Discovery of coordinately regulated pathways that provide innate protection against interbacterial antagonism
Figures
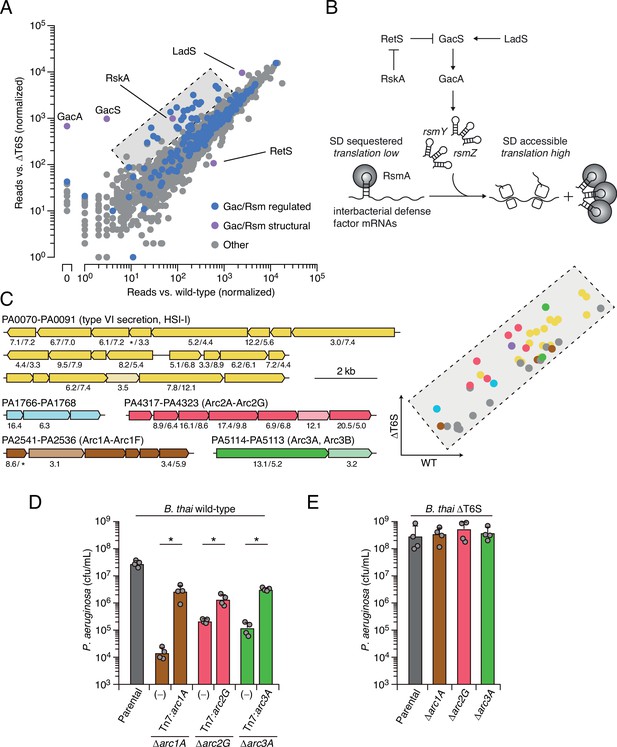
Multiple pathways under Gac/Rsm control contribute to P. aeruginosa defense against antagonism.
(A) Transposon library sequencing-based comparison of the fitness contribution of individual P. aeruginosa genes during growth competition with wild-type B. thai versus B. thai ∆T6S. Genes under Gac/Rsm control (blue) and those encoding core Gac/Rsm regulatory factors (purple, labeled) are indicated. (B) Overview of the Gac/Rsm pathway (SD, Shine–Dalgarno). The rsmY and rsmZ genes encode small RNA molecules that sequester the translational regulator RsmA. Hybrid sensor kinase PA1611 is here renamed RetS-regulating sensor kinase A (RskA). (C) Left: P. aeruginosa gene clusters hit (greater than threefold in replicate screens; Figure 1—source data 1) in this study. Numbers below genes indicate transposon insertion ratio (B. thai ∆T6S/wild-type) for screen in (A) followed by the replicate screen (Figure 1—figure supplement 1); light toned genes were hit in only one replicate. Asterisk indicates genes for which insertion was undetected in libraries obtained from B. thai wild-type competition. Right: zoom-in of boxed region of (A) with genes colored corresponding to clusters at left. (D, E) Recovery of P. aeruginosa cells with the indicated genotypes following growth competition against B. thai wild-type (D) or ∆T6S (E). For interbacterial competition assays, the mean ± SD of biological duplicates and associated technical duplicates is shown: *p<0.05 using unpaired two-tailed Student’s t-test.
-
Figure 1—source data 1
Transposon sequencing-based analysis of P. aeruginosa fitness determinants during antagonism by B. thai compared to B. thai with an inactive T6SS (∆icmF).
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig1-data1-v2.docx
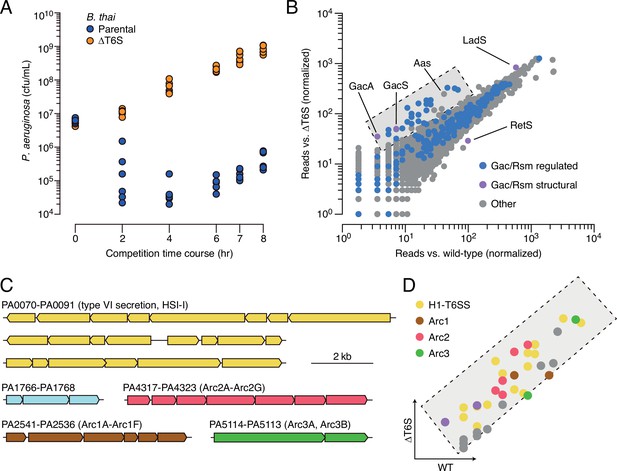
Multiple pathways under Gac/Rsm control contribute to P. aeruginosa defense against antagonism.
(A) Growth of wild-type P. aeruginosa in co-culture with the indicated B. thai strains. (B) Transposon library sequencing-based comparison of the fitness contribution of individual P. aeruginosa genes during growth competition with wild-type B. thai versus B. thai ∆T6S. Genes under Gac/Rsm control (blue) and those encoding core Gac/Rsm regulatory factors (purple, labeled) are indicated. This experiment is a biological replicate of that in Figure 1A. (C) P. aeruginosa gene clusters hit (Figure 1—source data 1) in this study. (D) Zoom-in of boxed region of (B) with genes colored corresponding to clusters at left. The interbacterial competition assay represents two biological replicates with two associated technical replicates.
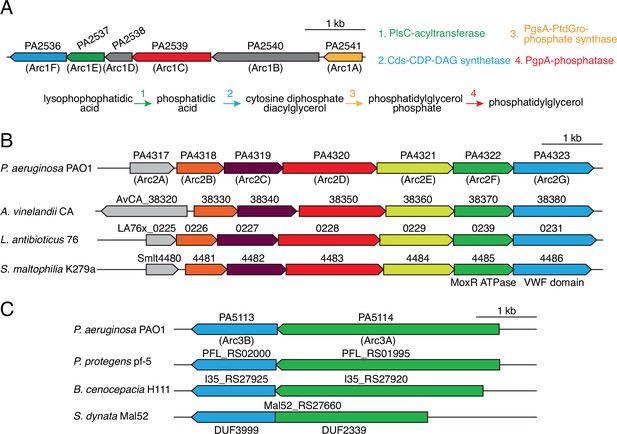
Antagonism resistance clusters possess functionally related genes and are in diverse bacteria.
(A) Schematic depicting the arc1 gene cluster of P. aeruginosa. Predicted enzymatic activity of Arc1A, B, D, F is indicated (right) and placed onto a phospholipid biosynthesis pathway (bottom). (B) Depiction of arc2 genes from the indicated species. Genes encoding the predicted MoxR-like ATPase and von Willebrand factor (VWF) domain protein are indicated below the respective genes. Orthologs are colored to highlight synteny. (C) Depiction of arc3 genes from the indicated species. S. dynata exemplifies an instance of translational fusion of arc3A,B. Domains of unknown function constituting the two proteins are indicated below.
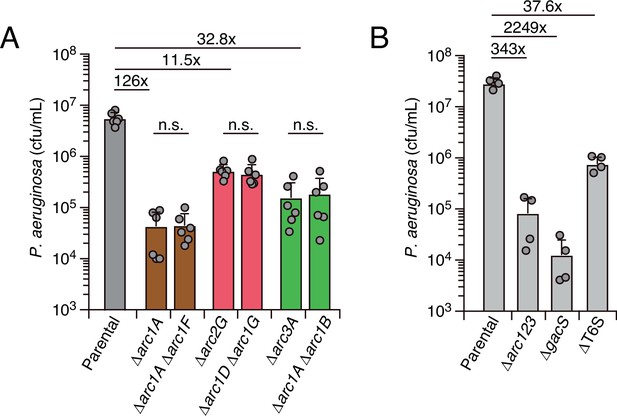
Interbacterial competition assays measure the contribution of arc1-3 to P. aeruginosa fitness during antagonism by B. thai.
