HLJ1 amplifies endotoxin-induced sepsis severity by promoting IL-12 heterodimerization in macrophages
Figures
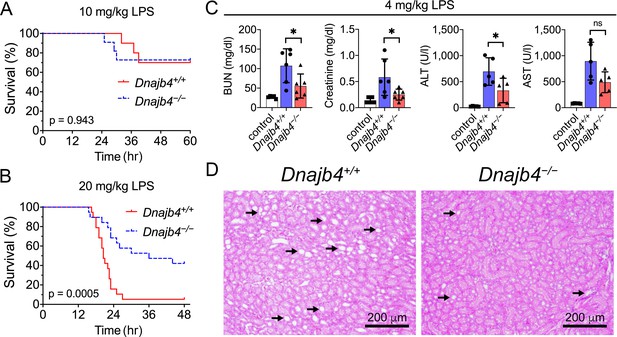
HLJ1 deletion protects against lipopolysaccharide (LPS)-induced organ injury and mortality.
Dnajb4−/− mice survive better than Dnajb4+/+ mice after high-dose LPS injection. Kaplan–Meier analysis of the overall survival of 6- to 8-week-old Dnajb4+/+ mice and Dnajb4−/− mice injected with (A) LD50 (10 mg/kg, n = 10–11 mice/group) or (B) high-dose (20 mg/kg, n = 19 mice/group) LPS. Log-rank Mantel-Cox test was used to compare survival curve. (C) Mice were i.p. injected with low-dose LPS (4 mg/kg) and after 24 hr serum levels of organ dysfunction markers BUN, creatinine, ALT, and AST were analyzed from n = 5–6 mice group. BUN, p = 0.037; creatinine, p = 0.048; ALT, p = 0.049; AST, p = 0.060. Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. *p < 0.05; ns, not significant. (D) Representative images of H&E staining of kidney sections from mice treated with 4 mg/kg LPS. Scale bar: 200 μm. Black arrows indicate kidney injury.
-
Figure 1—source data 1
Data for graphs depicted in Figure 1A–C.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig1-data1-v2.xlsx
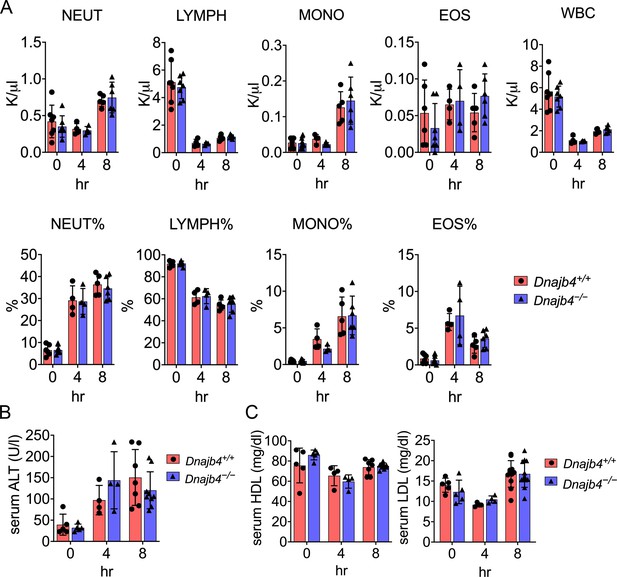
CBC counts, serum ALT, high-density lipoprotein (HDL), and low-density lipoprotein (LDL) levels of Dnajb4+/+ and Dnajb4−/− mice.
(A) Six to eight weeks Dnajb4+/+ and Dnajb4−/− mice were injected with lipopolysaccharide (LPS) of high dose (20 mg/kg). After 4 and 8 hr, blood was collected for analyzing percentage and counts of neutrophils (NEUT), lymphocytes (LYMPH), monocytes (MONO), eosinophils (EOS), and white blood cells (WBC) (n = 5–7 mice). (B) Serum from LPS-injected mice (n = 5–9) were analyzed for ALT levels. (C) Serum from n = 5–10 LPS-injected mice were analyzed for HDL and LDL levels. Data are mean ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test.
-
Figure 1—figure supplement 1—source data 1
Data for graphs depicted in Figure 1—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig1-figsupp1-data1-v2.xlsx
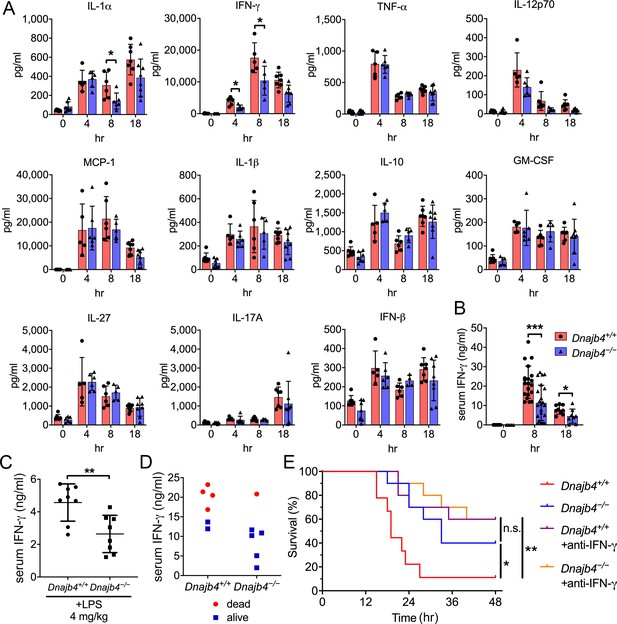
HLJ1 deletion alleviates IFN-γ-dependent sepsis death.
