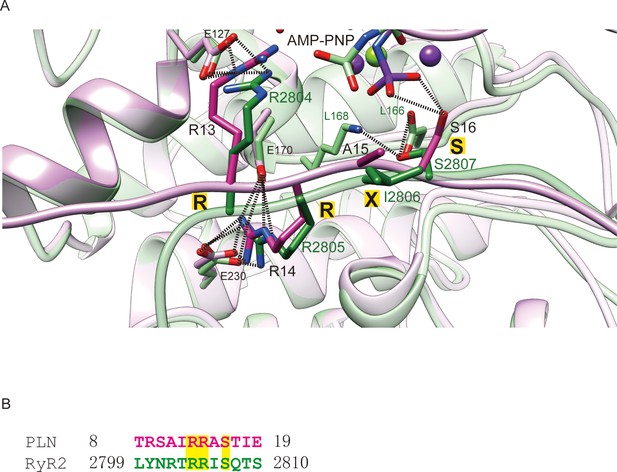
Structures of PKA–phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy
Figures
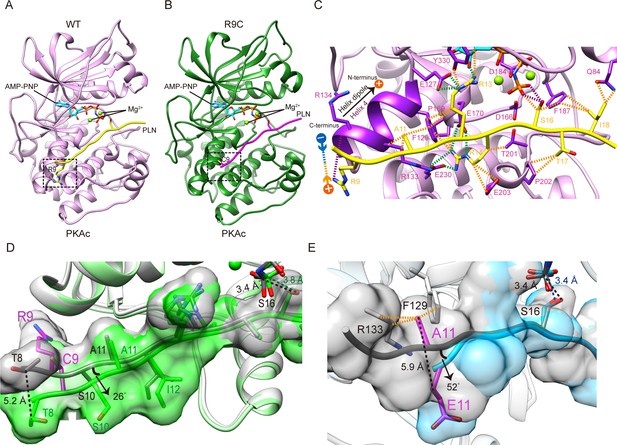
Crystal structures of PKAc-WT/R9C phospholamban (PLN) complex.
(A) Crystal structures of the ternary complex of PKAc, WT PLN, and AMP-PNP. Protein kinase A (PKA) is colored in pink, PLN in yellow, and AMP-PNP in cyan. (B) Crystal structures of the ternary complex of PKAc, R9C PLN, and AMP-PNP. PKA is colored in green, PLN in violet red, and AMP-PNP in cyan. (C) The interaction between PKAc and WT PLN. The van der Waals contacts (orange), the salt bridges (green), and the hydrogen bonds (purple) are indicated by the dash lines. (D) The superposition of PKAc-WT PLN (white-gray) with PKAc-R9C PLN (light green-green). R9C abolishes the electrostatic interaction between Arg9 and the helix dipole of helix 4, inducing conformational changes at the N-terminal region (NTR). (E) The superposition of PKAc:WT PLN (white-gray) with PKAc:PLN A11E (light blue-cyan). A11E forces the NTR to move away from PKAc without affecting the structure at the catalytic center.
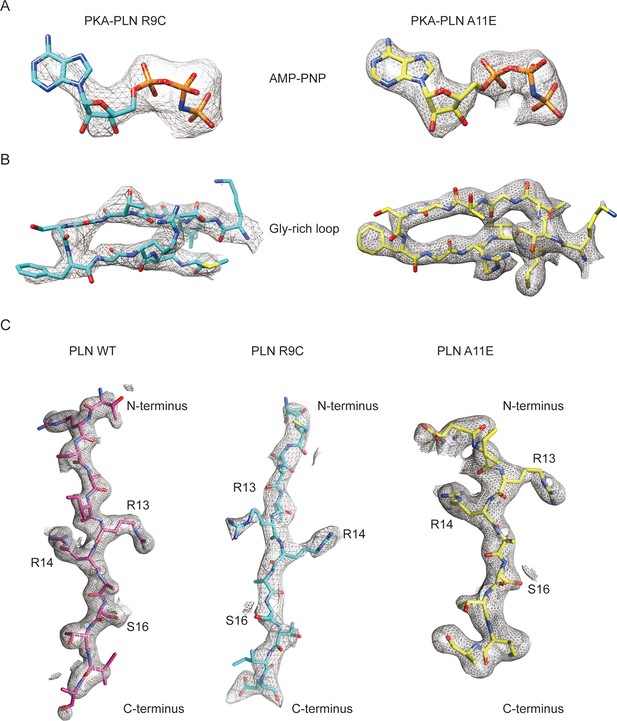
Electron density maps.