(A) Recovery of P. aeruginosa strains bearing the indicated single or double deletions within arc1-3 following growth competition against wild-type B. thai. (B) Recovery of P. aeruginosa cells with the indicated genotypes following growth competition against wild-type B. thai. The mean ± SD of biological duplicates and associated technical duplicates is shown: n.s. corresponds to p>0.05 by unpaired two-tailed Student’s t-test. The fold difference in growth yield between mutants and the parental strain is indicated above.
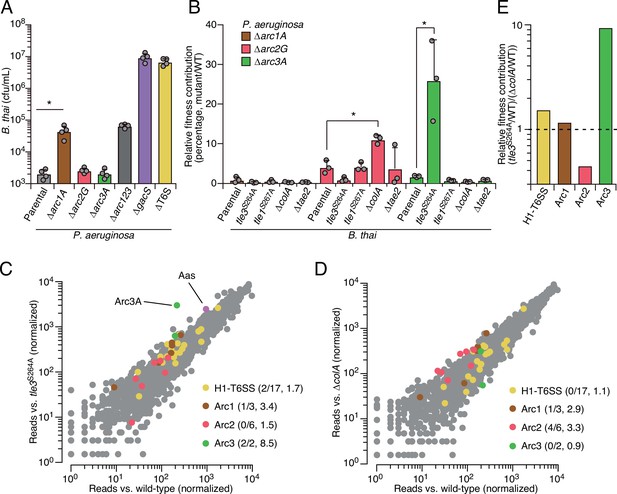
Arc pathways can provide toxin-specific antagonism defense that eclipses that of other cellular factors.
(A) Recovery of B. thai cells following growth competition against a relative abundance of P. aeruginosa containing the indicated gene deletions, colored according to Figure 1C. (B) Relative survival of P. aeruginosa Arc1-3-inactivated mutants following growth in competition with an excess of the indicated B. thai strains (mutant CFU/parental CFU × 100 for each B. thai competitor). The loss of ColA or Tle3 activity in B. thai increases the relative survival of P. aeruginosa lacking Arc2 or Arc3 activity, respectively. (C, D) Transposon library sequencing-based comparison of the fitness contribution of individual P. aeruginosa genes during growth competition with wild-type B. thai versus B. thai lacking Tle3 (C) or ColA (D) activity, colored according to Figure 1C. Values in parentheses correspond to the number of genes hit (greater than threefold change in transposon insertion frequency) within each cluster compared to the number within each cluster hit in the initial screens (B. thai wild-type versus B. thai ∆T6S) followed by the average transposon insertion frequency ratio (B. thai mutant/B. thai wild-type) of the genes in the depicted screen. (E) Fitness contribution of each P. aeruginosa Arc pathway and the H1-T6SS to defense against Tle3 versus ColA. For interbacterial competition assays, the mean ± SD of biological duplicates and two technical replicates is shown: *p<0.05 using unpaired two-tailed Student’s t-test.
-
Figure 2—source data 1
Transposon sequencing-based analysis of P. aeruginosa fitness determinants during antagonism by B. thai compared to B. thai tle3S264A.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-data1-v2.docx
-
Figure 2—source data 2
Transposon sequencing-based analysis of P. aeruginosa fitness determinants during antagonism by B. thai compared to B. thai ∆colA.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-data2-v2.docx
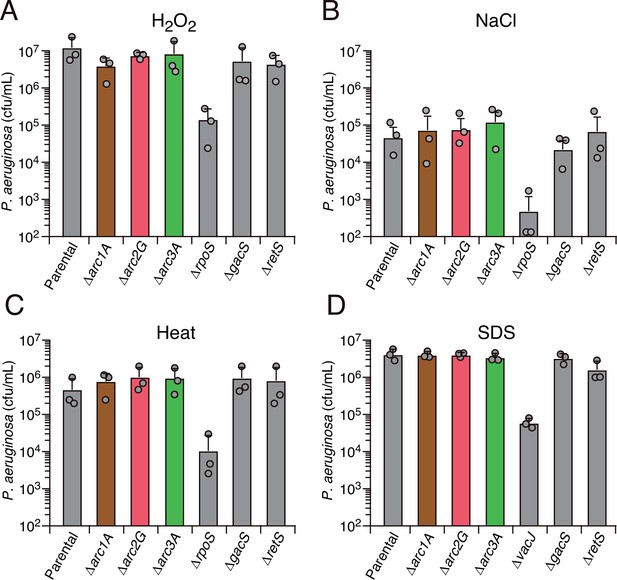
Arc1-3 do not contribute to the survival of P. aeruginosa exposed to common environmental stresses.
Survival of the indicated P. aeruginosa strains during exposure to hydrogen peroxide (50 mM, 30 min) (A), high salinity (3 M NaCl, 20 hr) (B), high temperature (55°C, 30 min) (C), or detergent (0.5% [w/v] SDS) (D). The mean ± SD of a representative biological replicate with three technical replicates is shown.
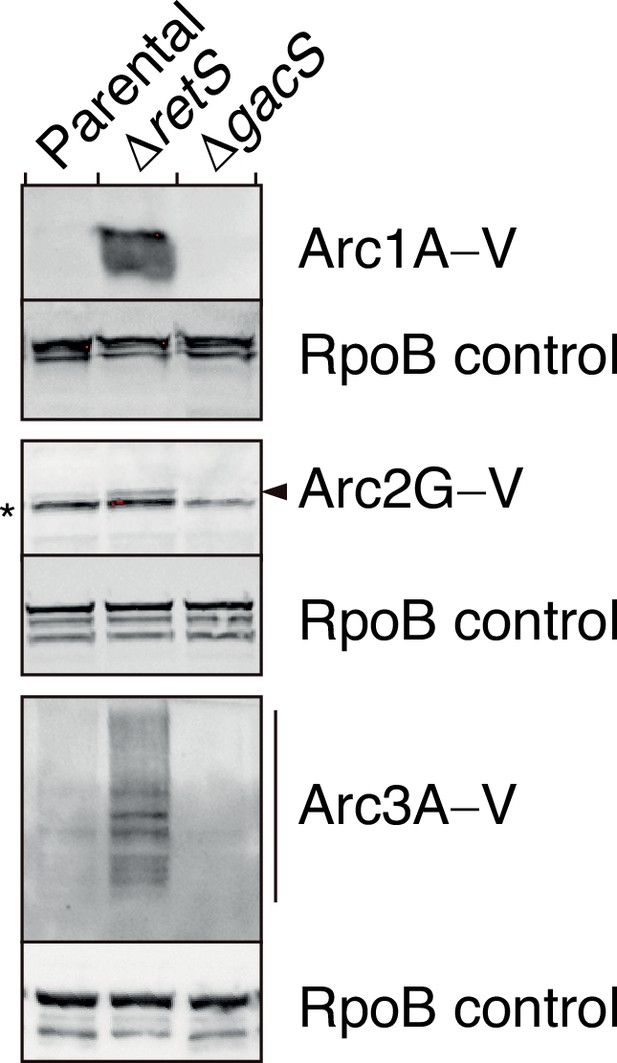
Arc1-3 are subject to tight regulation by the Gac/Rsm signaling pathway.
Anti-VSV-G immunoblot analysis was used to probe the levels of the indicated VSV–G-tagged (–V) Arc proteins encoded at their respective native genomic loci in P. aeruginosa wild-type, ∆retS, or ∆gacS backgrounds. The asterisk denotes a nonspecific VSV-G antibody-reactive band. The arrow highlights the position of Arc2G–V. Blot images are representative of at least two biological replicates. See Figure 2—figure supplement 2—source data 1–6.
