal complexes separated by sucrose density gradient.
-
Figure 2—source data 1
Full and unedited images corresponding to Figure 2A.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig2-data1-v1.zip
-
Figure 2—source data 2
Full and unedited blots corresponding to Figure 2B.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig2-data2-v1.zip
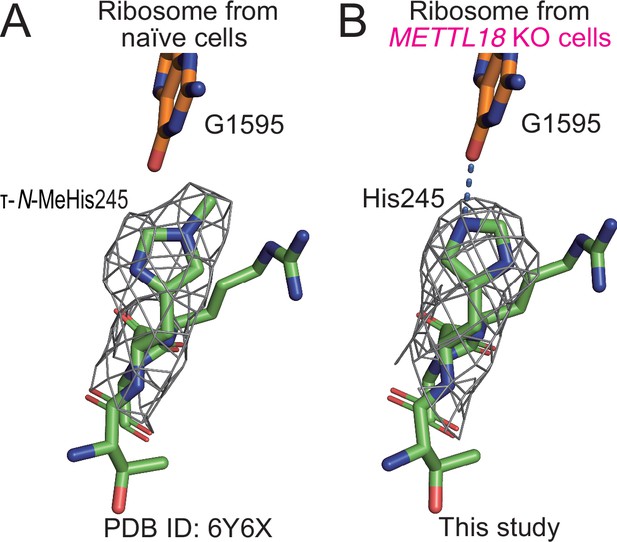
Structural differences in ribosomes upon methylation at His245.
(A) Stick models of 244GHR246 of RPL3 and G1595 of the 28S rRNA of the human ribosome are shown with the cryo-electron microscopy (cryo-EM) density map around His245. The τ-N-methyl group was manually added to the original model (PDB ID: 6Y6X) (Osterman et al., 2020) based on the cryo-EM density map. (B) The same model as in (A) of human ribosome from METTL18 knockout (KO) cells. A hydrogen bond between His245 and G1595 is indicated with a dotted blue line.
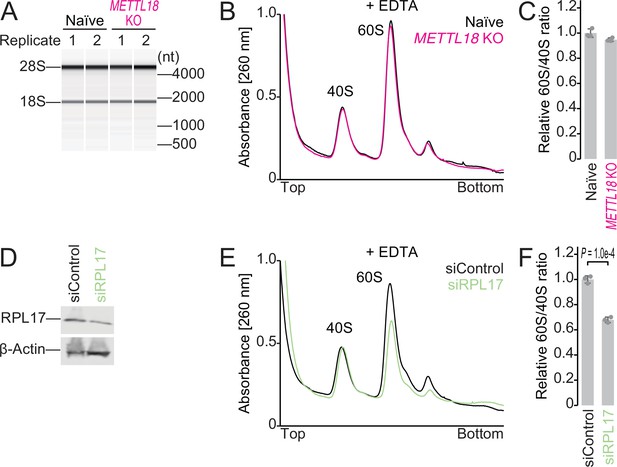
Ribosome subunit ratio in METTL18 cells.
(A) Electropherogram of ribosomal RNAs from naïve and METTL18 knockout (KO) HEK293T cells. Data from two replicates are shown. (B, C) Sucrose density gradient for ribosomal complexes from naïve and METTL18 KO HEK293T cells (B) and the quantification (C). The lysate was prepared with a buffer containing EDTA to dissociate 80S into 40S and 60S. In (C), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. (D) Western blot of the indicated proteins to confirm the knockdown of RPL17. β-Actin was probed for as a loading control. (E, F) Sucrose density gradient for ribosomal complexes from control siRNA (siControl) and RPL17 siRNA (siRPL17)-transfected cells (E) and the quantification (F). The lysate was prepared with a buffer containing EDTA to dissociate 80S into 40S and 60S. In (F), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. Significance was determined by Student’s t-test (unpaired, two-sided).
-
Figure 3—figure supplement 1—source data 1
Full and unedited blots corresponding to Figure 3—figure supplement 1D.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig3-figsupp1-data1-v1.tif
-
Figure 3—figure supplement 1—source data 2
Primary data for graphs in Figure 3—figure supplement 1C and F.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig3-figsupp1-data2-v1.xlsx
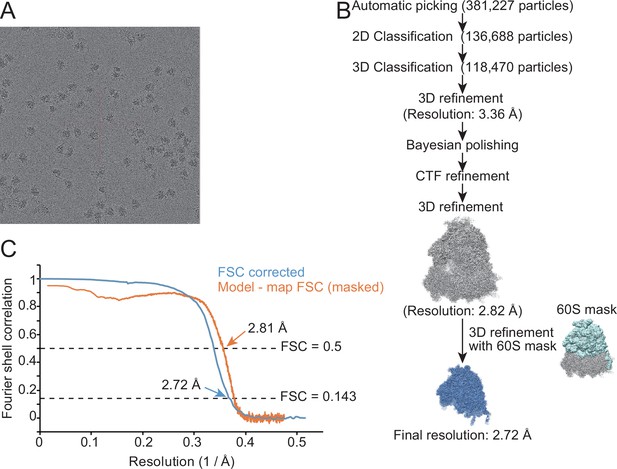
Characterization of the structure of the 60S subunit from METTL18 knockout (KO) cells.
