Development and evaluation of a machine learning-based in-hospital COVID-19 disease outcome predictor (CODOP): A multicontinental retrospective study
Figures
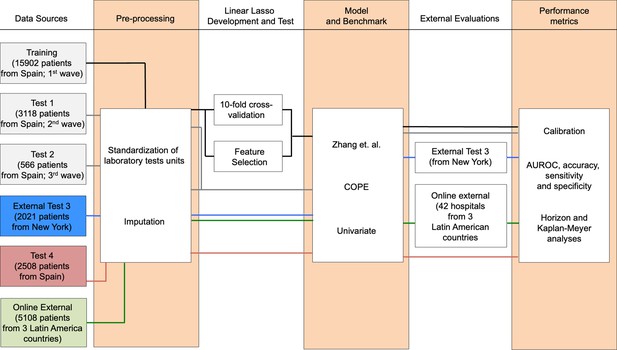
Flowchart depicting the different patient cohorts used in this study and the steps followed during the development, test, and independent evaluation of CODOP.
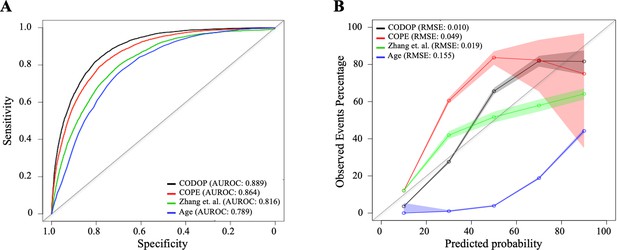
Discriminatory ability (using area under the receiver operating curves or AUROC; A) and calibration curves (B) for CODOP, COPE, Zhang et al., and Age in the training dataset.
-
Figure 2—source data 1
Prediction values for CODOP, COPE, Zhang et.
al in the training dataset.
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig2-data1-v2.xlsx
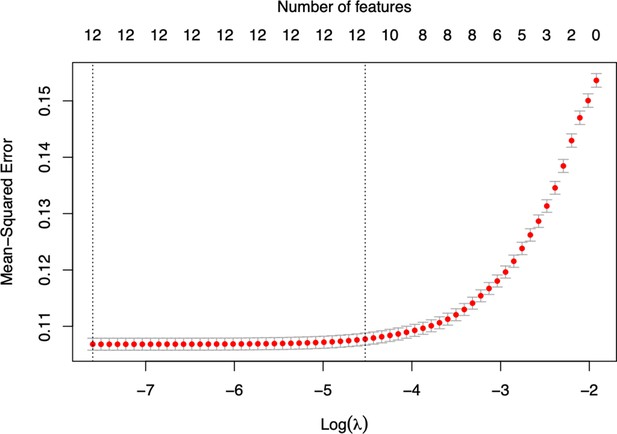
Optimisation of the final COPOD model by selecting predictors using the least absolute shrinkage and selection operator (LASSO) method.
The mean squared error is plotted versus log of the Penalty parameter (λ). Figure is produced by function cv.glmnet from R package glmnet.
-
Figure 2—figure supplement 1—source data 1
Mean squared error and the Penalty parameter (λ).
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig2-figsupp1-data1-v2.xlsx

Discriminatory ability (using area under receiver operating curves or AUROC) (A) and calibration curves (B) for CODOP, COPE, Zhang et al., and Age in the test datasets.
-
Figure 2—figure supplement 2—source data 1
Prediction values for CODOP, COPE, Zhang et.
al. in the Test 1, Test 2, External Test 3, and Test 4 datasets.
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig2-figsupp2-data1-v2.xlsx

Discriminatory ability of CODOP (using area under receiver operating curves or AUROC) taking into account the Delta and Omicron VOCs (A) and the vaccination status of the patients (B) in the Test 4 dataset.
-
Figure 2—figure supplement 3—source data 1
Prediction values for CODOP in vaccinated individuals and in patients infected with the Delta or Omicron virus variants.
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig2-figsupp3-data1-v2.xlsx
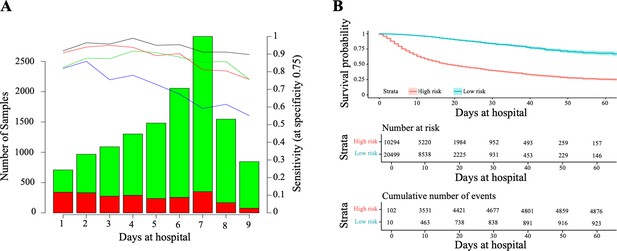
Horizon analysis (A) and survival analysis (B) in the training dataset.
In the horizon plot, x-axis represents the number of days at the hospital before clinical resolution, the bar plot is for the number of samples (the green colour is for survival and red for death), and lines are for sensitivity when the specificity was fixed at 75% in the training cohort (the black line is CODOP, the red line is COPE, the green line is Zhang et al., and the blue line is Age). In the survival analysis, the risk scores refer to the probability provided by CODOP.
-
Figure 3—source data 1
Prediction values for CODOP, COPE, Zhang et.
al. for the horizon analysis in the training dataset.