(A) Serum from Dnajb4+/+ and Dnajb4−/− mice administered with lipopolysaccharide (LPS) was analyzed at the indicated time points to quantify 11 cytokines via a cytokine bead array. IL-1α, p = 0.03; 4 hr IFN-γ, p = 0.027; 8 hr IFN-γ, p = 0.04; n = 5–8 per group. (B) Serum IFN-γ levels were quantified using ELISA 8 hr (n = 20–22) and 18 hr (n = 8–9) after LPS injection. 8 hr IFN-γ, p < 0.001; 18 hr IFN-γ, p = 0.039. (C) Mice (n = 8 biological replicates) were injected with lower dose 4 mg/kg LPS and after 8 hr serum was collected for quantification of IFN-γ levels. p = 0.005. (D) Correlation between survival status and serum IFN-γ levels in Dnajb4+/+ and Dnajb4−/− mice injected with 20 mg/kg LPS (n = 6 mice/group). (E) Kaplan–Meier analysis of overall survival of Dnajb4+/+ and Dnajb4−/− mice (n = 9–10) injected with 100 μg anti-IFN-γ neutralizing antibodies 1 hr before LPS (20 mg/kg) challenge. Dnajb4+/+ versus Dnajb4−/− mice, p = 0.015; Dnajb4+/+ versus Dnajb4+/++anti-IFN-γ, p = 0.007. Data presented are means ± standard deviation (SD). Significance was calculated by using two-tailed, unpaired Student’t t-test. Log-rank Mantel-Cox test was used to compare survival curve. *p < 0.05, **p < 0.01, ***p < 0.001.
-
Figure 2—source data 1
Data for graphs depicted in Figure 2A–E.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig2-data1-v2.xlsx
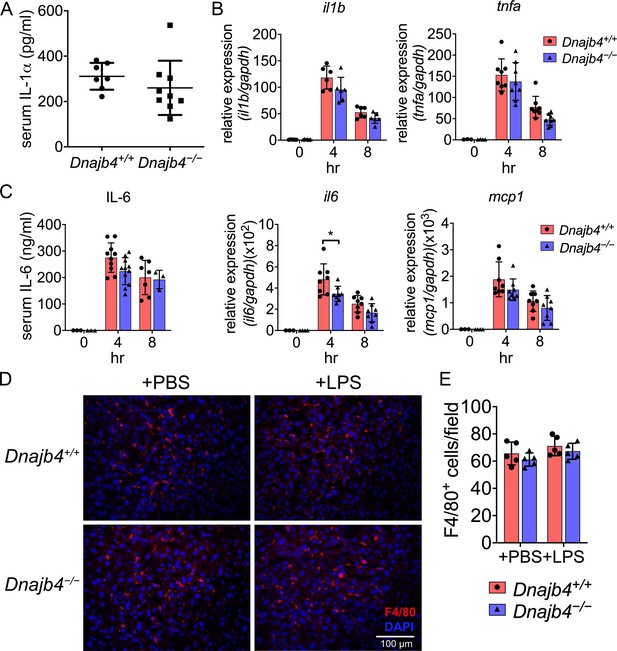
Inflammatory cytokine expression and macrophage numbers were unchanged in the liver of sepsis mice.
(A) Serum IL-1α levels from n = 8–9 mice were quantified 8 hr after 20 mg/kg lipopolysaccharide (LPS) injection. (B) After 4 or 8 hr, the RNA from n = 6–8 total livers were isolated and gene expression levels were quantified via quantitative real-time PCR (qRT-PCR). IL-6, p = 0.033. (C) Serum levels of IL-6 in Dnajb4+/+ and Dnajb4−/− mice were quantified via ELISA 4 hr (n = 11–14) and 8 hr (n = 4–7) after LPS administration. (D) Quantification of liver-resident macrophages in LPS-challenged mice. Representative photographs of F4/80 immunofluorescence staining from liver sections of phosphate-buffered saline (PBS) or LPS-injected Dnajb4+/+ and Dnajb4−/− mice. Eight hours after LPS injection, mice were sacrificed and liver was fixed, dehydrated, embedded, cryosectioned into 8 μm thickness, and incubated with anti-F4/80 antibodies to stain mature macrophages (red). (E) Quantitation of F4/80+ macrophages. Positively stained cells were counted at ×400 magnification in 6 fields from 3 sections/mouse and from 5 mice/group. Data are mean ± standard deviation (SD). Significance was calculated by using two-tailed, unpaired Student’t t-test. *p < 0.05.
-
Figure 2—figure supplement 1—source data 1
Data for graphs depicted in Figure 2—figure supplement 1A–C, E.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig2-figsupp1-data1-v2.xlsx
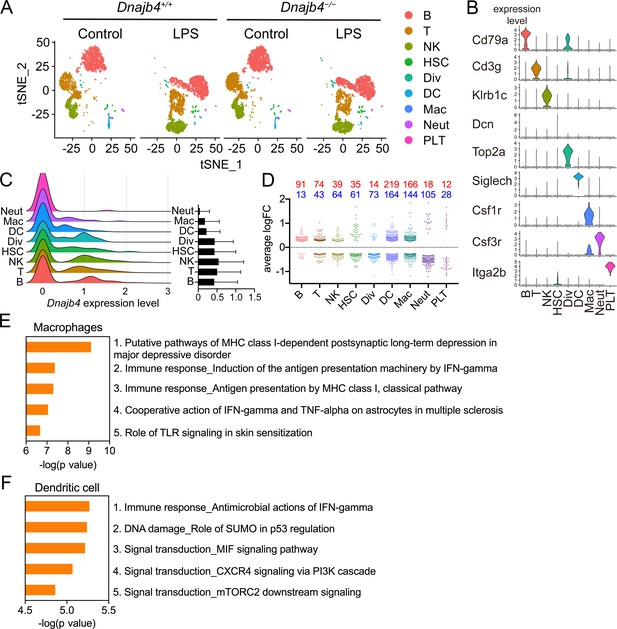
Single-cell RNA sequencing (scRNA-seq) reveals activated IFN-γ-mediated signaling pathways in macrophages and dendritic cells.