(A) Representative cryo-electron microscopy (cryo-EM) micrographs of human ribosomes isolated from METTL18 KO cells. (B) Flow of the cryo-EM structural analysis of the human 60S subunit from METTL18 KO cells. (C) Resolution curves of the reconstituted cryo-EM structure of the human 60S subunit from METTL18 KO cells.
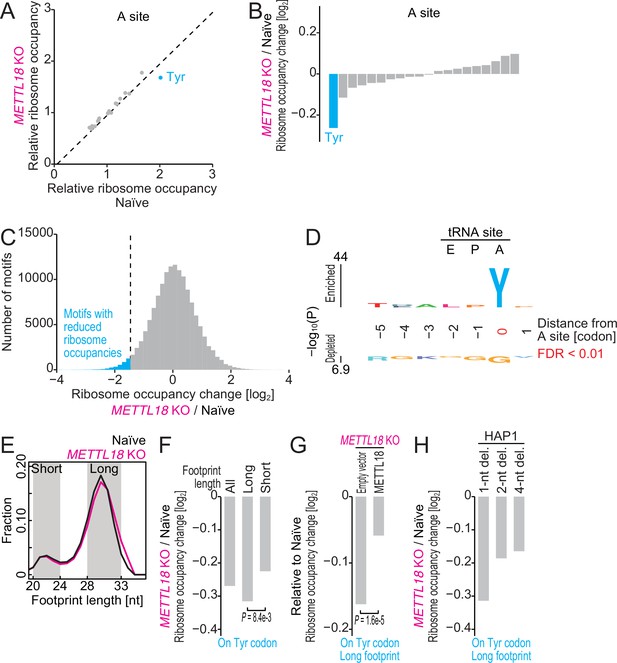
Ribosome profiling reveals Tyr codon-specific translation retardation by RPL3 methylation.
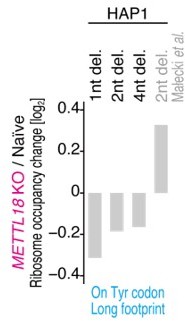
(A) Ribosome occupancy at A-site codons in naïve and METTL18 knockout (KO) HEK293T cells. Data were aggregated into codons with each amino acid species. (B) Ribosome occupancy changes at A-site codons caused by METTL18 KO. (C) Histogram of ribosome occupancy changes in METTL18 KO cells across motifs around A-site codons (seven amino acid motifs). Cyan: motifs with reduced ribosome occupancy (defined by ≤ mean – 2 SD). (D) Amino acid motifs associated with reduced ribosome occupancy in METTL18 KO cells (defined in C) are shown relative to the A-site (at the 0 position). (E) Distribution of footprint length in naïve and METTL18 KO HEK293T cells. (F) Ribosome occupancy changes on Tyr codons by METTL18 KO along all, long (28–33 nt), and short (20–24 nt) footprints. Significance was determined by the Mann–Whitney U-test. (G) The recovery of long footprint reduction in METTL18 KO cells by ectopic expression of METTL18 protein. Significance was determined by the Mann–Whitney U-test. (H) Changes in ribosome occupancy on Tyr codons by METTL18 KO in HAP1 cells along long (28–33 nt) footprints. Del., deletion. In (A–C) and (E–H), the means of two independent experiments are shown.
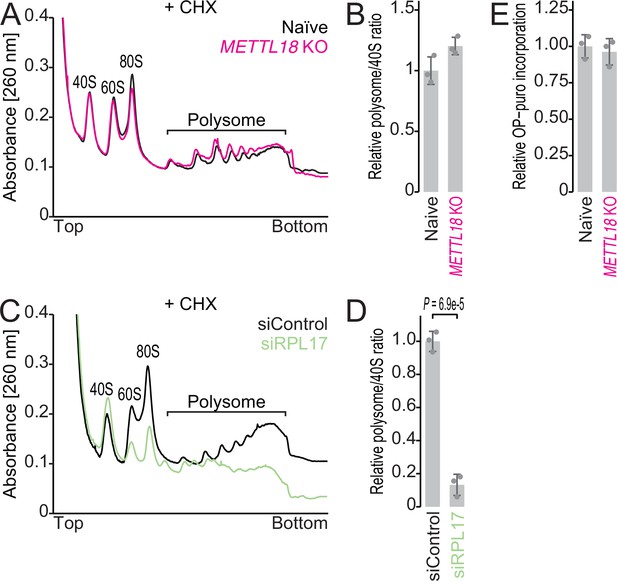
Basal translation activity in METTL18 cells.
(A, B) Sucrose density gradient for ribosomal complexes from naïve and METTL18 knockout (KO) HEK293T cells (A) and the quantification (B). The 80S ribosome and polysomes were stabilized by Mg ion and cycloheximide. In (B), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. (C, D) Sucrose density gradient for ribosomal complexes from control siRNA (siControl)- and RPL17 siRNA (siRPL17)-transfected cells (C) and the quantification (D). 80S and polysomes were stabilized by Mg ion and cycloheximide. In (D), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. Significance was determined by Student’s t-test (unpaired, two-sided). (E) Newly synthesized proteins in naïve and METTL18 KO HEK293T cells were labeled with OP-puro and then conjugated with infrared 800 (IR800) dye with a click reaction. The signal was normalized to total proteins stained with Coomassie brilliant blue (CBB). Data from three replicates (points) and the mean (bar) with SD (error bar) are shown.