Density maps for the AMP-PNP region (A), PKAc Gly-rich loop region (B), and phospholamban (PLN) peptides (C) from the crystal structures of PKAc–PLNs.
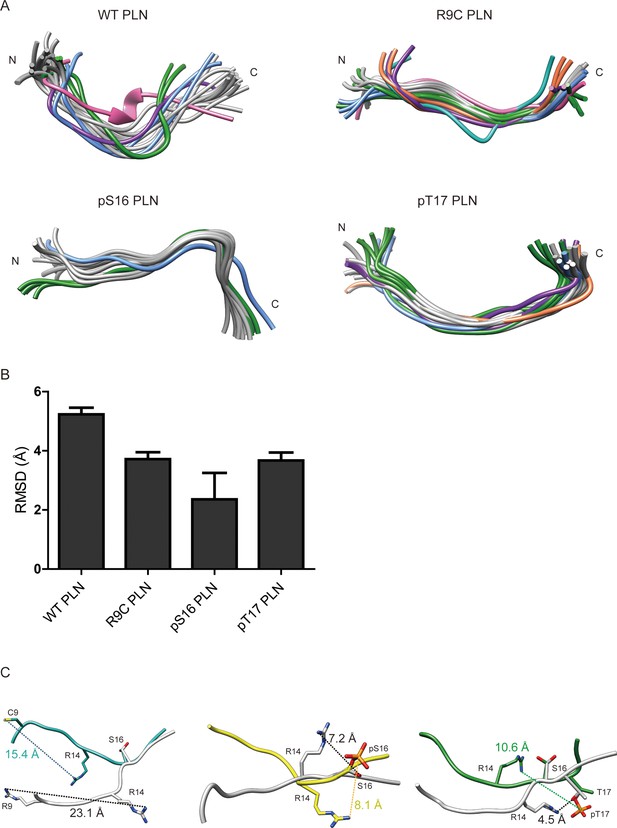
Nuclear magnetic resonance (NMR) structures of phospholamban (PLN)/ALN peptides in the absence of PKAc.
(A) Ensemble cluster views of the superposition of the top 20 lowest energy structures of WT PLN, R9C PLN, pS16 PLN, and pT17 PLN. The top 7 ranked clusters are colored in gray, forest green, cornflower blue, purple, sandy brown, light sea green, and hot pink, respectively. (B) The root mean square deviation (RMSD) between the representative structures from each ensemble cluster. WT PLN clearly shows a higher RMSD than the other four peptides, suggesting its higher flexibility. (C) A comparison of the top structures of WT PLN (white) with the ones from R9C PLN (light sea green), pS16 PLN (yellow), and pT17 PLN PLN (forest green). S16 is used for the superposition.

Sequence alignment between phospholamban (PLN) and ALN.
Identically conserved residues are colored in green, and weakly similar residues are coloured in grey. The protein kinase A (PKA)/calmodulin-dependent protein kinase II (CaMKII) phosphorylation site is colored in yellow and orange, respectively.
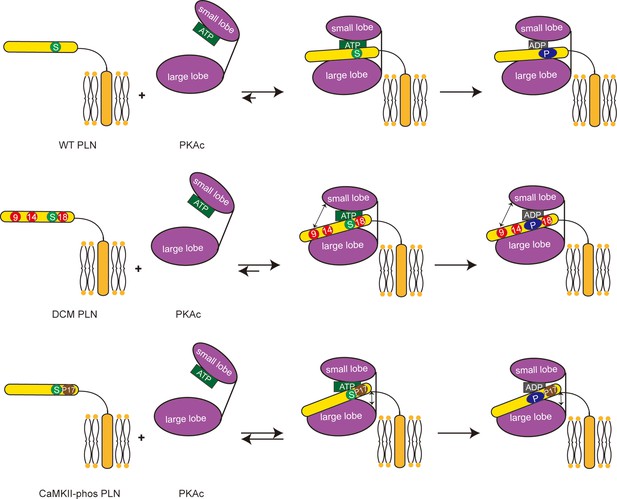
Schematic representation showing the impacts of dilated cardiomyopathy (DCM) mutations and calmodulin-dependent protein kinase II (CaMKII) phosphorylation on the phosphorylation of phospholamban (PLN) by protein kinase A (PKA).
Both DCM mutations at the positions 9, 14, and 18 (red circles) and the CaMKII phosphorylation (brown circle) reduce the interaction between PLN (yellow) and PKAc (purple), and subsequently decrease the phosphorylation level of PLN.