(A) Mice were injected with 20 mg/kg lipopolysaccharide (LPS), or phosphate-buffered saline (PBS) as a control, and after 8 hr hepatic nonparenchymal cells were isolated for scRNA-seq analysis. The plot shows the t-distributed stochastic neighbor embedding (t-SNE) visualization of liver nonparenchymal cell clusters based on 11,651 single-cell transcriptomes. B, B cells; T, T cells; NK, NK cells; HSC, hepatic stellate cells; Div, dividing cells; DC, dendritic cells; Mac, macrophages; Neut, neutrophils; PLT, platelets. (B) Expression levels of representative known marker genes for each cluster. (C) Visualization of expression distribution of the Dnajb4 gene in each cluster of cells in PBS-treated Dnajb4+/+ mice. Data presented are means ± standard deviation (SD). (D) Cell-type distribution and log-transformed expression fold change (logFC) for upregulated (red) and downregulated (blue) genes from a comparison of LPS-treated Dnajb4+/+ mice with LPS-treated Dnajb4−/− mice. The Wilcoxon rank-sum test was used to identify differentially expressed genes (p < 0.05, |logFC| > 0.25). Enrichment analysis showing ranked pathway signatures associated with up- and downregulated genes (p < 0.05, |logFC| > 0.25) from a comparison of macrophages (E) and dendritic cells (F) from LPS-injected Dnajb4+/+ mice with those from Dnajb4−/− mice.
-
Figure 3—source data 1
Data for graphs depicted in Figure 3D–F.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig3-data1-v2.xlsx
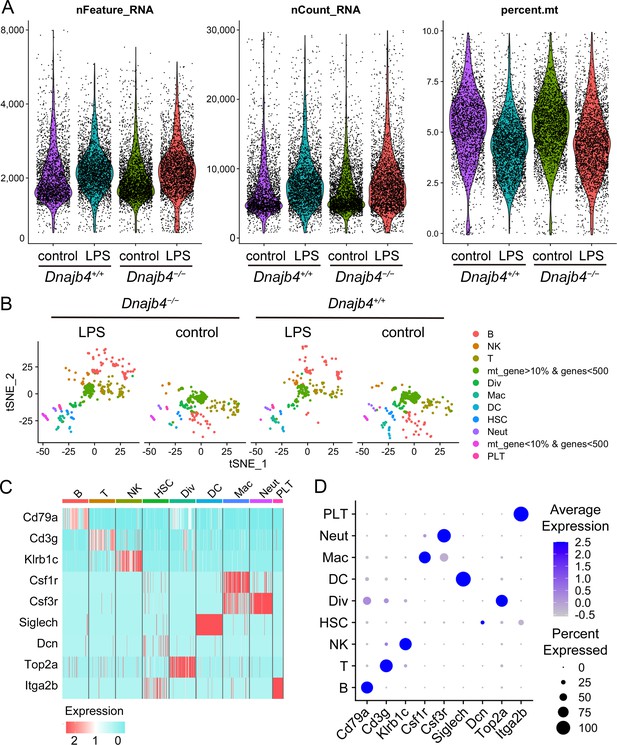
Quality control of single-cell RNA sequencing (scRNA-seq) data.
(A) Cells with unique molecular identifier (UMI) count of greater than 30,000, fewer than 500, or greater than 6000 genes, and >10% of total expression from mitochondrial genes were excluded. (B) t-Distributed stochastic neighbor embedding (t-SNE) visualization of 1917 excluded cells in which UMI count of greater than 30,000, fewer than 500, or greater than 6000 genes, and >10% of total expression from mitochondrial genes. (C) Heat map and (D) dot plot of known marker genes expression in each cell cluster.
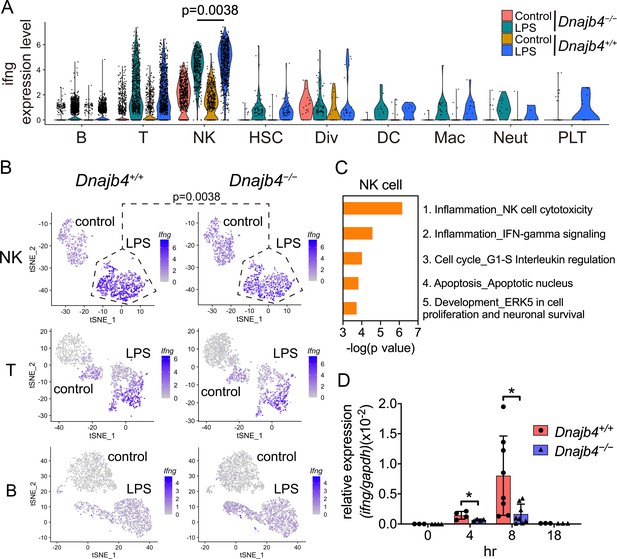
HLJ1 deficiency leads to altered IFN-γ-related signatures in natural killer (NK) cells under lipopolysaccharide (LPS) stress.
(A) Violin plot showing IFN-γ expression levels in each type of cell. Significance was calculated using the Wilcoxon rank-sum test. (B) t-Distributed stochastic neighbor embedding (t-SNE) visualization of IFN-γ expression profiles in NK, T, and B cells isolated from Dnajb4+/+ and Dnajb4−/− mice injected with LPS. Significance was calculated using the Wilcoxon rank-sum test. (C) Enrichment analysis showing ranked network signatures associated with up- and downregulated genes (p < 0.05, |logFC| > 0.25) from a comparison of NK cells from LPS-injected Dnajb4+/+ mice with NK cells from Dnajb4−/− mice. (D) Dnajb4+/+ and Dnajb4−/− mice (n = 4-9 mice/group) were injected with LPS and, at the indicated time points, whole liver mRNA was extracted for the measurement of hepatic IFN-γ expression levels via quantitative real-time PCR (qRT-PCR). p = 0.026 and p = 0.014 for the 4 and 8 hr groups, respectively. Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. *p < 0.05.