-
Figure 2—figure supplement 2—source data 1
Original uncropped image and original uncropped image with relevant bands indicated for Figure 2—figure supplement 2, Arc1A–V.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-figsupp2-data1-v2.pdf
-
Figure 2—figure supplement 2—source data 2
Original uncropped image and original uncropped image with relevant bands indicated for Figure 2—figure supplement 2, Arc1A–V RpoB control.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-figsupp2-data2-v2.pdf
-
Figure 2—figure supplement 2—source data 3
Original uncropped image and original uncropped image with relevant bands indicated for Figure 2—figure supplement 2, Arc2G–V.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-figsupp2-data3-v2.pdf
-
Figure 2—figure supplement 2—source data 4
Original uncropped image and original uncropped image with relevant bands indicated for Figure 2—figure supplement 2, Arc2G–V RpoB control.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-figsupp2-data4-v2.pdf
-
Figure 2—figure supplement 2—source data 5
Original uncropped image and original uncropped image with relevant bands indicated for Figure 2—figure supplement 2, Arc3A–V.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-figsupp2-data5-v2.pdf
-
Figure 2—figure supplement 2—source data 6
Original uncropped image and original uncropped image with relevant bands indicated for Figure 2—figure supplement 2, Arc3A–V RpoB control.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig2-figsupp2-data6-v2.pdf
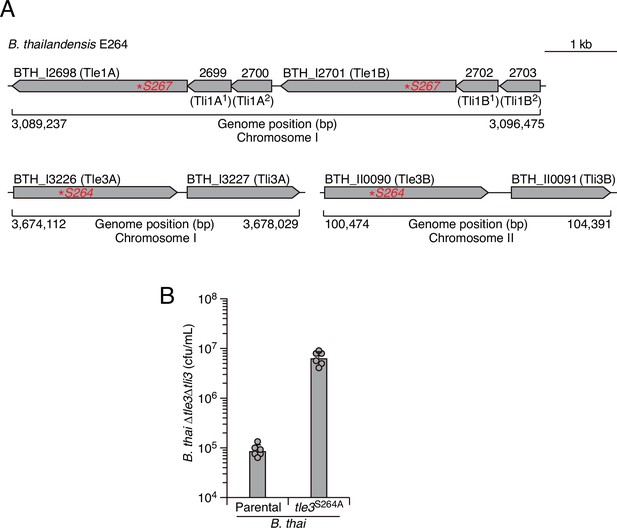
The loci encoding Tle1 and Tle3 are duplicated in B. thai, and Tle3 induces potent self-intoxication.
(A) Schematic depicting the tle1/tli1 (top) and tle3/tli3 (bottom) loci in the genome B. thai E264 strain. Predicted catalytic serine residues mutagenized in this study are indicated by red asterisks. (B) Recovery of B. thai strain sensitized to Tle3 intoxication (∆tle3∆tli3) following growth competition against B. thai wild-type or a strain in which both orthologs of Tle3 contain an inactivating point mutation. The mean ± SD of two biological replicates with two technical replicates is shown.
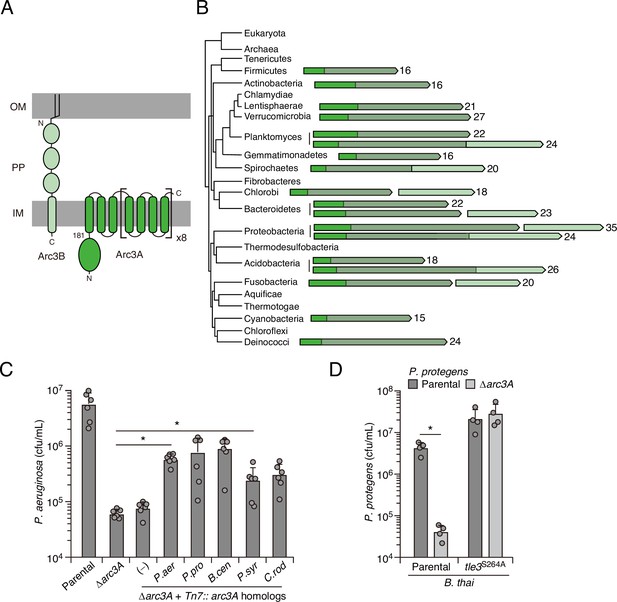
Arc3B is a predicted massive polytopic membrane protein with functionally complementing family members that occur widely in Gram-negative and -positive bacterial phyla.
(A) Schematic depiction of Arc3A and Arc3B based on bioinformatic predictions. (B) Phylogeny of bacterial phyla with representative arc genes depicted and colored as in (A). Membrane-associated regions (grayed) and the number of predicted transmembrane segments are indicated. (C) Recovery of P. aeruginosa strains bearing the indicated mutations and containing a control chromosomal insertion (–) or insertion constructs expressing Arc3B proteins derived from assorted bacteria (P. aer, P. aeruginosa; P. pro, P. protegens; B. cen, B. cenocepacia; P. syr, P. syringae; C. rod, Citrobacter rodentium) following growth competition against B. thai wild-type. (D) Recovery of the indicated P. protegens strains following growth competition against B. thai wild-type or a strain lacking Tle3 activity. For interbacterial competition assays, the mean ± SD of biological duplicates and two or three associated technical replicates is shown: *p<0.05 using unpaired two-tailed Student’s t-test.
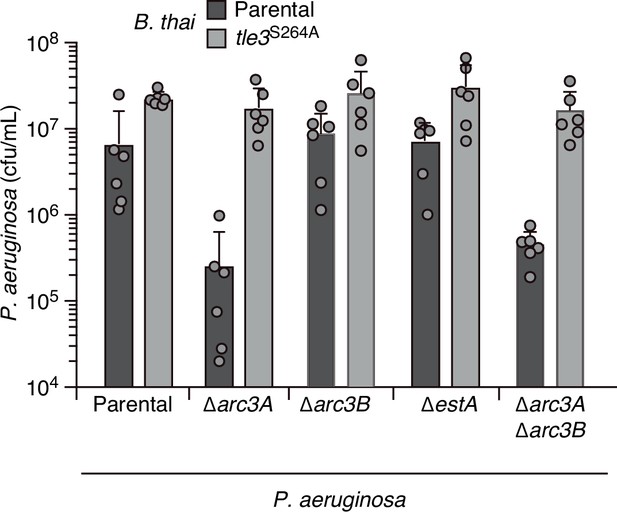
EstA and Arc3B inactivation does not significantly impact P. aeruginosa fitness in pairwise growth competition experiments with B. thai.
Recovery of the indicated P. aeruginosa strains following growth in competition with B. thai wild-type or a strain lacking Tle3 activity (tle3S264A). Means ± SD of biological duplicates with three technical replicates s shown.
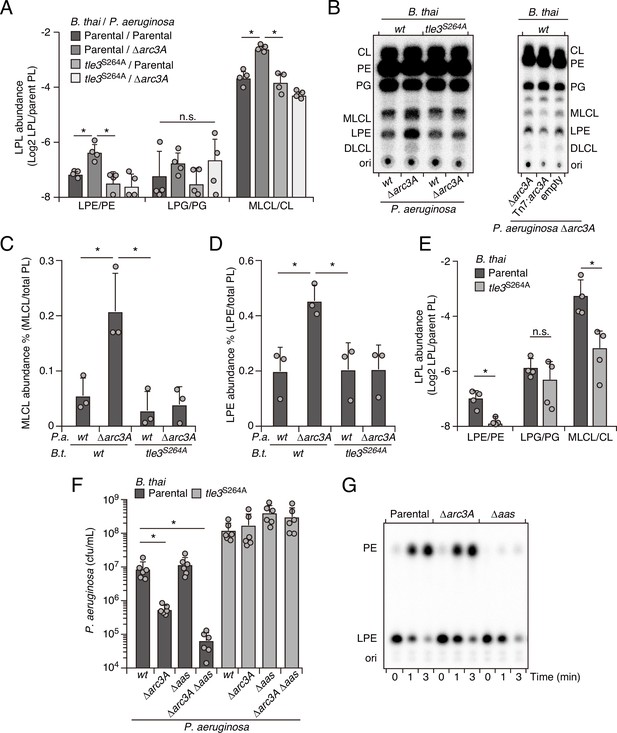
Arc3 prohibits Tle3-catalyzed lysophospholipid accumulation by a mechanism distinct from known pathways.