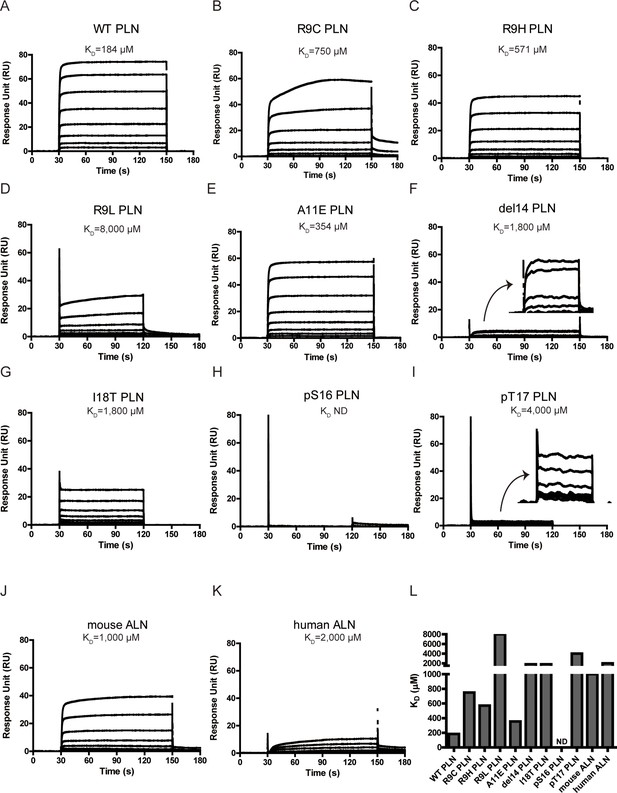
Surface plasmon resonance (SPR) analysis of PKAc–phospholamban (PLN)/ALN interactions.
(A–K) SPR sensorgrams of the binding of the PLN/ALN peptides onto immobilized PKAc. The calculated KD values are displayed above the corresponding sensorgrams. (L) The relative KD values of PKAc with different peptide substrates measured by SPR assay.
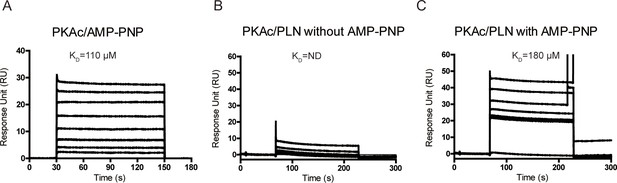
The surface plasmon resonance (SPR) analysis of PKAc/AMP-PNP interactions.
(A) SPR sensorgrams for the binding of AMP-PNP with immobilized PKAc. SPR sensorgrams for the binding of WT phospholamban (PLN) with immobilized PKAc in the absence (B) or presence (C) of AMP-PNP.
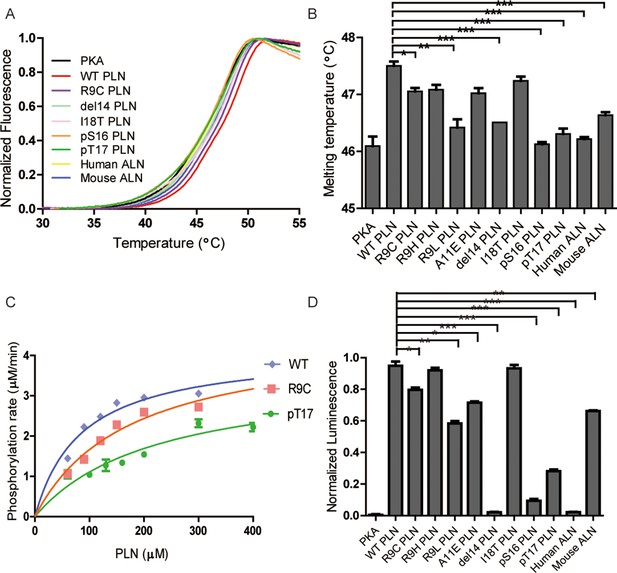
Thermal melt analysis of PKAc–phospholamban (PLN)/ALN complexes and the activities of PKAc measured by ADP-Glo assay.
(A) The averaged thermal melt curves from four replicates each of PKAc complexed with different peptides. (B) A bar graph comparing the melting temperatures of PKAc complexed with different peptides. Error bars show the standard deviation. ***p < 0.0001, **p < 0.001, *p < 0.01 (one-tail Student’s t-test). (C) A plot of vi vs. [PLN peptide] for PKAc-catalyzed phosphorylation of WT-, R9C-, and pT17-PLN substrates. The data were fit to the Michaelis–Menten equation. (D) The relative activities of PKAc with different peptide substrates measured by ADP-Glo assay.