-
Figure 4—figure supplement 1—source data 1
Primary data for graphs in Figure 4—figure supplement 1B, D, and E.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig4-figsupp1-data1-v1.xlsx
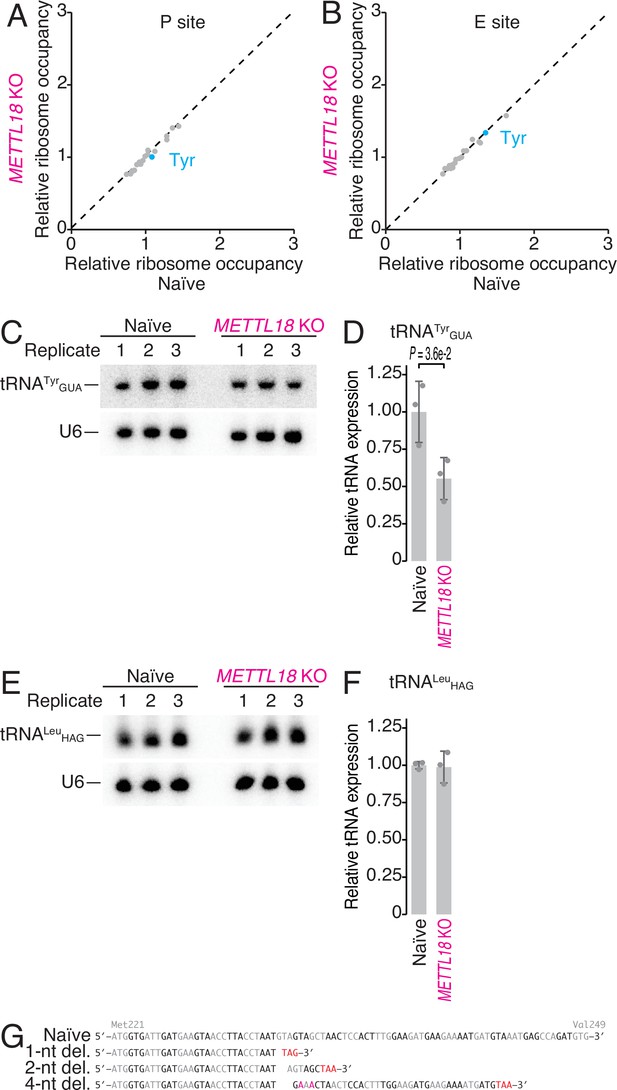
Characterization of ribosome occupancy monitored by ribosome profiling.
(A, B) Ribosome occupancy at P-site (A) and E-site (B) codons. Data were aggregated into codons with each amino acid species. The means of two independent experiments are shown. (C, D) Northern blot for tRNATyrGUA (C) and its quantification (D). U6 snRNA was used as loading control. In (D), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. Significance was determined by Student’s t-test (unpaired, two-sided). (E, F) Same as (C) and (F) but for tRNALeuHAG. In (F), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. H stands for A, C, or U. (G) Schematic representation of mutations in METTL18 KO HAP1 cells. Del., deletion.
-
Figure 4—figure supplement 2—source data 1
Full and unedited blots corresponding to Figure 4—figure supplement 2C.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig4-figsupp2-data1-v1.zip
-
Figure 4—figure supplement 2—source data 2
Full and unedited blots corresponding to Figure 4—figure supplement 2E.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig4-figsupp2-data2-v1.zip
-
Figure 4—figure supplement 2—source data 3
Primary data for graphs in Figure 4—figure supplement 2D and F.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig4-figsupp2-data3-v1.xlsx
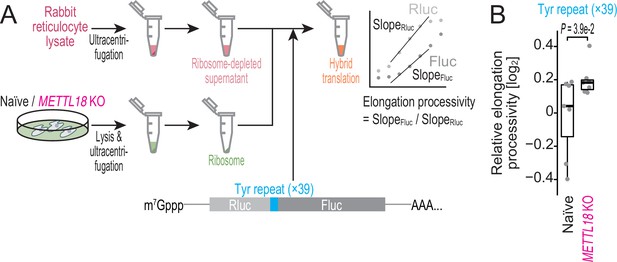
Ribosome without RPL3 methylation shows higher processivity on Tyr codons in vitro.
(A) Schematic representation of the hybrid translation system and the processivity reporter. (B) The box plot for the relative ratio of SlopeFluc to SlopRluc for the reporter with Tyr repeat insertion. Data from seven replicates (points) are shown. Significance was determined by Brunner–Munzel test (unpaired, two-sided).
-
Figure 5—source data 1
Primary data for graphs in Figure 5B.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig5-data1-v1.xlsx
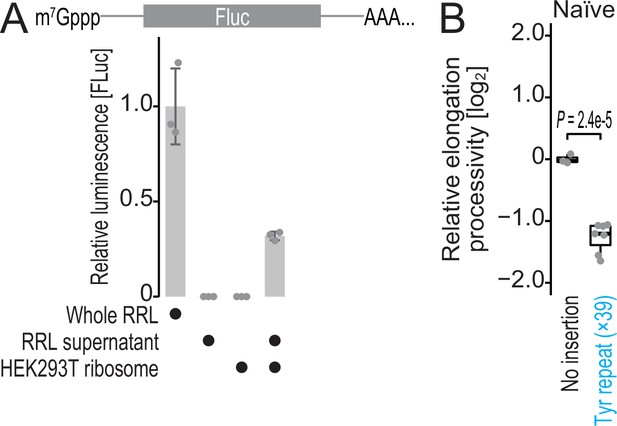
Characterization of hybrid translation system and Renilla-firefly fused reporter.