(A–D) Mass spectrometric (A) and radiographic thin layer chromatography (TLC) (B) analysis of major phospholipid and lysophospholipid species within lipid extracts derived from the indicated mixtures of P. aeruginosa and B. thai strains. P. aeruginosa were grown in 32PO42- prior to incubation with B. thai. Strains were allowed to interact for 1 hr before phospholipids were harvested. Radiolabeled molecules of interest within biological triplicate experiments resolved by TLC were quantified by densitometry (C, D) (E) Mass spectrometric analysis of major phospholipid and lysophospholipid species within lipid extracts derived from mixtures containing B. thai lacking Tle3-specific immunity factors competing with the indicated B. thai strains. (F) Recovery of the indicated P. aeruginosa strains following growth competition against B. thai wild-type or a strain lacking Tle3 activity. (G) Radiographic TLC analysis of products extracted after incubation of P. aeruginosa-derived spheroplasts with purified radiolabeled lysophosphatidylethanolamine (LPE). For interbacterial competition assays, the mean ± SD of biological replicates and associated technical replicates is shown: *p<0.05 using unpaired two-tailed Student’s t-test. TLC images shown are representative of at least two biological replicates. See also Figure 4—source data 1–3.
-
Figure 4—source data 1
Original uncropped image and original uncropped image with relevant spots indicated for Figure 4B, left TLC.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig4-data1-v2.pdf
-
Figure 4—source data 2
Original uncropped image and original uncropped image with relevant spots indicated for Figure 4B, right TLC.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig4-data2-v2.pdf
-
Figure 4—source data 3
Original uncropped image and original uncropped image with relevant spots indicated for Figure 4G, right TLC.
- https://cdn.elifesciences.org/articles/74658/elife-74658-fig4-data3-v2.pdf
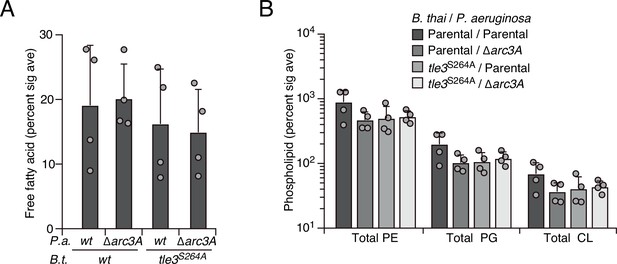
Tle3 intoxication does not impact free fatty acids nor intact parent phospholipids in P. aeruginosa.
Mass spectrometric analysis of free fatty acid (A) and parent phospholipid species (B) within lipid extracts derived from the indicated mixtures. Data represent the mean ± SD of biological duplicates with two technical replicates.
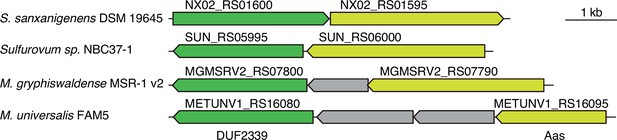
Arc3B genes are encoded adjacent to aas genes in diverse bacteria.
Schematic depicting arc3B and aas loci in the indicated bacterial species.
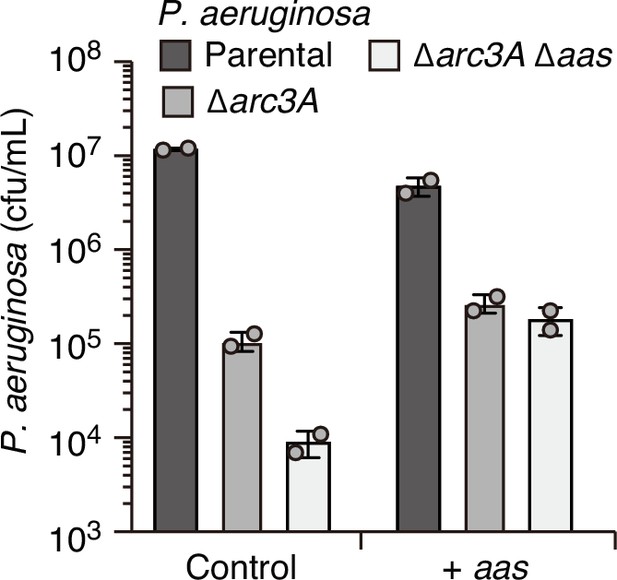
Overexpression of aas fails to complement the competitive defect associated with Arc3A inactivation.
Recovery of the indicated P. aeruginosa strains carrying empty pPSV39 (control) or pPSV39-aas grown in competition with an excess of B. thai with Isopropyl β-D-1-thiogalactopyranoside (IPTG) for induction of expression from the plasmid. Data represent mean and standard deviation from technical duplicates from one biological replicate representative of two conducted.
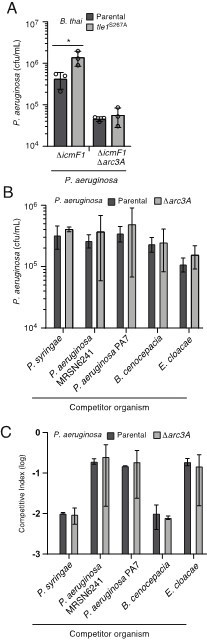
Inactivation of Arc3A does not detectably impact P. aeruginosa growth in competition with assorted phospholipase producing species.
(A) Recovery of P. aeruginosa with the indicated genotypes following growth in competition with an excess of wild-type or Tle1-inactivated B. thai. (B and C) Recovery of P. aeruginosa (B) or competitive index obtained (C; final donor:recipicient ratio divided by initial donor:recipienct ratio) following growth in competition with the indicated organisms.
Tables
Mass spectrometric analysis of P. aeruginosa Arc3A and Arc3B immunoprecipitation samples.
| Locus ID* | Peptide counts of sample (%)† | Fold change (VSV-G/Ctrl ‡) |
|---|---|---|
| Arc3B-VSV-G sample | ||
| PA5113 (Arc3B) | 5.86 | > 200 |
| PA4385 | 1.62 | 2.29 |
| PA5016 | 1.45 | 0.82 |
| PA0090 | 1.17 | 0.72 |
| PA3729 | 1.12 | 1.30 |
| PA4761 | 1.12 | 0.71 |
| PA5114 (Arc3A) | 1.12 | N.D. § |
| PA5239 | 1.06 | 1.10 |
| PA3861 | 1.00 | 1.10 |
| PA4269 | 1.00 | 1.10 |
| PA1092 | 0.95 | 0.78 |
| PA0141 | 0.89 | 0.71 |
| PA0963 | 0.84 | 1.27 |
| PA2976 | 0.84 | 0.83 |
| PA4260 | 0.84 | 0.69 |
| PA4265 | 0.84 | 0.61 |
| PA4270 | 0.84 | 1.04 |
| PA1803 | 0.78 | 0.74 |
| PA2151 | 0.78 | 0.77 |
| PA3950 | 0.78 | 1.19 |
| PA3001 | 0.73 | 1.10 |
| PA3656 | 0.73 | 0.80 |
| PA5173 | 0.73 | 0.76 |
| PA5554 | 0.73 | 1.20 |
| PA5556 | 0.73 | 2.05 |
| Arc3A-VSV-G sample | ||
| PA5554 | 2.92 | 4.81 |
| PA5113 (Arc3B) | 2.22 | > 200 |
| PA5114 (Arc3A) | 1.44 | N.D. |
| PA4751 | 1.30 | 12.87 |
| PA5556 | 1.26 | 3.55 |
| PA3729 | 1.10 | 1.28 |
| PA5016 | 1.10 | 0.62 |
| PA4429 | 0.99 | N.D. |
| PA4942 | 0.96 | 6.36 |
| PA2493 | 0.92 | 2.60 |
| PA3160 | 0.90 | N.D. |
| PA4246 | 0.87 | 0.79 |
| PA0090 | 0.85 | 0.53 |
| PA1552 | 0.83 | N.D. |
| PA0077 | 0.81 | 5.33 |
| PA2494 | 0.81 | ND |
| PA2976 | 0.72 | 0.71 |
| PA0659 | 0.70 | N.D. |
| PA4761 | 0.70 | 0.44 |
| PA4941 | 0.65 | 2.57 |
| PA4265 | 0.63 | 0.46 |
| PA3656 | 0.61 | 0.67 |
| PA3794 | 0.61 | N.D. |
| PA2151 | 0.56 | 0.55 |
| PA3821 | 0.56 | N.D. |
-
*
Proteins containing at least two peptides identified in a given immunoprecipitation sample are included. Arc3A and Arc3B are bolded.