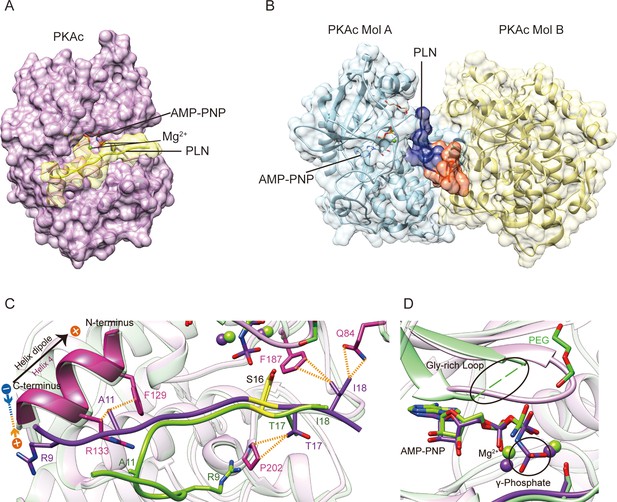
Crystal structures of PKAc–phospholamban (PLN) complex.
Crystal structures of the ternary complex of PKAc, PLN, and AMP-PNP from the current study (A) and a previous study (PDB ID 3O7L)58 (B). PLN interacts with a single PKAc in our structure and with two PKAc molecules in 3O7L. The N-terminal region (NTR) of PLN interacts extensively with the second PKAc (Mol B) in 3O7L. (C, D) Superposition of our structure (purple) and 3O7L (green) shows that PLNs adopt different conformation at both ends. The γ-phosphate group in AMP-PNP and Gly-rich Loop is missing from 3O7L. The electrostatic interaction between Arg9 and the helix dipole is indicated. The PKA phosphorylation sites are highlighted in yellow.
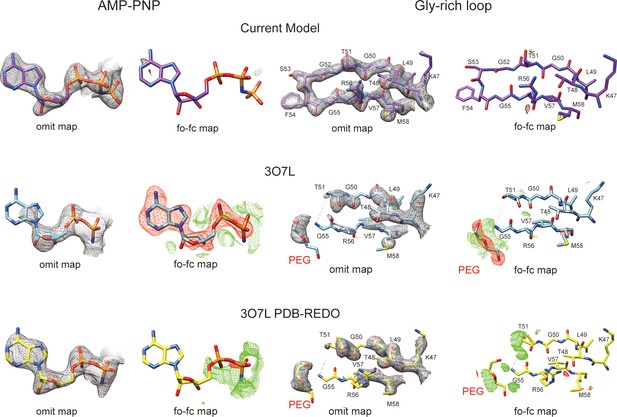
Comparison of two structures of PKAc–phospholamban (PLN) complex.
Comparison of the electron density maps for AMP-PNP region and PKA Gly-rich loop region from our structure (violet red), 3O7L (blue), and 3O7L re-refined by PDB-REDO (yellow) (https://pdb-redo.eu). The omit maps are displayed at contour level of 2σ, and the mFo-DFc difference maps are displayed at contour levels of 3σ.
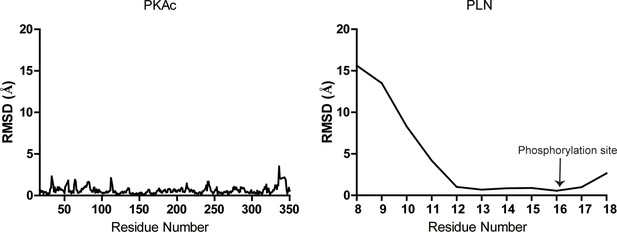
Root mean square deviation (RMSD) plots.
Plots showing per residue RMSD for the PKAc (left) and phospholamban (PLN; right) from our structure relative to 3O7L.