-
Figure 4—source data 1
Data for graphs depicted in Figure 4C, D.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig4-data1-v2.xlsx
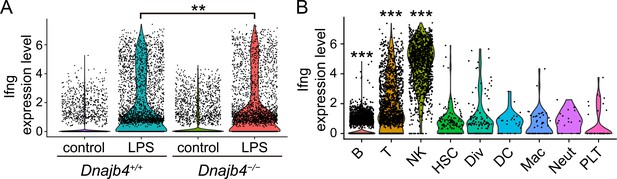
IFN-γ gene expression analysis was split according to treatment, genotype or cell type.
(A) IFN-γ gene expression levels in each cell. Significance was calculated using the Wilcoxon rank-sum test; p = 0.009. (B) IFN-γ expression patterns in B (p < 0.001), T (p < 0.001), and natural killer (NK) (p < 0.001) cells and other clusters in lipopolysaccharide (LPS)-treated mice. Significance was calculated using the Wilcoxon rank-sum test. **p < 0.01, ***p < 0.001.
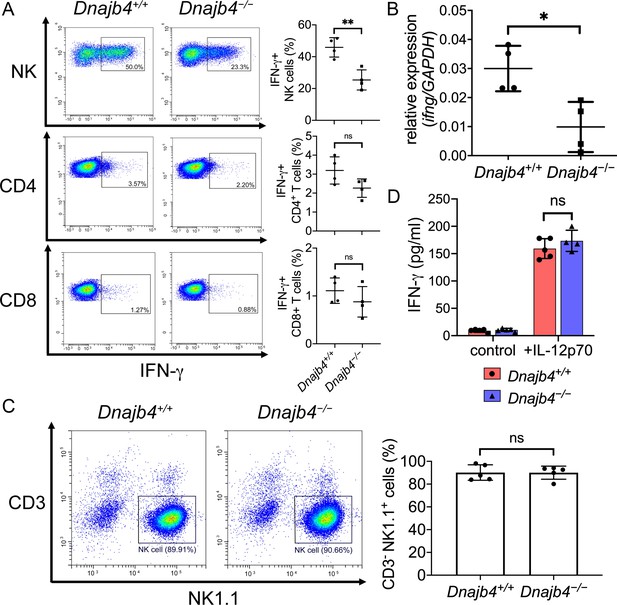
Intracellular IFN-γ levels decreased in splenic natural killer (NK) cells after HLJ1 deletion.
(A) Dnajb4+/+ and Dnajb4−/− mice (n = 4 per group) were injected intraperitoneally with 20 mg/kg lipopolysaccharide (LPS), and splenocytes were isolated after 2.5 hr. Expression of intracellular IFN-γ levels in Dnajb4+/+ and Dnajb4−/− NK, CD4+ T, and CD8+ T cells were detected via flow cytometry analysis. NK cells, p = 0.004. Representative samples are shown. (B) RNA from n = 4 mice spleens were isolated 4 hr after LPS administration, and transcriptional levels of IFN-γ were quantified via quantitative real-time PCR (qRT-PCR); p = 0.014. (C) Expression of NK1.1 in primary NK cells isolated from Dnajb4+/+ and Dnajb4−/− mice (n = 5 per group) was detected via flow cytometry. Representative samples are shown. (D) Primary NK cells purified from Dnajb4+/+ and Dnajb4−/− mice spleens were treated with 10 ng/ml IL-12p70 for 24 hr and supernatant IFN-γ was quantified using ELISA (n = 4–5 biological replicates). Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. *p < 0.05, **p < 0.01, n.s., not significant.
-
Figure 5—source data 1
Data for graphs depicted in Figure 5A–D.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig5-data1-v2.xlsx
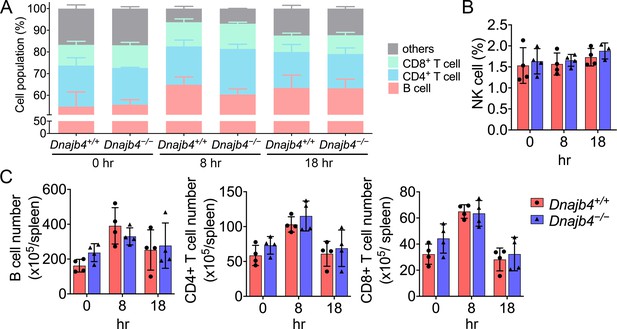
Splenic immune cell population identification of Dnajb4+/+ and Dnajb4−/− mice.
(A) CD4+, CD8+ T cell, and B cell population and (B) natural killer (NK) cell population are presented as percentage. Splenic immune cells were isolated from lipopolysaccharide (LPS)-treated Dnajb4+/+ and Dnajb4−/− mice (n = 4 mice/group) and were identified with surface markers of B, CD4+ T, and CD8+ T cells by flow cytometry. (C) Immune cells population are presented as total number. Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test.
-
Figure 5—figure supplement 1—source data 1
Data for graphs depicted in Figure 5—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig5-figsupp1-data1-v2.xlsx
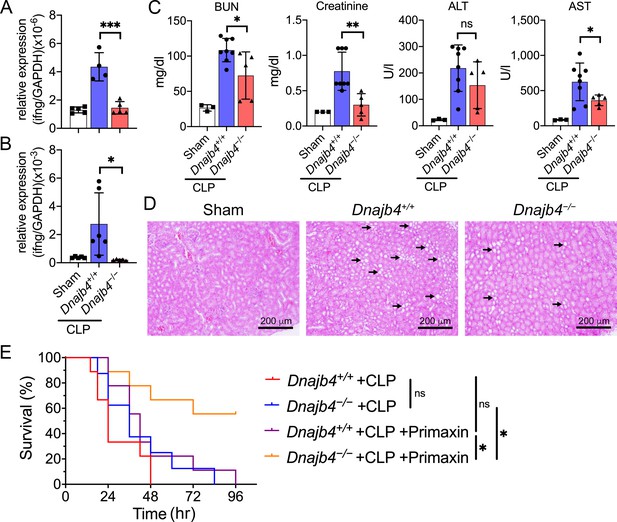
HLJ1 deletion protect mice from CLP-induced organ dysfunction and septic death.