-
†
Value corresponds to the abundance (peptide counts) of the protein within the total immunoprecipitation sample. Only the 25 most abundant proteins in each sample are shown.
-
‡
Immunoprecipitation from a P. aeruginosa strain lacking a VSV-G-tagged protein served as the control sample (Ctrl).
-
§
The protein was not detected in the control sample.
Internal standards and parameters used in lipidomic analysis.
| Internal standards used | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compound formula | Compound Name | Alternative name | ||||||
| C20H41O9P | LysoPG(14:0) | |||||||
| C24H49O9P | LysoPG(18:0) | |||||||
| C34H67O10P | PG(14:0/14:0) | |||||||
| C46H91O10P | PG(20:0/20:0), i.e. diphytanoyl PG | |||||||
| C19H40O7PN | LysoPE(14:0) | |||||||
| C23H48O7PN | LysoPE(18:0) | |||||||
| C29H58O8PN | PE(12:0/12:0) | |||||||
| C45H90O8PN | PE(20:0/20:0), i.e. diphytanoyl PE | |||||||
| C66H120O17P2 | CL(57:4) | 14:1(3)–15:1 CA | ||||||
| C70H134O17P2 | CL(61:1) | 15:0(3)–16:1 CA | ||||||
| C89H166O17P2 | CL(80:4) | 22:1(3)–14:1 CA | ||||||
| Mass spectrometry global parameters on Waters Xevo TQS mass spectrometer | ||||||||
| Parameter | Value | |||||||
| Cycle time | Automatic | |||||||
| Source temperature (°C) | 150 | |||||||
| Desolvation temperature (°C) | 250 | |||||||
| Cone gas flow (L/h) | 150 | |||||||
| Desolvation gas fow (L/h) | 650 | |||||||
| Collision gas flow (mL/min) | 0.14 | |||||||
| Nebuliser gas (Bar) | 7 | |||||||
| LM 1 Resolution | 2.8 | |||||||
| HM 1 Resolution | 14.8 | |||||||
| Mass spectral acquisition and data processing parameters | ||||||||
| Acquisition parameters | ||||||||
| Compound | Function | m/z range | Start/end time (min) | Scan duration (s) | Ionization mode | Cone voltage (V) | Collision energy (V) | Total scan time (min) |
| PG | Neutral Loss of 189.04 | 680–880 | 1.6–5 | 2 | ES+ | 30 | 8 | 3.4 |
| LPE, PE | Neutral Loss of 141.02 | 422–922 | 1.6–5 | 5 | ES+ | 40 | 20 | 3.4 |
| LPG | Parents of 152.9 | 420–570 | 5.2–10 | 0.75 | ES- | 30 | 38 | 4.8 |
| CL, MLCL | Parents of 153 | 865–1,572 | 1.6–30 | 4.7 | ES- | 40 | 75 | 28.4 |
| Data processing parameters | ||||||||
| Subtraction | Smooth mean | Centroid mid peak width at half height (all top 50%) | ||||||
| 1, 40, 0.01 | 2 × 0.4 | 2 | ||||||
| 1, 40, 0.01 | 2 × 0.4 | 2 | ||||||
| 1, 40, 0.01 | 1 × 0.6 | 5 | ||||||
| 1, 40, 0.01 | 2 × 0.6 | 5 | ||||||
Transposon sequencing data summary.
| Growth experiment | Mutant pool pre-competition | Pool with WT B. thai - rep 1 | Pool with δt6s B. thai - rep 1 |
|---|---|---|---|
| fastq file name | tn_mutant_pool.fastq | pool_v_wt_rep1.fastq | pool_v_deltaT6_rep1.fastq |
| Total reads | 515,434 | 5,788,728 | 6,452,899 |
| Trimmed (valid Tn prefix) | 515,293 | 5,786,427 | 6,450,594 |
| Mapped | 350,923 | 4,666,597 | 5,283,645 |
| Mapped to TA sites | 349,915 | 4,634,942 | 5,241,672 |
| TA_sites | 94,404 | 94,404 | 94,404 |
| TAs_hit | 50,348 | 57,644 | 60,331 |
| Max reads per TA site | 324 | 9,519 | 5,886 |
| Mean reads per hit TA site | 6.9 | 80.4 | 86.9 |
| Pool with WT B.thai - rep 2 | Pool with ΔT6S B.thai - rep 2 | Pool with tle3S264A B. thai | Pool with ΔcolA B. thai |
| pool_v_wt_rep2.fastq | pool_v_deltaT6_rep2.fastq | pool_v_tle3.fastq | pool_v_colA.fastq |
| 411,515 | 720,133 | 3,226,978 | 3,130,545 |
| 411,401 | 719,895 | 3,225,816 | 3,129,574 |
| 266,466 | 481,466 | 3,024,047 | 2,938,318 |
| 265,661 | 479,769 | 2,997,466 | 2,922,995 |
| 94,404 | 94,404 | 94,404 | 94,404 |
| 46,500 | 53,148 | 59,145 | 58,101 |
| 669 | 447 | 3,484 | 3,568 |
| 5.7 | 9.0 | 50.7 | 50.3 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Pseudomonas aeruginosa) | PAO1 | Stover et al., 2000 | ||
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa2541 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa2541∆pa2536 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa4323 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa4323∆pa4320 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114∆pa5113 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa2541, Tn7::AraE::pa2541 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa4323, Tn7::AraE::pa4323 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114, Tn7::AraE::pa5114 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa2541∆pa4323∆pa5114 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆gacS (∆pa0928) | LeRoux et al., 2015a | ||
| Strain, strain background (P. aeruginosa) | PAO1 ∆icmF1 (∆pa0077) | Silverman et al., 2011 | ||
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5112 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114,Tn7::AraE::empty | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114,Tn7::AraE:: pfl_rs01995 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114,Tn7::AraE:: i35_rs27920 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114, Tn7::AraE::pspto_rs26695 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114, Tn7::AraE::ta05_rs00690 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa3267 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆pa5114∆pa3267 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆retS (∆pa4856) | Mougous et al., 2006 | ||
| Strain, strain background (P. aeruginosa) | PAO1 ∆retS ∆pa3267 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆retS ∆pa5114 | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆rpoS (∆pa3622) | Jørgensen et al., 1999 | ||
| Strain, strain background (P. aeruginosa) | PAO1 ∆vacJ (∆pa2800) | Munguia et al., 2017 | ||