(A) In vitro translation from firefly luciferase was conducted in the indicated materials. Data from three replicates (points) and the mean (bar) with SD (error bar) are shown. RRL, rabbit reticulocyte lysate. (B) The box plot for the relative ratio of SlopeFluc to SlopRluc for the reporters with and without Tyr repeat insertion. Data from three replicates for no insertion reporter and seven replicates for Tyr repeat-inserted reporter (points) are shown. Significance was determined by the Student’s t-test (unpaired, two-sided).
-
Figure 5—figure supplement 1—source data 1
Primary data for graphs in Figure 5—figure supplement 1A, B.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig5-figsupp1-data1-v1.xlsx
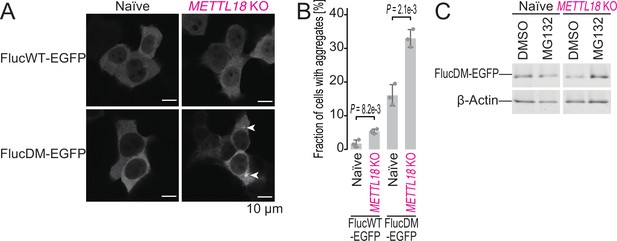
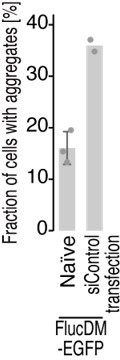
METTL18 deletion leads to cellular proteotoxicity.
(A) Microscopic images of FlucWT-EGFP or FlucDM-EGFP in naïve and METTL18 knockout (KO) HEK293T cells. Arrowhead, protein aggregation; scale bar, 10 μm. (B) Quantification of cells with Fluc-EGFP aggregates. Data from three replicates (points) and the mean (bar) with SD (error bar) are shown. Significance was determined by Student’s t-test (unpaired, two-sided). (C) Western blot for FlucDM-EGFP (probed by anti-GFP antibody) expressed in naïve and METTL18 KO HEK293T cells treated with MG132 (0.25 μM for 24 hr). β-Actin was probed as a loading control.
-
Figure 6—source data 1
Full and unedited images corresponding to Figure 6A.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig6-data1-v1.zip
-
Figure 6—source data 2
Full and unedited blots corresponding to Figure 6C.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig6-data2-v1.tif
-
Figure 6—source data 3
Primary data for graphs in Figure 6B.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig6-data3-v1.xlsx
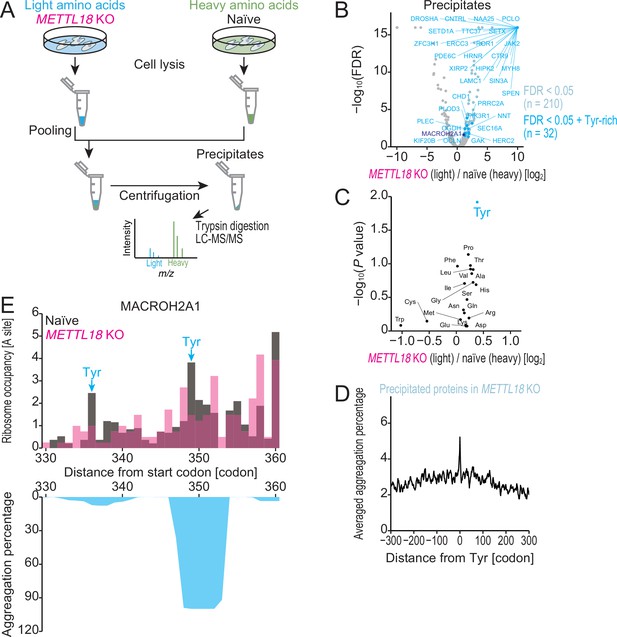
METTL18 deletion aggregates Tyr-rich proteins.
(A) Schematic representation of SILAC-MS for precipitated proteins. (B) Volcano plot for precipitated proteins in METTL18 knockout (KO) cells, assessed by SILAC-MS (n = 2). Tyr-rich proteins were defined as proteins with 30 or more Tyr residues. (C) Amino acids associated with protein precipitation in METTL18 KO cells. Precipitated proteins enriched with each amino acid were compared to the total precipitated proteome. The mean fold change and the significance (Mann–Whitney U-test) were plotted. (D) Metagene plot for aggregation percentage, calculated with TANGO (Fernandez-Escamilla et al., 2004), around Tyr codons of precipitated proteins in METTL18 KO cells (defined in B). (E) Distribution (at the A-site) of ribosome footprint occupancy (the mean of two independent experiments) along the MACROH2A1 gene in naïve (gray) and METTL18 KO (magenta) HEK293T cells, depicted with the aggregation percentage (light blue) calculated by TANGO (Fernandez-Escamilla et al., 2004). Tyr codon positions are highlighted with arrows.