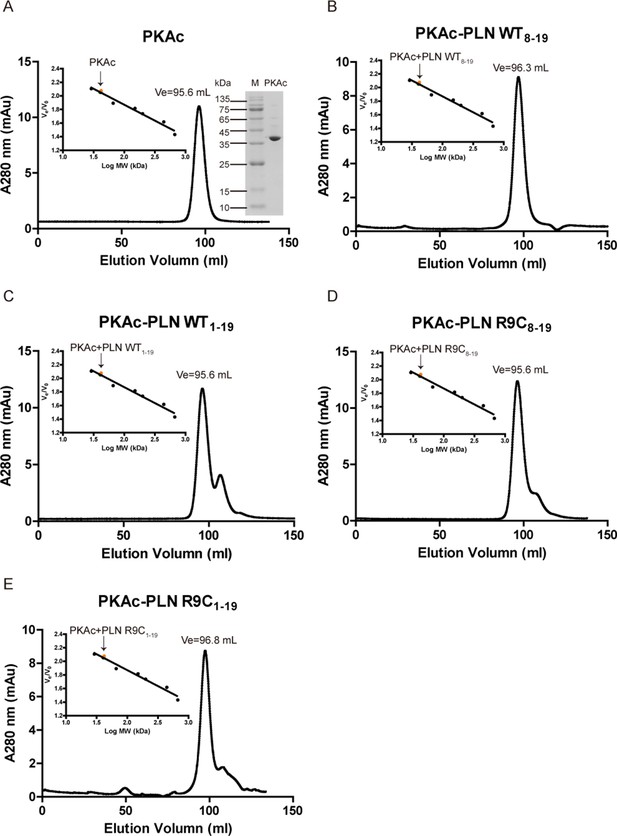
Oligomerization states of PKAc.
Elution profiles of PKAc (A) or PKAc–phospholamban (PLN) complexes (B–E) filtration chromatography using a Superdex 200 16/600 column (GE Healthcare, USA). The right inset in A is a 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel of purified PKAc. The left inset in A–E shows the plotted standard curve for this column. The molecular weight (MW) estimated from their elution volumes are ~40 kDa, suggesting monomeric forms in solution.
Tables
Data collection and refinement statistics for the PKAc–phospholamban (PLN) crystals.
| Crystal | PKAc-WT PLN | PKAc-A11E PLN | PKAc-R9C PLN |
|---|---|---|---|
| λ for data collection (Å) | 0.9795 | 0.9795 | 0.9795 |
| Data collection | |||
| Space group | P 2 21 21 | P 1 21 1 | C 2 2 21 |
| Cell dimension (Å) | |||
| a, b, c (Å) | 52.57, 70.49, 99.03 | 49.56, 69.37, 56.24 | 50.91, 104.88, 168.10 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 101.97, 90.00 | 90.00, 90.00, 90.00 |
| Resolution | 42.14–2.16 | 33.12–2.80 | 44.19–3.43 |
| Rmerge | 0.194 (0.907) | 0.139 (0.527) | 0.165 (0.738) |
| Average I/σ(I) | 9.9 (1.8) | 8.5 (1.8) | 10.3 (2.5) |
| Completeness (%) | 91.42 (68.22) | 99.11 (94.08) | 96.35 (95.02) |
| Redundancy | 6.9 (6.1) | 3.3 (3.1) | 5.4 (5.7) |
| Refinement | |||
| Resolution (Å) | 42.14–2.16 | 33.12–2.80 | 44.19–3.43 |
| Highest resolution shells (Å) | 2.24 (2.16) | 2.78 (2.83) | 3.56 (3.43) |
| No. of reflections | 18,585 | 9242 | 6088 |
| Average B-factor | 30.88 | 37.93 | 95.81 |
| Protein | 30.52 | 37.97 | 95.92 |
| Ligands | 24.40 | 34.94 | 86.85 |
| Water | 36.01 | 37.30 | – |
| Rwork | 0.179 (0.230) | 0.203 (0.294) | 0.257 (0.339) |
| Rfree | 0.227 (0.290) | 0.252 (0.415) | 0.317 (0.360) |
| RMSD length (Å) | 0.008 | 0.002 | 0.001 |
| RMSD angle (°) | 1.20 | 0.490 | 0.370 |
| No. of atoms | |||
| Protein | 2780 | 2672 | 2775 |
| Ligands | 33 | 33 | 33 |
| Water | 238 | 23 | 0 |
| Ramachandran plot (%) | |||
| Most favored | 95.85 | 95.18 | 94.07 |
| Additionally allowed | 4.15 | 4.82 | 5.64 |
Enzyme kinetic parameters for PKAc-catalyzed phosphorylation of different phospholamban (PLN) substrates measured by ADP-Glo assay.
| WT PLN | PLN R9C | pThr17 PLN | |
|---|---|---|---|
| Vmax (μM/min) | 4.1 ± 0.23 | 4.5 ± 0.33 | 3.6 ± 0.46 |
| KM (μM) | 85 ± 13 | 173 ± 25 | 223 ± 57 |
| kcat (s−1) | 6.9 ± 0.38 | 7.6 ± 0.6 | 6.0 ± 0.7 |
| kcat/KM (s−1 M−1) | 8.1 × 104 | 4.4 × 104 | 2.7 × 104 |