| Strain, strain background (P. aeruginosa) | PAO1 pa2541_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆retS, pa2541_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆gacS, pa2541_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 pa4323_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆retS, pa4323_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆gacS, pa4323_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 pa5114_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆retS, pa5114_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆gacS, pa5114_vsv-g | This study | See Materials and methods | |
| Strain, strain background (P. aeruginosa) | PAO1 ∆retS, pa5113_vsv-g | This study | See Materials and methods | |
| Strain, strain background (Burkholderia thailandensis) | E264 (ATCC 700388) | Yu et al., 2006 | ||
| Strain, strain background (B. thailandensis) | E264 ∆icmF1 (∆BTH_I2954) | LeRoux et al., 2015a | ||
| Strain, strain background (B. thailandensis) | E264 tle3S264A(BTH_II0090S264A + BTH_I3226S264A) | This study | See Materials and methods | |
| Strain, strain background (B. thailandensis) | E264 tle1S267A(BTH_I2698S267A + BTH_I2701S267A) | This study | See Materials and methods | |
| Strain, strain background (B. thailandensis) | E264 ∆colA (∆BTH_I2691) | This study | See Materials and methods | |
| Strain, strain background (B. thailandensis) | E264 ∆tae2 (∆BTH_I0068) | Russell et al., 2012 | ||
| Strain, strain background (B. thailandensis) | E264 Tn7::Tp-PS12-mCherry | LeRoux et al., 2012 | ||
| Strain, strain background (B. thailandensis) | E264 tle3 sensitized strain (∆BTH_I3225-8, ∆BTH_II0089-94) | This study | See Materials and methods | |
| Strain, strain background (B. thailandensis) | E264 Tn7::Tp-PS12-mCherry tle3 sensitized strain (∆BTH_I3225-8, ∆BTH_II0089-94) | This study | See Materials and methods | |
| Strain, strain background (Pseudomonas protegens) | pf-5 (ATCC BAA-477) | Paulsen et al., 2005 | ||
| Strain, strain background (P. protegens) | Pseudomonas protegens pf-5 ∆pfl_rs01995 | This study | See Materials and methods | |
| Strain, strain background (Escherichia coli) | DH5α | Thermo Fisher ScientificCat# 18258012 | ||
| Strain, strain background (E. coli) | SM10 | Biomedal Lifescience Cat# BS-3303 | ||
| Strain, strain background (E. coli) | UE54MG1655 lpp-2Δara714 rcsF::mini-Tn10 cam pgsA::FRT-kan-FRT | Harvat et al., 2005 | ||
| Recombinant DNA reagent | pEXG2-∆pa2541 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa2536 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa4323 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa4320 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa5114 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa5114/pa5113 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa3267 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa5113 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pa5112 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-∆pfl_rs01995 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-pa2541_vsv-g (plasmid) | This study | Construct to introduce VsvG tag, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-pa4323_vsv-g (plasmid) | This study | Construct to introduce VsvG tag, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-pa5114_vsv-g (plasmid) | This study | Construct to introduce VsvG tag, see Materials and methods | |
| Recombinant DNA reagent | pEXG2-pa5113_vsv-g (plasmid) | This study | Construct to introduce VsvG tag, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE (plasmid) | Hoang et al., 2000 | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE-pa2541 (plasmid) | This study | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE-pa4323 (plasmid) | This study | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE-pa5114 (plasmid) | This study | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE-pfl_rs01995 (plasmid) | This study | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE-i35_rs27920 (plasmid) | This study | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE-pspto_rs26695 (plasmid) | This study | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pUC18-Tn7t-pBAD-araE-ta05_rs00690 (plasmid) | This study | Arabinose-inducible expression system, see Materials and methods | |
| Recombinant DNA reagent | pTNS3 (plasmid) | Choi et al., 2008 | ||
| Recombinant DNA reagent | pJRC115-BTH_II0090S264A (plasmid) | This study | Point mutation construct, see Materials and methods | |
| Recombinant DNA reagent | pJRC115-BTH_I3226S264A (plasmid) | This study | Point mutation construct, see Materials and methods | |
| Recombinant DNA reagent | pJRC115-BTH_I2698S267A (plasmid) | This study | Point mutation construct, see Materials and methods | |
| Recombinant DNA reagent | pJRC115-BTH_I2701S267A (plasmid) | This study | Point mutation construct, see Materials and methods | |
| Recombinant DNA reagent | pJRC115-∆BTH_I2691 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pJRC115-∆BTH_I3225-8 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pJRC115-∆BTH_II0089-94 (plasmid) | This study | Deletion construct, see Materials and methods | |
| Recombinant DNA reagent | pUC18T-miniTn7T-Tp-PS12-mCherry (plasmid) | LeRoux et al., 2012 | Construct for mCherry expression | |
| Software, algorithm | Seqmagick | Matsen Group, 2020, https://github.com/fhcrc/seqmagick | ||
| Software, algorithm | TRANSIT TPP | https://transit.readthedocs.io/en/latest/tpp.html | ||
| Antibody | Anti-VSV-G (rabbit polyclonal) | MilliporeSigma | Cat# V4888-200UG | Western blot (1:5000 dilution) |
| Antibody | Anti-rabbit IgG HRP conjugated (goat) | MilliporeSigma | Cat# A6154-1ML | Western blot (1:5000 dilution) |
| Antibody | Anti-ribosome polymerase β (mouse monoclonal) | BioLegend | Cat# 663903; RRID: AB_2564524 | Western blot (1:1000 dilution) |
| Antibody | Anti-mouse IgG HRP conjugated (sheep) | MilliporeSigma | Cat# AC111P | Western blot (1:4000 dilution) |
Oligonucleotides used in this study.