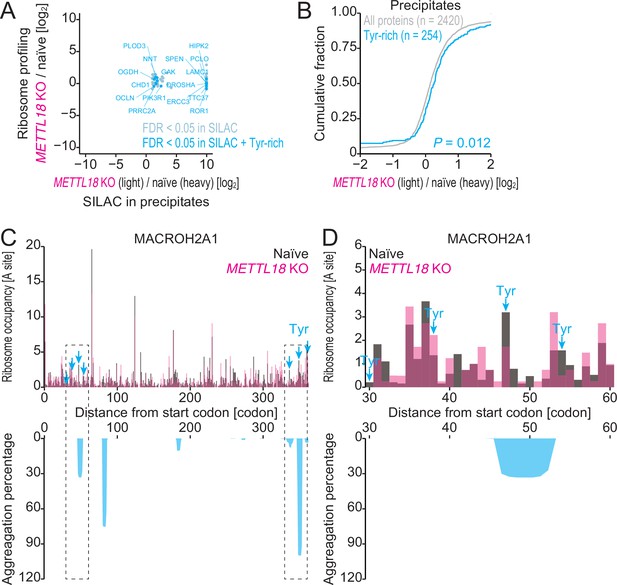
Characterization of precipitated proteins identified by SILAC-MS.
(A) Comparison of fold change in protein precipitates (n = 2) assessed by SILAC and that in ribosome profiling by METTL18 depletion. (B) Cumulative distribution of Tyr-rich proteins along the fold change in protein precipitates by METTL18 depletion. Significance was calculated by the Mann–Whitney U-test. (C, D) Distribution (at the A-site) of ribosome footprint occupancy (the mean of two independent experiments) along the MACROH2A1 gene in naïve (gray) and METTL18 KO (magenta) HEK293T cells, depicted with the aggregation percentage (light blue) calculated with TANGO (Fernandez-Escamilla et al., 2004). Tyr codon positions are highlighted with arrows. Data on the entire CDS (C) and the 30–60 amino acid region (D) are depicted.
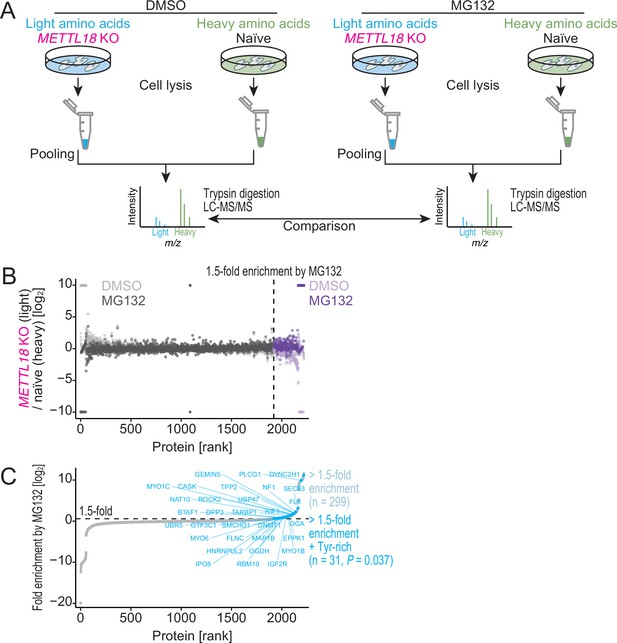
METTL18 deletion degrades Tyr-rich proteins by proteasome.
(A) Schematic representation of SILAC-MS for total proteins. (B, C) Cellular protein abundance changes in METTL18 knockout (KO) cells with the treatment of proteasome inhibitor MG132 and the control DMSO, assessed by SILAC-MS (n = 2). The relative abundance in METTL18 KO HEK293T cells compared to naïve HEK293T cells is calculated. Data with DMSO or MG132 treatment (B) and fold enrichment (MG132 compared to DMSO) (C) are shown ranked by the fold enrichment in (C). Proteins with 1.5 or higher enrichment by MG132 treatment are highlighted. Tyr-rich proteins are defined as proteins with 30 or more Tyr residues.
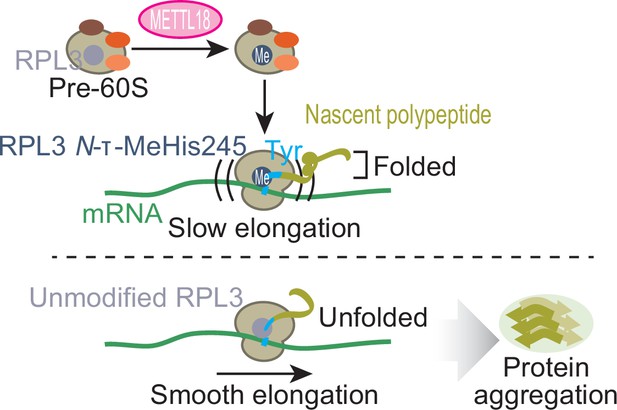
Schematic representation of METTL18-mediated control of translation and proteostasis.
METTL18 adds a methyl moiety at the τ-N position of His245 in RPL3 in the form of an early 60S biogenesis intermediate. Methylated ribosomes slow elongation at Tyr codons and extend the duration of nascent peptide folding, ensuring proteostatic integrity. Without RPL3 methylation, the accumulation of unfolded and ultimately aggregated proteins in cells was induced.