(A) CLP or sham surgery were performed on Dnajb4+/+ and Dnajb4−/− mice, and after 18 hr whole liver mRNA was extracted for the measurement of IFN-γ expression levels via quantitative real-time PCR (qRT-PCR) (n = 5–6). p < 0.001. (B) Spleen mRNA was also extracted for the measurement of IFN-γ expression levels via qRT-PCR (n = 5–6). p = 0.031. (C) Serum levels of BUN, creatinine, ALT, and AST were analyzed 18 hr after sham (n = 3) or CLP surgery (n = 5–8). BUN, p = 0.024; creatinine, p = 0.005; ALT, p = 0.225; AST, p = 0.048. (D) Representative images of H&E staining of sham and CLP mouse kidney sections. Scale bar: 200 μm. Black arrows indicate kidney injury. (E) Kaplan–Meier analysis of overall survival of Dnajb4+/+ and Dnajb4−/− mice. Mice were i.p. injected with 25 mg/kg imipenem/cilastatin (Primaxin) immediately after CLP. Antibiotic treatment was continued twice per day throughout the observation period (n = 8–11 per group). Dnajb4−/− + CLP + Primaxin versus Dnajb4−/− + CLP + Primaxin, p = 0.013; Dnajb4−/− + CLP versus Dnajb4−/− + CLP + Primaxin, p = 0.010. Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. Log-rank Mantel-Cox test was used to compare survival curve. *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant.
-
Figure 6—source data 1
Data for graphs depicted in Figure 6A–C, E.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig6-data1-v2.xlsx
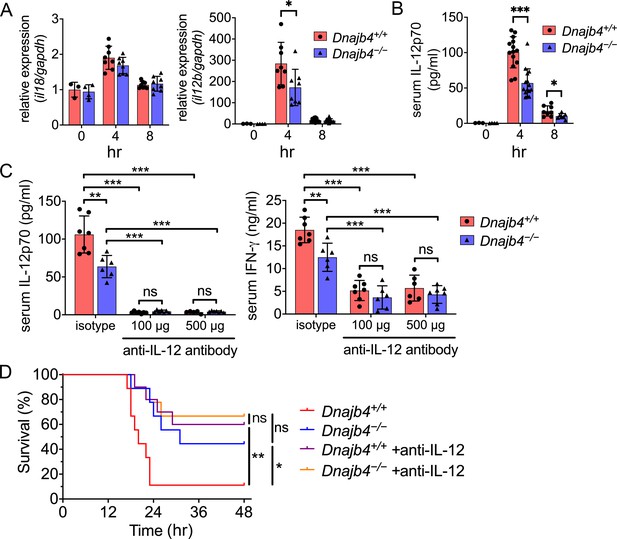
HLJ1 deletion alleviates IL-12-dependent septic death.
Dnajb4+/+ and Dnajb4−/− mice were intraperitoneally injected with 20 mg/kg lipopolysaccharide (LPS). (A) After 4 or 8 hr, the RNA from n = 6–8 total livers were isolated and gene expression levels were quantified via qRT-PCR. IL-12b, p = 0.029. (B) Serum levels of IL-12p70 in LPS-treated Dnajb4+/+ and Dnajb4−/− mice were quantified via ELISA 4 hr (n = 11–14) and 8 hr (n = 4–7) after LPS administration. 4 hr IL-12p70, p < 0.001; 8 hr IL-12p70, p = 0.033. (C) Dnajb4+/+ and Dnajb4−/− mice were intraperitoneally injected with anti-IL-12 neutralizing antibodies 1 hr prior to the injection of 20 mg/kg LPS. After the administration of LPS and anti-IL-12 antibodies, the serum was collected at the indicated time points and analyzed for IL-12 and IFN-γ levels. (D) Kaplan–Meier analysis of overall survival of Dnajb4+/+ and Dnajb4−/− mice injected with 100 μg anti-IL-12 neutralizing antibodies 1 hr before the 20 mg/kg LPS challenge (n = 9–11 per group). Dnajb4+/+ versus Dnajb4−/− mice, p = 0.014; Dnajb4+/+ versus Dnajb4+/+ + anti-IL-12, p = 0.007. Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. Log-rank Mantel-Cox test was used to compare survival curve. *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant.
-
Figure 7—source data 1
Data for graphs depicted in Figure 7A–D.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig7-data1-v2.xlsx
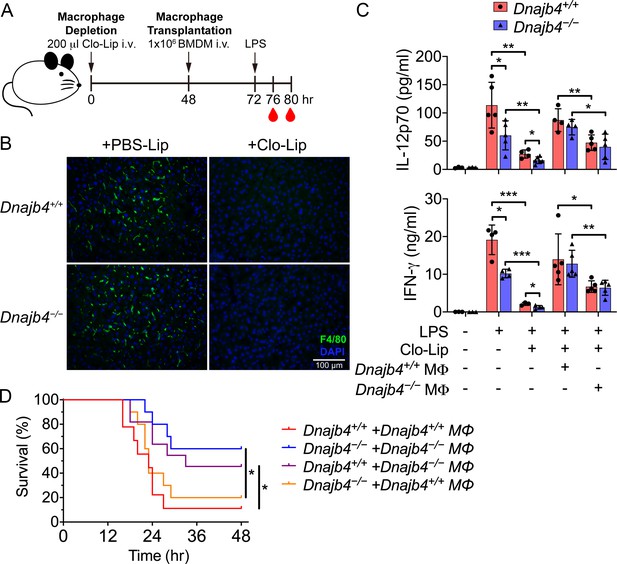
HLJ1 deletion in macrophages reduced serum levels of IL-12 and IFN-γ and mitigated septic death in vivo.