| Oligonucleotides 5′–3′ | Source |
|---|---|
| pEXG2_∆PA2541_F1CAAGCTTCTGCAGGTCGACTCTAGAAGGTGGAACCGGACCTGAAG | Integrated DNA Technology |
| pEXG2_∆PA2541_R1CTCAGGCCTGGGAAATCATGTCAGCCAGTCC | Integrated DNA Technology |
| pEXG2_∆PA2541_F2CATGATTTCCCAGGCCTGAGAGGGAACG | Integrated DNA Technology |
| pEXG2_∆PA2541_R2TAAGGTACCGAATTCGAGCTCGTATGACCCAGGCGGTCG | Integrated DNA Technology |
| pEXG2_∆PA4323_F1AGCTCGAGCCCGGGGATCCTCTAGAGACGCAGAACCCGATCGAG | Integrated DNA Technology |
| pEXG2_∆PA4323_R1AAAGCACGCCCGATGGCTTCATACGCGG | Integrated DNA Technology |
| pEXG2_∆PA4323_F2GAAGCCATCGGGCGTGCTTTGACGGATC | Integrated DNA Technology |
| pEXG2_∆PA4323_R2GGAAGCATAAATGTAAAGCAAGCTTGAGCGATGCCGAGCTGGA | Integrated DNA Technology |
| pEXG2_∆PA5114_F1CAAGCTTCTGCAGGTCGACTCTAGAGCCGCATGCCCTTGACCT | Integrated DNA Technology |
| pEXG2_∆PA5114_R1CGACCTCGGCAATCCATTGCATGCGTCGAATC | Integrated DNA Technology |
| pEXG2_∆PA5114_F2GCAATGGATTGCCGAGGTCGCATCGGAG | Integrated DNA Technology |
| pEXG2_∆PA5114_R2AATTCGAGCTCACCAGATCATCGGCGCCG | Integrated DNA Technology |
| pEXG2_∆PA2536_F1CAAGCTTCTGCAGGTCGACTCTAGACTCGGCCTGGCCCGAGC | Integrated DNA Technology |
| pEXG2_∆PA2536_R1CAGGCGAACCTCAGGGCGTTTCGTTCATCTCAGGCCTCC | Integrated DNA Technology |
| pEXG2_∆PA2536_F2GGAGGCCTGAGATGAACGAAACGCCCTGAGGTTCGCCTG | Integrated DNA Technology |
| pEXG2_∆PA2536_R2TAAGGTACCGAATTCGAGCTCCACATCGGTCTCAGCGAAGC | Integrated DNA Technology |
| pEXG2_∆PA4320_F1AGCTCGAGCCCGGGGATCCTCTAGATGTTCGGCTTCTACATCATGAAC | Integrated DNA Technology |
| pEXG2_∆PA4320_R1TGCGCCAGCCCACGCTGGCGTCAGTCAG | Integrated DNA Technology |
| pEXG2_∆PA4320_F2CGCCAGCGTGGGCTGGCGCAGCCTTTTC | Integrated DNA Technology |
| pEXG2_∆PA4320_R2GGAAGCATAAATGTAAAGCAAGCTTGTCGGCGGTTTCCTCGCT | Integrated DNA Technology |
| pEXG2_∆PA5114/PA5113_F1CAAGCTTCTGCAGGTCGACTCTAGAGCCGCATGCCCTTGACCT | Integrated DNA Technology |
| pEXG2_∆PA5114/PA5113_R1TGTTCGGCGAAATCCATTGCATGCGTCGAATC | Integrated DNA Technology |
| pEXG2_∆PA5114/PA5113_F2GCAATGGATTTCGCCGAACAAGCCATGAG | Integrated DNA Technology |
| pEXG2_∆PA5114/PA5113_R2TAAGGTACCGAATTCGAGCTCAACAGCCAGACCACGATGTAGC | Integrated DNA Technology |
| pEXG2_∆PA5113_F1CAAGCTTCTGCAGGTCGACTCTAGATTGCTAGGGGTGCTGGCG | Integrated DNA Technology |
| pEXG2_∆PA5113_R1ATGGCTTGTTGGAGCGCGTCATGGCTGC | Integrated DNA Technology |
| pEXG2_∆PA5113_F2GACGCGCTCCAACAAGCCATGAGCCGGTTC | Integrated DNA Technology |
| pEXG2_∆PA5113_R2TAAGGTACCGAATTCGAGCTCAACAGCCAGACCACGATGTAG | Integrated DNA Technology |
| pEXG2_∆PA5112_F1CAAGCTTCTGCAGGTCGACTCTAGATACCGCTGGCAGTTGCCG | Integrated DNA Technology |
| pEXG2_∆PA5112_R1AGAAGTCCAGCGCCATTCTGATCATTCTCTTACTC | Integrated DNA Technology |
| pEXG2_∆PA5112_F2CAGAATGGCGCTGGACTTCTGAAACGGCGGC | Integrated DNA Technology |
| pEXG2_∆PA5112_R2TAAGGTACCGAATTCGAGCTCCGCGCAACCGCCGGTTGG | Integrated DNA Technology |
| pEXG2_∆PA3267_F1CAAGCTTCTGCAGGTCGACTCTAGATCTACATCGACTTCGACG | Integrated DNA Technology |
| pEXG2_∆PA3267_R1ATCAGCGAGCTTGGGTCATCGTCCTTGTTAC | Integrated DNA Technology |
| pEXG2_∆PA3267_F2GATGACCCAAGCTCGCTGATCGATCCGC | Integrated DNA Technology |
| pEXG2_∆PA3267_R2TAAGGTACCGAATTCGAGCTCTTCGCCGGCCTGTTCGAAG | Integrated DNA Technology |
| pEXG2_∆PFL_RS01995_F1CAAGCTTCTGCAGGTCGACTCTAGAAACTACAACGTCAGCCTG | Integrated DNA Technology |
| pEXG2_∆PFL_RS01995_R1TTCATTCAACGGTTTGCCTATCCATTGCATGTGTCG | Integrated DNA Technology |
| pEXG2_∆PFL_RS01995_F2CGACACATGCAATGGATAGGCAAACCGTTGAATGAA | Integrated DNA Technology |
| pEXG2_∆PFL_RS01995_R2TAAGGTACCGAATTCGAGCTCCTGCGACCACACCAGCG | Integrated DNA Technology |
| pEXG2_PA2541_VSVG_F1CAAGCTTCTGCAGGTCGACTCTAGAGCGCTGATACGGACCATGC | Integrated DNA Technology |
| pEXG2_PA2541_VSVG_R1TTTTCCTAATCTATTCATTTCAATATCTGTATAGGCCTGGCCGTCGCTGCCCCG | Integrated DNA Technology |
| pEXG2_PA2541_VSVG_F2TATACAGATATTGAAATGAATAGATTAGGAAAATGAGAGGGAACGGGCGAAC | Integrated DNA Technology |
| pEXG2_PA2541_VSVG_R2TAAGGTACCGAATTCGAGCTCCTCCTGCGGCGCACGATG | Integrated DNA Technology |
| pEXG2_PA4323_VSVG_F1CAAGCTTCTGCAGGTCGACTCTAGATGGAGTTCCACCAGTTGCGCG | Integrated DNA Technology |
| pEXG2_PA4323_VSVG_R1TGATCCGTCATTTTCCTAATCTATTCATTTCAATATCTGTATAAAGCACGCCGGCGCGCT | Integrated DNA Technology |
| pEXG2_PA4323_VSVG_F2TATACAGATATTGAAATGAATAGATTAGGAAAATGACGGATCAGACCGACTG | Integrated DNA Technology |
| pEXG2_PA4323_VSVG_R2TAAGGTACCGAATTCGAGCTCAAGGTACACTTCTCCGCC | Integrated DNA Technology |
| pEXG2_PA5114_VSVG_F1CAAGCTTCTGCAGGTCGACTCTAGACTGGAGCTGCGCTACCTGTTCG | Integrated DNA Technology |
| pEXG2_PA5114_VSVG_R1TAATCTATTCATTTCAATATCTGTATATGGCTGCTCGGCCTCCGA | Integrated DNA Technology |
| pEXG2_PA5114_VSVG_F2GATATTGAAATGAATAGATTAGGAAAATCGGAGGCCGAGCAGCCA | Integrated DNA Technology |
| pEXG2_PA5114_VSVG_R2TAAGGTACCGAATTCGAGCTCGCTCGGACCAGATCATCGGC | Integrated DNA Technology |
| pEXG2_PA5113_VSVG_F1CAAGCTTCTGCAGGTCGACTCTAGACTGCGTCTGGCCTGGCCG | Integrated DNA Technology |
| pEXG2_PA5113_VSVG_R1TTTTCCTAATCTATTCATTTCAATATCTGTATATGGCTTGTTCGGCGAGGAAC | Integrated DNA Technology |