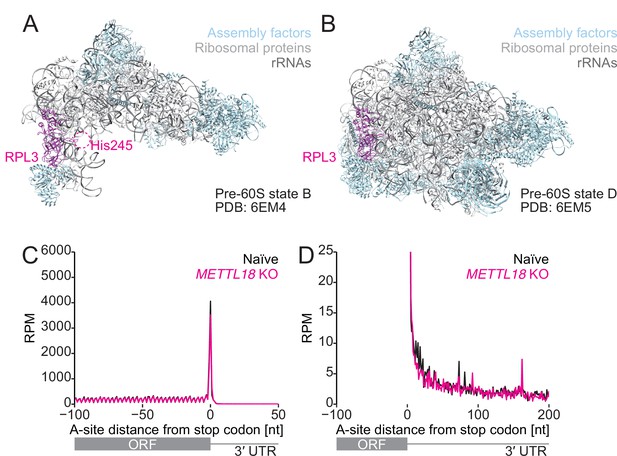
Comparison of the structure of the early and late pre-60S as a possible RPL3 methylation target.
(A, B) Structures of early (state B, PDB 6EM4:) and late (state D, PDB: 6EM5) pre-60S (Kater et al., 2017). A possible region of the protein fragment containing His245 in RPL3 is highlighted in a dashed circle. RPL3, magenta; assembly factor, light blue; ribosomal proteins, light gray; rRNA, dark gray. (C, D) Metagene analysis of ribosome footprints around the stop codon. A-site potion of footprints is depicted. In (D), a zoomed-in view of the plot is shown. The mean of two independent experiments is shown.
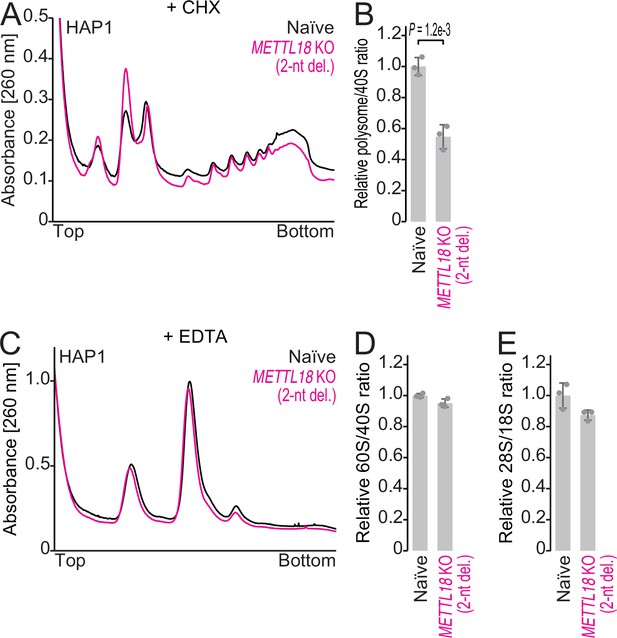
The impacts of METTL18 deletion in HAP1 cells on ribosomal complex formation.
(A, B) Sucrose density gradient for ribosomal complexes from naïve and METTL18 knockout (KO) (2-nt del.) HAP1 cells (A) and the quantification (B). 80S and polysomes were stabilized by Mg ion and cycloheximide. In (B), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. Significance was determined by Student’s t-test (unpaired, two-sided). (C, D) Sucrose density gradient for ribosomal complexes from naïve and METTL18 KO (2-nt del.) HAP1 cells (C) and the quantification (D). The lysate was prepared with a buffer containing EDTA to dissociate 80S into 40S and 60S. In (D), data from three replicates (points) and the mean (bar) with SD (error bar) are shown. (E) Quantification of ribosomal RNAs from naïve and METTL18 KO (2-nt del.) HAP1 cells by fragment analyzer. Data from three replicates (points) and the mean (bar) with SD (error bar) are shown.
-
Figure 9—figure supplement 2—source data 1
Primary data for graphs in Figure 9—figure supplement 2B, D, and E.
- https://cdn.elifesciences.org/articles/72780/elife-72780-fig9-figsupp2-data1-v1.xlsx
Tables
Data collection, model building, refinement, and validation statistics for cryo-electron microscopy (cryo-EM) data obtained in this study.