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig3-data1-v2.xlsx
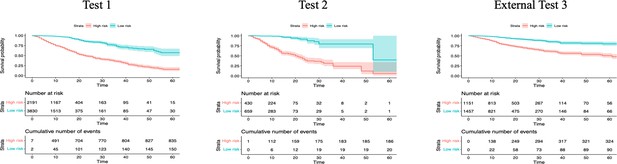
Survival analysis in the test datasets.
Shade areas indicate 95% confidence intervals. The risk scores refer to the probability provided by CODOP.
-
Figure 3—figure supplement 1—source data 1
Prediction values for CODOP, COPE, Zhang et.
al. for the risk stratification analysis in the Test 1, Test 2, and External Test 3 datasets.
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig3-figsupp1-data1-v2.xlsx
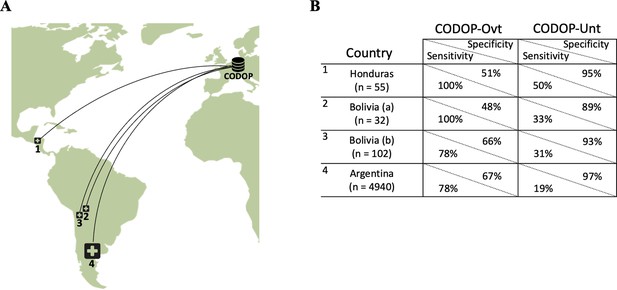
The geographical location of the external cohorts from 42 different Latin American hospitals used during the online evaluations (A) and performance of web calculators CODOP-Ovt and CODOP-Unt in these external cohorts number of patients from each institution are indicated in parenthesis; (B).
-
Figure 4—source data 1
Prediction values for CODOP in the Latin American cohort.
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig4-data1-v2.xlsx
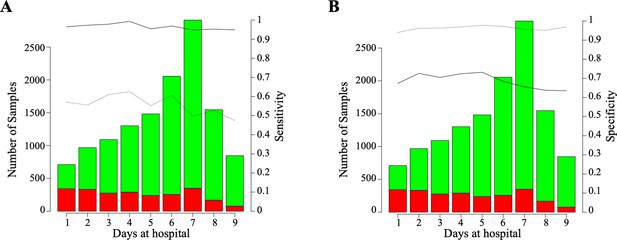
Horizon analysis in the training dataset for sensitivity (A) and specificity (B).
The black solid line is CODOP-Ovt and black dotted line is CODOP-Unt.
-
Figure 4—figure supplement 1—source data 1
Prediction values for CODOP-Ovt and CODOP-Unt for the horizon analysis in the training dataset.
- https://cdn.elifesciences.org/articles/75985/elife-75985-fig4-figsupp1-data1-v2.xlsx
Tables
Features used during CODOP development with the training cohort, the values used for imputation, and the percentage of missing values.
Numerical variables are reported by median (Md) and interquartile range (IQR).
| Variable | Imputed value | Md (IQR) | Missing % |
|---|---|---|---|
| Age (years) | 66·67,911 | 68 (56–79) | 0·0 |
| Sex (male, female) | none | 6 775 females and 9 127 males | 0·0 |
| Hemoglobin (g/dL) | 13·33,201 | 13 (12–15) | 1·7 |
| Platelet Count (x 106 /L) | 250 097·7 | 223,000 (164 000-311 000) | 1·8 |
| Eosinophils (x 106 /L) | 63·81,817 | 10 (0–100) | 3·0 |
| Lymphocytes (x 106 /L) | 1 243·575 | 1,000 (700-1 420) | 1·9 |
| Neutrophils (x 106 /L) | 5 525·894 | 4 490 (3 090-6 800) | 2·2 |
| Monocytes (x 106 /L) | 535·8,804 | 470 (300–660) | 2·7 |
| C-Reactive Protein (mg/L) | 74·48,964 | 41 (12–108) | 4·6 |
| Creatinine (mg/dL) | 1·156,574 | 1 (1–1) | 2·0 |
| Lactate Dehydrogenase (U/L) | 363·9,083 | 306 (234–424) | 13·0 |
| Aspartate aminotransferase (U/L) | 49·27,098 | 35 (24–53) | 18·4 |
| Alanine aminotransferase (U/L) | 48·99,699 | 32 (20–54) | 7·4 |
| Total bilirrubin (mg/dL) | 0·6429202 | 1 (0–1) | 26·5 |
| Serum Sodium (mmol/L) | 138·4,268 | 138 (136–141) | 2·6 |
| Serum Potassium (mmol/L) | 4·178,441 | 4 (4–4) | 3·7 |
| Glucose (mg/dL) | 124·2,852 | 108 (92–135) | 5·2 |
| Prothrombin time (s) | 19·99,798 | 13 (12–14) | 35·8 |
| Fibrinogen (mg/dL) | 608·0043 | 601 (497–713) | 37·0 |
| Dimer (ng/mL) | 2 122·158 | 672 (370–1 320) | 21·7 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75985/elife-75985-transrepform1-v2.docx
-
Supplementary file 1
Supplementary Tables.
- https://cdn.elifesciences.org/articles/75985/elife-75985-supp1-v2.docx
-
Supplementary file 2
List of Collaborators.
- https://cdn.elifesciences.org/articles/75985/elife-75985-supp2-v2.xlsx