(A) 200 μl clodronate liposomes (Clo-Lip) were administered intravenously to Dnajb4+/+ and Dnajb4−/− mice to deplete their endogenous macrophages. After 48 hr, the mice were intravenously injected with 1 × 106 bone marrow-derived macrophages (BMDMs) isolated from Dnajb4+/+ or Dnajb4−/− mice. After BMDM transplantation, Dnajb4+/+ and Dnajb4−/− mice were administered with 20 mg/kg lipopolysaccharide (LPS) and serum was collected at 4 or 8 hr for IL-12 or IFN-γ quantification, respectively. (B) Representative photographs of F4/80 immunofluorescence staining of liver sections from phosphate-buffered saline (PBS) liposome or clodronate liposome-injected Dnajb4+/+ and Dnajb4−/− mice. The liver was fixed, dehydrated, embedded, cryosectioned into slices 8 μm thick, and incubated with anti-F4/80 antibodies to stain the mature macrophages (green). The scale bar represents 100 μm. (C) Mice transplanted with Dnajb4+/+ BMDMs (Dnajb4+/+ MΦ) or Dnajb4−/− BMDMs (Dnajb4−/− MΦ) were administered LPS, and serum from n = 4–5 mice was analyzed for IL-12p70 and IFN-γ levels via ELISA. Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. (D) Kaplan–Meier analysis of the overall survival of LPS-injected Dnajb4+/+ and Dnajb4−/− mice transplanted with Dnajb4+/+ and Dnajb4−/− BMDMs (n = 9–10 per group). For Dnajb4+/++Dnajb4+/+ MΦ versus Dnajb4+/++Dnajb4−/− MΦ, p = 0.037. For Dnajb4−/−+Dnajb4−/− MΦ versus Dnajb4−/−+Dnajb4+/+ MΦ, p = 0.036. Log-rank Mantel-Cox test was used to compare survival curve. *p < 0.05, **p < 0.01, ***p < 0.001.
-
Figure 8—source data 1
Data for graphs depicted in Figure 8C, D.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig8-data1-v2.xlsx
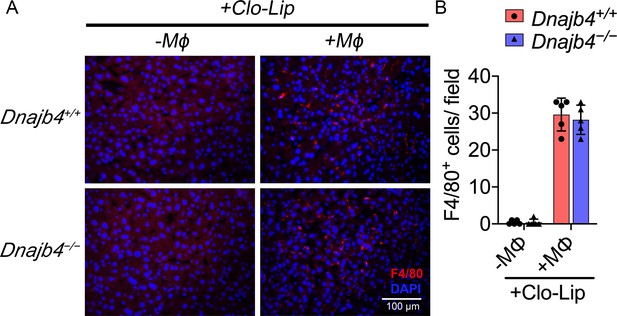
Quantification of liver macrophages in bone marrow-derived macrophage (BMDM)-transplanted mice.
(A) Representative photographs of F4/80 immunofluorescence staining from liver sections of Dnajb4+/+ and Dnajb4−/− mice with BMDMs adoptive transfer or with Clo-Lip alone. 24 hr after i.v. injection of BMDMs, mice were sacrificed and liver was fixed, dehydrated, embedded, cryosectioned into 8 μm thickness, and incubated with anti-F4/80 antibodies to stain mature macrophages (red). (B) Quantitation of F4/80+ macrophages. Positively stained cells were counted at ×400 magnification in 6 fields from 3 sections/mouse and from 5 mice/group. Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test.
-
Figure 8—figure supplement 1—source data 1
Data for graphs depicted in Figure 8—figure supplement 1B.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig8-figsupp1-data1-v2.xlsx
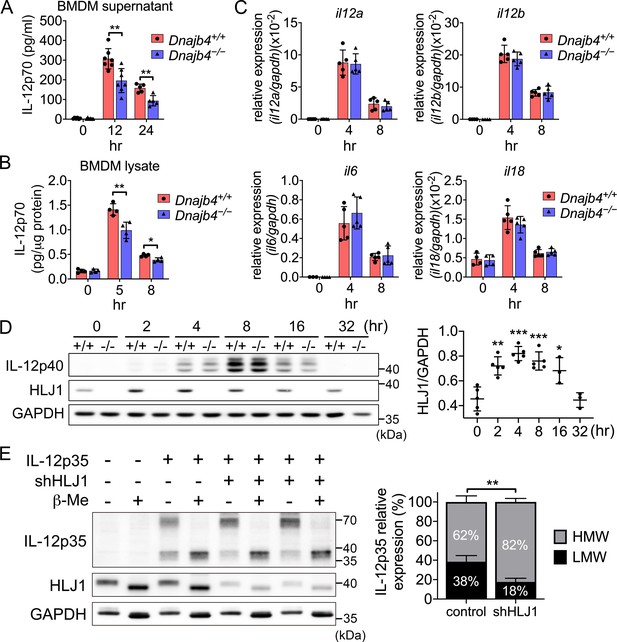
HLJ1 deletion leads to the accumulation of homodimeric IL-12p35 and reduced levels of heterodimeric IL-12p70.