| Human large ribosomal subunit (obtained from METTL18 KO cells) (PDB: 7F5S, EMD-31465) | |
|---|---|
| Data collection and processing | |
| Microscope | Tecnai Arctica |
| Camera | K2 Summit |
| Magnification | 39,000 |
| Voltage (kV) | 200 |
| Electron exposure (e-/Å2) | 50 |
| Exposure per frame | 1.25 |
| Number of frames collected | 40 |
| Defocus range (μm) | –1.5 to –3.1 |
| Micrographs (no.) | 5,517 |
| Pixel size (Å) | 0.97 |
| 3D processing package | RELION-3.1 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 381,227 |
| Final particle images (no.) | 118,470 |
| Initial reference map | EMD-9701 (40 Å) |
| RELION estimated accuracy | |
| Rotations (°) | 0.162 |
| Translations (pixel) | 0.287 |
| Map resolution | |
| masked (FSC = 0.143, Å) | 2.72 |
| Map sharpening B-factor | –63.0 |
| Refinement | |
| Model refinement package | phenix.real_space_refine |
| Initial model used | 6QZP |
| Model composition | |
| Chains | 45 |
| Non-hydrogen atoms | 138,634 |
| Residues | Protein: 6509; nucleotide: 3991 |
| Ligands | ZN: 5, MG: 297 |
| B factors (Å2) | |
| Protein | 62.85 |
| Nucleotide | 81.15 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 0.834 |
| Validation | |
| Molprobity score | 1.92 |
| Clashscore | 9.61 |
| Poor rotamers (%) | 0.13 |
| CaBLAM outliers (%) | 3.35 |
| Ramachandran plot | |
| Favored (%) | 93.79 |
| Allowed (%) | 6.08 |
| Disallowed (%) | 0.12 |
| Map CC (CCmask) | 0.90 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | METTL18 | GenBank | NM_033418 | |
| Gene (H. sapiens) | RPL3 | GenBank | NM_000967 | |
| Gene (Mus musculus) | METTL18 | GenBank | NM_027279 | cDNA clone AK139786 (FANTOM) was used |
| Gene (M. musculus) | RPL3 | GenBank | NM_013762 | |
| Cell line (H. sapiens) | Naïve HEK293T | RIKEN BRC | RCB2202 | Female |
| Cell line (H. sapiens) | METTL18 KO HEK293T | This paper | Female; CRISPR/Cas9-edited cell line, knocking out METTL18 | |
| Cell line (H. sapiens) | METTL18 KO HEK293T with stable METTL18 expression | This paper | Female; exogenous METTL18 expression was induced in METTL18 KO HEK293T | |
| Cell line (H. sapiens) | SETD3 KO HEK293T | This paper | Female; CRISPR/Cas9-edited cell line, knocking out SETD3 | |
| Cell line (H. sapiens) | SETD3-METTL18 DKO HEK293T | This paper | Female; CRISPR/Cas9-edited cell line, knocking out SETD3 and METTL18 simultaneously | |
| Cell line (H. sapiens) | Naïve HAP1 | Horizon Discovery | Cat# C631 | Male |
| Cell line (H. sapiens) | HAP1 1-nt deletion | Horizon Discovery | Cat# HZGHC000541c009 | Male; CRISPR/Cas9-edited cell line containing a 1-nt deletion in a coding exon of METTL18 |
| Cell line (H. sapiens) | HAP1 2-nt deletion | Horizon Discovery | Cat# HZGHC000541c012 | Male; CRISPR/Cas9-edited cell line containing a 2-nt deletion in a coding exon of METTL18 |
| Cell line (H. sapiens) | HAP1 4-nt deletion | Horizon Discovery | Cat# HZGHC000541c002 | Male; CRISPR/Cas9-edited cell line containing a 4-nt deletion in a coding exon of METTL18 |
| Transfected construct (H. sapiens) | PX330-B/B-gMETTL18 | This paper | Guide RNA expression | |
| Transfected construct (H. sapiens) | pL-CRISPR.EFS.tRFP-gSETD3 | This paper | Guide RNA expression | |
| Transfected construct (H. sapiens) | pcDNA3-hRPL3-FLAG (WT and His245Ala) | This paper | Protein expression | |
| Transfected construct (H. sapiens) | pcDNA3-mRPL3-FLAG (WT and His245Ala) | This paper | Protein expression | |
| Transfected construct (H. sapiens) | pQCXIP-hMETTL18-HA | This paper | Protein expression | |
| Transfected construct (H. sapiens) | hMETTL18-Asp193Lys-Gly195Arg-Gly197Arg-HA | This paper | Protein expression | |
| Transfected construct (H. sapiens) | siRNA to RPL17 | Horizon Discovery | L-013633- 01-0005 | |
| Transfected construct (H. sapiens) | Control siRNA | Horizon Discovery | D-001810- 10-50 | |
| Transfected construct (H. sapiens) | pCI-neo Fluc-EGFP | Addgene | RRID:Addgene_90170 | Protein expression |
| Transfected construct (H. sapiens) | pCI-neo FlucDM-EGFP | Addgene | RRID:Addgene_90172 | Protein expression |
| Antibody | Anti-α-tubulin (mouse monoclonal) | Sigma-Aldrich | Cat# T5168; RRID:AB_477579 | WB 1:1000 |
| Antibody | Anti-METTL18 (rabbit polyclonal) | Proteintech Group | Cat# 25553-1-AP; RRID:AB_2503968 | WB (1:1000) |
| Antibody | Anti-SETD3 (rabbit polyclonal) | Abcam | Cat# ab174662; RRID:AB_2750852 | WB (1:1000) |
| Antibody | Anti-RPL3 (mouse monoclonal) | Proteintech Group | Cat# 66130-1-lg; RRID:AB_2881529 | WB (1:1000) |
| Antibody | Anti-RPL3 (rabbit polyclonal) | Proteintech Group | Cat# 11005-1-AP; RRID:AB_2181760 | WB (1:1000) |
| Antibody | Anti-PES1 (rat monoclonal) | Abcam | Cat# ab252849; RRID:AB_2915993 | WB (1:1000) |