| pEXG2_PA5113_VSVG_F2TATACAGATATTGAAATGAATAGATTAGGAAAATGAGCCGGTTCCGCGCTATG | Integrated DNA Technology |
| pEXG2_PA5113_VSVG_R1TAAGGTACCGAATTCGAGCTCGTCGGGCAACAGCCAGAC | Integrated DNA Technology |
| pUC18_PA2541_FAGC GAATTCGAGCTCGGTACCACGGGAGGAAAG ATGATTTCCGTCTATCAACTC | Integrated DNA Technology |
| pUC18_PA2541_RCTCATCCGCCAAAACAGCCAAGCTTTCAGGCCTGGCCGTCGC | Integrated DNA Technology |
| pUC18_PA4323_FAGCGAATTCGAGCTCGGTACCACGGGAGGAAAGATGAAGCCATCGCGCGCCCTGCTGG | Integrated DNA Technology |
| pUC18_PA4323_RCTCATCCGCCAAAACAGCCAAGCTTTCAAAGCACGCCGGCGCGCTTCCAG | Integrated DNA Technology |
| pUC18_PA5114_FAGCGAATTCGAGCTCGGTACCACGGGAGGAAAGATGCAATGGATTTTCATGCTGG | Integrated DNA Technology |
| pUC18_PA5114_RCTCATCCGCCAAAACAGCCAAGCTTTCATGGCTGCTCGGCCTC | Integrated DNA Technology |
| pUC18_pfl_rs01995_FAGCGAATTCGAGCTCGGTACCACGGGAGGAAAGATGCAATGGATATTCATGCTGC | Integrated DNA Technology |
| pUC18_pfl_rs01995_RCTCATCCGCCAAAACAGCCAAGCTTTCACGATGACACTCCTTCATTC | Integrated DNA Technology |
| pUC18_i35_rs27920_FAGCGAATTCGAGCTCGGTACCACGGGAGGAAAGATGAACTGGGCATTCGCCG | Integrated DNA Technology |
| pUC18_i35_rs27920_RCTCATCCGCCAAAACAGCCAAGCTTTCATGGCTGGCCGTCCTG | Integrated DNA Technology |
| pUC18_pspto_rs26695_FAGCGAATTCGAGCTCGGTACCACGGGAGGAAAGATGCTTTGGATTTGTCTGGTAG | Integrated DNA Technology |
| pUC18_pspto_rs26695_RCTCATCCGCCAAAACAGCCAAGCTTTCATGGCGTTTCAGGCGCTG | Integrated DNA Technology |
| pUC18_ta05_rs00690_FAGCGAATTCGAGCTCGGTACCACGGGAGGAAAGATGGACGACCTTTTAATCCTG | Integrated DNA Technology |
| pUC18_ta05_rs00690_RCTCATCCGCCAAAACAGCCAAGCTTTCATTTGTTTTCTCCAGCTTTG | Integrated DNA Technology |
| pJRC115_ BTH_II0090S264A_F1TAAAACGACGGCCAGTGCCAAGCTTCGACGGATCAGCGTTTCAAGCTGC | Integrated DNA Technology |
| pJRC115_ BTH_II0090S264A_R1ACCTTGGGCATGACCCATCACCGTGATCGTTTCGTG | Integrated DNA Technology |
| pJRC115_ BTH_II0090S264A_F2GGTCATGCCCAAGGTACGATCATCACGCTGCTCG | Integrated DNA Technology |
| pJRC115_ BTH_II0090S264A_R2GCTCGGTACCCGGGGATCCTCTAGACGGCTCGGCACGATGCGC | Integrated DNA Technology |
| pJRC115_ BTH_I3226S264A_F1TAAAACGACGGCCAGTGCCAAGCTTCGACGGATCAGCGTTTCAAGCTGC | Integrated DNA Technology |
| pJRC115_ BTH_I3226S264A_R1ACCTTGGGCATGACCCATCACCGTGATCGTTTCGTG | Integrated DNA Technology |
| pJRC115_ BTH_I3226S264A_F2GGTCATGCCCAAGGTACGATCATCACGCTGCTCG | Integrated DNA Technology |
| pJRC115_ BTH_I3226S264A_R2GCTCGGTACCCGGGGATCCTCTAGACGGCTCGGCACGATGCGC | Integrated DNA Technology |
| pJRC115_ BTH_I2698S267A_F1TCAATCAGTATCTAGAGGGACACCTTTCTCAAGCGAAATC | Integrated DNA Technology |
| pJRC115_ BTH_I2698S267A_R1GCCGCGCGCGAAACCAAACACATAAAGGCGAATGCG | Integrated DNA Technology |
| pJRC115_ BTH_I2698S267A_F2GGTTTCGCGCGCGGCGCAGCGGAAGCTCGCACGTTTTC | Integrated DNA Technology |
| pJRC115_ BTH_I2698S267A_R2TGTTAAGCTAGAATTCCATCAAACCCGCTGTCCCATGCTC | Integrated DNA Technology |
| pJRC115_ BTH_I2701S267A_F1TAAAACGACGGCCAGTGCCAAGCTTGAAAGACGAGCAGGACGCG | Integrated DNA Technology |
| pJRC115_ BTH_I2701S267A_R1ACCCCGCGCGAAACCGAATACGTATAGCCGAATGCG | Integrated DNA Technology |
| pJRC115_ BTH_I2701S267A_F2GGTTTCGCGCGGGGTGCAGCGGAGGCTCGCAC | Integrated DNA Technology |
| pJRC115_ BTH_I2701S267A_R2GCTCGGTACCCGGGGATCCTCTAGACTCGACCTCCAGCAGATCG | Integrated DNA Technology |
| pJRC115_ ∆BTH_I2691_F1TAAAACGACGGCCAGTGCCAAGCTTATTTCAAGCGCGGCCAGTC | Integrated DNA Technology |
| pJRC115_ ∆BTH_I2691_R1ATCTATGCGAGCTTCCCGCCATTTTTATTCC | Integrated DNA Technology |
| pJRC115_ ∆BTH_I2691_F2GGCGGGAAGCTCGCATAGATGAGTGATG | Integrated DNA Technology |
| pJRC115_ ∆BTH_I2691_R2GCTCGGTACCCGGGGATCCTCTAGATTCTGTCAATACTTAAAATACAATTTTC | Integrated DNA Technology |
| pJRC115_ ∆BTH_II0089-94_F1TAAAACGACGGCCAGTGCCAAGCTTGAATCAGTGCATCGCTGTAC | Integrated DNA Technology |
| pJRC115_ ∆BTH_II0089-94_R1ATGTTTTGCTTAGCGTATCGTTCAAATTGG | Integrated DNA Technology |
| pJRC115_ ∆BTH_II0089-94_F2CGATACGCTAAGCAAAACATGAGATTATTGAAGAG | Integrated DNA Technology |
| pJRC115_ ∆BTH_II0089-94_R2GCTCGGTACCCGGGGATCCTCTAGATATGCAACGCATTGCCGAAAC | Integrated DNA Technology |
| pJRC115_ ∆BTH_I3225-8_F1TAAAACGACGGCCAGTGCCAAGCTTCCGGTCAATATACCACCATC | Integrated DNA Technology |
| pJRC115_ ∆BTH_I3225-8_R1ATGTGCGCCCCGTATCGTTCAAATTGGTCAC | Integrated DNA Technology |
| pJRC115_ ∆BTH_I3225-8_F2GAACGATACGGGGCGCACATAATAAGACTTG | Integrated DNA Technology |
| pJRC115_ ∆BTH_I3225-8_R2GCTCGGTACCCGGGGATCCTCTAGAATTTGCTGTTTCTGCTGATG | Integrated DNA Technology |
| PCR_1AGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGGGGGGGGGGGGGGG | Integrated DNA Technology |
| PCR_1BTCATCGGCTCGTATAATGTGTGG | Integrated DNA Technology |
| PCR_2ACAAGCAGAAGACGGCATACGAGATTCGCCTTAGTCTCGTGGGCTCGG | Integrated DNA Technology |
| PCR_2BCAAGCAGAAGACGGCATACGAGATCTAGTACGGTCTCGTGGGCTCGG | Integrated DNA Technology |
| PCR_2CAATGATACGGCGACCACCGAGATCTACACCTAGAGACCGGGGACTTATCAGCCAACCTGTTA | Integrated DNA Technology |
| Seq_primerCTAGAGACCGGGGACTTATCAGCCAAC | Integrated DNA Technology |