(A) Bone marrow-derived macrophages (BMDMs) isolated from n = 6–7 Dnajb4+/+ and Dnajb4−/− mice were treated with 10 ng/ml lipopolysaccharide (LPS) and 20 ng/ml IFN-γ. Supernatant was collected at the indicated time points, and IL-12p70 was quantified via ELISA. 12 hr, p = 0.003; 24 hr, p = 0.003. (B) LPS/IFN-γ-treated BMDMs from n = 4–5 mice were lysed at the indicated time points and intracellular IL-12p70 was quantified via ELISA. 5 hr, p = 0.006; 8 hr, p = 0.012. (C) IL-12a, IL-12b, IL-6, and IL-18 expression was determined via quantitative real-time PCR (qRT-PCR) in LPS/IFN-γ-treated BMDMs isolated from n = 5 mice. (D) Intracellular IL-12p40 and HLJ1 expression levels were analyzed in LPS/IFN-γ-treated BMDMs isolated from Dnajb4+/+ (+/+) and Dnajb4−/− (−/−) mice. Representative samples of n = 3–5 biological replicates are shown. GAPDH served as a loading control. In comparisons with the 0 hr group (right panel): 2 hr, p = 0.001; 4 hr, p < 0.001; 8 hr, p = <0.001; 16 hr, p = 0.02. (E) The influence of human HLJ1 knockdown on the redox state of human IL-12p35 was analyzed via non-reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). 293T cells were (co-)transfected with the indicated IL-12p35 subunits and shRNA targeting HLJ1. The percentage of high-molecular-weight (HMW) and low-molecular-weight (LMW) IL-12p35 species in the presence or absence of shHLJ1 was quantified (right panel, n = 4 biological repeats for shHLJ1- and control-transfected cultures; p = 0.001). Where indicated, samples were treated with β-mercaptoethanol (β-Me) after cell lysis to provide a standard for completely reduced protein. GAPDH served as a loading control. Data presented are means ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
-
Figure 9—source data 1
Data for graphs depicted in Figure 9A–E.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig9-data1-v2.xlsx
-
Figure 9—source data 2
Original and labeled blots images of Figure 9D, E.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig9-data2-v2.zip
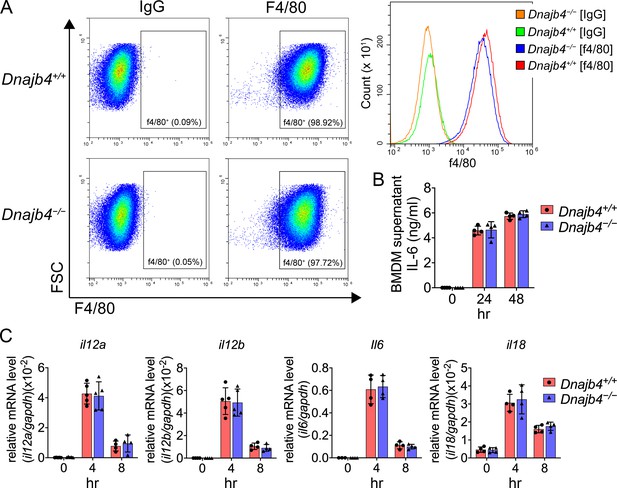
Transcriptional levels of proinflammatory cytokines in lipopolysaccharide (LPS)-treated bone marrow-derived macrophages (BMDMs).
(A) F4/80+ BMDMs analyzed with flow cytometry. Bone marrow cells were isolated from Dnajb4+/+ and Dnajb4−/− mouse and differentiated with M-CSF (10 ng/ml) for 7 days. (B) Dnajb4+/+ and Dnajb4−/− BMDMs isolated from n = 4 mice were treated with 100 ng/ml LPS and supernatants were collected at the indicated time points and IL-6 was analyzed by ELISA. (C) Transcriptional levels of il12a, il12b, il6, and il18 in BMDMs isolated from n = 4–5 mice were quantified by quantitative real-time PCR (qRT-PCR). Data are mean ± standard deviation (SD). Statistical analysis was performed by using the two-tailed, unpaired Student’s t-test.
-
Figure 9—figure supplement 1—source data 1
Data for graphs depicted in Figure 9—figure supplement 1B, C.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig9-figsupp1-data1-v2.xlsx
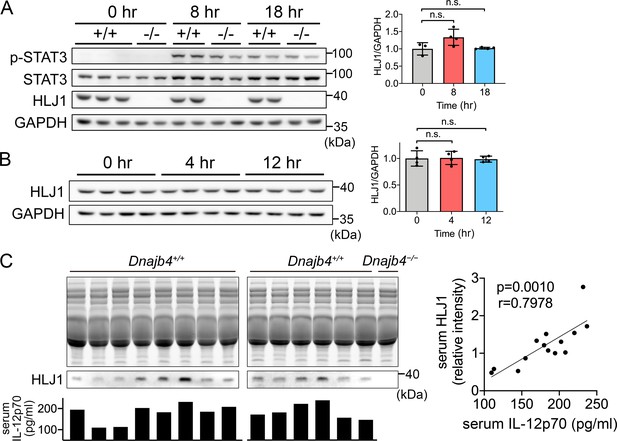
Expression levels of HLJ1 in the liver and serum from lipopolysaccharide (LPS)-injected mice.
(A, B) HLJ1 and STAT3 protein levels and phosphorylation levels in LPS-treated 6–8 weeks Dnajb4+/+ (+/+) and Dnajb4−/− (−/−) mouse liver were determined. Representative samples are showed from n = 4 per group. Data are mean ± standard deviation (SD). Image J software was used to quantify the intensity and statistical analysis was performed by using the two-tailed, unpaired Student’s t-test. n.s., not significant. (C) Dnajb4+/+ and Dnajb4−/− mice were administrated with 20 mg/kg LPS and after 4 hr serum was collected for analysis of IL-12p70 and HLJ1 expression by ELISA and western blotting, respectively. For western blotting, 4 μl serum were used for each lane and total proteins in the acrylamide gel were stained as internal control. p value and r value were calculated from 14 samples by Spearman’s correlation coefficient testing.
-
Figure 9—figure supplement 2—source data 1
Data for graphs depicted in Figure 9—figure supplement 2A–C.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig9-figsupp2-data1-v2.zip
-
Figure 9—figure supplement 2—source data 2
Original and labeled blots images of Figure 9—figure supplement 2A–C.