| Antibody | Anti-NMD3 (rabbit monoclonal) | Abcam | Cat# ab170898; RRID:AB_2915994 | WB (1:1000) |
| Antibody | Anti-HA (mouse monoclonal) | MBL | Cat# M180-3; RRID:AB_10951811 | WB (1:1000) |
| Antibody | Anti-GFP (rabbit polyclonal) | Abcam | Cat# ab6556; RRID:AB_305564 | WB (1:1000) |
| Antibody | Anti-β-actin (mouse monoclonal) | MBL | Cat# M177-3; RRID:AB_10697039 | WB (1:1000) |
| Antibody | Anti-mouse IgG, conjugate with HRP (sheep polyclonal) | Cytiva | Cat# NA931V; RRID:AB_772210 | WB (1:5000) |
| Antibody | Anti-rabbit IgG, conjugated with HRP (donkey polyclonal) | Cytiva | Cat# NA934V; RRID:AB_772206 | WB (1:5000) |
| Antibody | Anti-mouse IgG, conjugated with IRDye 680RD (goat polyclonal) | LI-COR Biosciences | Cat# 925-68070; RRID:AB_2651128 | WB (1:10,000) |
| Antibody | Anti-rabbit IgG, conjugated with IRDye 680RD (goat polyclonal) | LI-COR Biosciences | Cat# 925-68071; RRID:AB_2721181 | WB (1:10,000) |
| Antibody | Anti-mouse IgG, conjugated with IRDye 800CW (goat polyclonal) | LI-COR Biosciences | Cat# 926-32210; RRID:AB_621842 | WB (1:10,000) |
| Antibody | Anti-rabbit IgG, conjugated with IRDye 800CW (goat polyclonal) | LI-COR Biosciences | Cat# 926-32211; RRID:AB_621843 | WB (1:10,000) |
| Antibody | Anti-rat IgG, conjugated with IRDye 800CW (goat polyclonal) | LI-COR Biosciences | Cat# 926-32219; RRID:AB_1850025 | WB (1:10,000) |
| Antibody | Anti-GFP (mouse monoclonal) | Abcam | Cat# ab1218; RRID:AB_298911 | IF (1:1000) |
| Antibody | Anti-mouse IgG, conjugated with Alexa Fluor 488 (goat polyclonal) | Thermo Fisher Scientific | Cat# R37120; RRID:AB_2556548 | IF (1:1000) |
| Recombinant DNA reagent | pET19b-mMETTL18 | This paper | Expression of N-terminally His-tagged mouse METTL18 in Escherichia coli | |
| Recombinant DNA reagent | pCold-GST-mMETTL18 | This paper | Expression of N-terminally His- and GST-tagged mouse METTL18 in E. coli | |
| Recombinant DNA reagent | Salmonella MTAN | Addgene | RRID:Addgene_64041 | Expression of Salmonella MTAN in E. coli |
| Recombinant DNA reagent | pGL3 basic | Promega | Cat# E1751 | |
| Recombinant DNA reagent | psiCHECK2 | Promega | Cat# C8021 | |
| Recombinant DNA reagent | psiCHECK2-Y0× | This study | Encoding Rluc-Fluc fusion | |
| Recombinant DNA reagent | psiCHECK2-Y39× | This study | Encoding Rluc-Fluc fusion with Tyr repeat insertion | |
| Sequence-based reagent | Probe for tRNATyrGUA | This paper | 5′-ACAGTCCTCCGCTCTACCAGCTGA-3′ | |
| Sequence-based reagent | Probe for tRNALeuHAG | This paper | 5′-CAGCGCCTTAGACCGCTCGGCCA-3′ | |
| Sequence-based reagent | Probe for U6 | This paper | 5′-CACGAATTTGCGTGTCATCCTT-3′ | |
| Commercial assay or kit | QuikChange Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200518 | |
| Commercial assay or kit | PEI transfection reagent | Polysciences | ||
| Commercial assay or kit | TransIT-293 | Mirus | Cat# MIR2700 | |
| Commercial assay or kit | TransIT-X2 Dynamic Delivery System | Mirus | Cat# MIR6000 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Cat# E1910 | |
| Commercial assay or kit | Rabbit Reticulocyte Lysate, Nuclease-Treated | Promega | Cat# L4960 | |
| Commercial assay or kit | Click-iT Cell Reaction Buffer Kit | Thermo Fisher Scientific | Cat# C10269 | |
| Commercial assay or kit | T7-Scribe Standard RNA IVT kit | CELLSCRIPT | Cat# C-MSC11610 | |
| Commercial assay or kit | ScriptCap m7G Capping system | CELLSCRIPT | Cat# C-SCCE0625 | |
| Commercial assay or kit | A-Plus poly(A) polymerase Tailing kit | CELLSCRIPT | Cat# C-PAP5104H | |
| Chemical compound, drug | IRdye800CW azide | LI-COR Biosciences | Cat# 929-65000 | |
| Chemical compound, drug | MG132 | FUJIFILM Wako Chemicals | Cat# 139-18451 | |
| Software, algorithm | Proteome Discoverer | Thermo Fisher Scientific | Version 2.3 | LC-MS/MS for methylated peptide |
| Software, algorithm | Proteome Discoverer | Thermo Fisher Scientific | Version 2.4 | SILAC-MS |
| Software, algorithm | MASCOT | Matrix Science | Version 2.7 | LC-MS/MS for methylated peptide and SILAC-MS |
| Software, algorithm | RELION-3.1 | https://doi.org/10.1107/S2052252520000081 | ||
| Software, algorithm | CTFFIND-4.1 | https://doi.org/10.1016/j.jsb.2015.08.008 | ||
| Software, algorithm | PHENIX | https://doi.org/10.1107/S0907444909052925 | ||
| Software, algorithm | Coot | https://doi.org/10.1107/S0907444910007493 | ||
| Software, algorithm | Image Studio | LI-COR Biosciences | Version 5.2 | |
| Software, algorithm | STAR | https://doi.org/10.1093/bioinformatics/bts635 | Version 2.7.0a | |
| Software, algorithm | kpLog | https://doi.org/10.1093/nar/gkx323 | http://kplogo.wi.mit.edu |