- https://cdn.elifesciences.org/articles/76094/elife-76094-fig9-figsupp2-data2-v2.zip
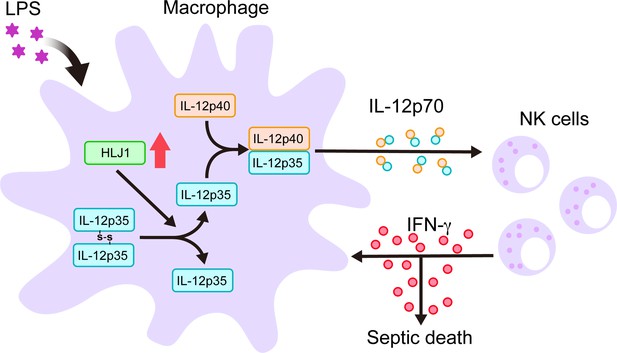
Schematic diagram delineates how HLJ1 functions and controls IL-12 biosynthesis, IFN-γ production, and subsequent sepsis-related mortality.
HLJ1 protein, which can be induced when macrophages are stimulated with lipopolysaccharide (LPS), helps the conversion of high-molecular-weight (HMW) misfolded IL-12p35 homodimers to IL-12p35 monomers. Bioactive IL-12p70 heterodimers, composed of IL-12p35 and IL-12p40 subunits, are released into the circulation by macrophages and thereby stimulates natural killer (NK) cells. Eventually, activated NK cells in the liver and spleen release IFN-γ in sufficient quantities to lead to organ damage and even death during sepsis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | HLJ1 (Dnajb4) | GenBank | MGI:1914285 | |
| Strain, strain background (Mus musculus) | HLJ1 knockout mice (Dnajb4−/−) in C57BL/6 background | Medical College, National Taiwan University | N/A | National Core Facility for Biopharmaceuticals – A4, Ministry of Science and Technology, Taiwan (https://ncfb.nycu.edu.tw/en/a4.html) |
| Cell line (Homo sapiens) | Epithelial Kidney; Embryo | ATCC | 293T | Transfected with HLJ1-shRNA containing vectors |
| Transfected construct (Homo sapiens) | HLJ1 shRNA | National RNAi Core Facility (Academia Sinica, Taiwan) | TRCN0000419874 NM_007034 | Transfected construct to express the shRNA |
| Transfected construct (Homo sapiens) | IL-12p35 overexpression | Origene | RC211224 NM_000882 | Transfected construct to express the human IL12A |
| Biological sample (Mus musculus) | Primary NK cells | This paper | Freshly isolated from Mus musculus | |
| Biological sample (Mus musculus) | Bone marrow-derived macrophages | This paper | Freshly isolated from Mus musculus | |
| Antibody | Anti-IL-12 (Clone: C17.8) (Rat monoclonal) | Biolegend | Cat# 505310 | WB (1:1000) Neutralization (100 and 500 μg/mouse) |
| Antibody | Anti-IFN-γ (Clone: XMG1.2) (Rat monoclonal) | BioXCell | Cat# BE0055 | Neutralization (100 μg/mouse) |
| Antibody | Anti-IL-12A (Clone: EPR5736) (Rabbit monoclonal) | Abcam | Cat# Ab133751 | WB (1:1000) |
| Antibody | Anti-Dnajb4 (HLJ1) (Rabbit polyclonal) | Proteintech | Cat# 13064-1-AP | WB (1:5000) |
| Antibody | Anti-F4/80 [CI:A3-1] (Rat monoclonal) | Abcam | Cat# ab6640 | ICC/IF (1:100) |
| Sequence-based reagent | Ifng_F | Arterioscler Thromb Vasc Biol. 2005 Apr;25(4):791–6. | qRT-PCR primer | AGCAACAGCAAGGCGAAAA |
| Sequence-based reagent | Ifng_R | Arterioscler Thromb Vasc Biol. 2005 Apr;25(4):791–6. | qRT-PCR primer | CTGGAC CTGTGGGTTGTTGA |
| Sequence-based reagent | Il12a_F | PNAS July 10, 2012 109 (28) 11200–11205 | qRT-PCR primer | AAGAACGAGAGTTGCCTGGCT |
| Sequence-based reagent | IL12a_R | PNAS July 10, 2012 109 (28) 11200–11205 | qRT-PCR primer | TTGATGGCCTGGAACTCTGTC |
| Sequence-based reagent | Il12b_F | J Immunol March 1, 2019, 202 (5) 1406–1416 | qRT-PCR primer | GAAGTTCAACATCAAGAGCAGTAG |
| Sequence-based reagent | Il12b_R | J Immunol March 1, 2019, 202 (5) 1406–1416 | qRT-PCR primer | AGGGAGAAGTAGGAATGGGG |
| Peptide, recombinant protein | IFN-γ | Peprotech | Cat# 315-05 | 20 ng/ml |
| Peptide, recombinant protein | M-CSF | Peprotech | Cat# 315-02 | 10 ng/ml |
| Commercial assay or kit | Mouse IFN-γ ELISA | Biolegend | Cat# 430804 | |
| Commercial assay or kit | Mouse IL-12 ELISA | Biolegend | Cat# 433604 | |
| Commercial assay or kit | LEGENDplex | Biolegend | Cat# 740446 | |
| Chemical compound, drug | Liposome-encapsulated clodronate | Liposoma | Car# C-025 | 100 μl/10 gbw |
| Software, algorithm | MetaCore software | Clarivate https://portal.genego.com/ | Pathway analysis | |
| Software, algorithm | GraphPad Prism software | GraphPad Prism | Version 8.0.0 | |
| Software, algorithm | Loupe browser | 10× genomics https://www.10xgenomics.com/products/loupe-browser | ||
| Other | Lipopolysaccharide (LPS) from E. coli O111:B4 | Sigma-Aldrich | Cat# L2630 | Low-dose 4 mg/kg LD50 10 mg/kg High-dose 20 mg/kg |





