Cell Culture: Implementing robotics and artificial intelligence
Most animals, including humans, are made up of different organs and tissues formed by specialized cells that perform specific roles. When tissues degenerate or become damaged, the affected cells have to be replaced so the tissues can keep performing their roles. This regenerative potential exists thanks to populations of stem cells in each tissue, which can divide to produce more stem cells – maintaining a constant pool of stem cells for repair – or differentiate into specialized cells to replace damaged cells.
The division and differentiation of stem cells needs to be in balance: if too many stem cells differentiate, the pool of stem cells could become exhausted, but if the stem cells divide uncontrollably this could lead to cancer. However, this balance often fails with age, or due to environmental or genetic reasons. One of the goals of regenerative medicine is to produce differentiated cells in the laboratory that can be used to heal tissues when they have lost the ability to regenerate for themselves (Pizevska et al., 2022). These treatments could have many potential long-term health benefits, including extending life expectancy, and research into these treatments is increasing both in the laboratory and in the clinic.
In order to use cells differentiated in the laboratory in a clinical setting, it is essential that the protocols used to produce the desired cell types reproducibly, and in high enough numbers. This is difficult to do because differentiating cells are highly sensitive to stimuli in their environments, meaning that their culture conditions have to be carefully controlled (Figure 1). Human operators introduce an added layer of variability that is difficult to control for, since each person does cell culture a little differently, and the protocols used to manufacture regenerative medicine products are often complex (Sebastian et al., 2019). Additionally, the requirements that cells and cell-derived products need to meet to be used in the clinic are changing rapidly, and the tests used to assess these requirements can also propagate variability (Simon et al., 2016). As a result of these issues, the quality of the cells produced for regenerative medicine treatments is often unreliable.
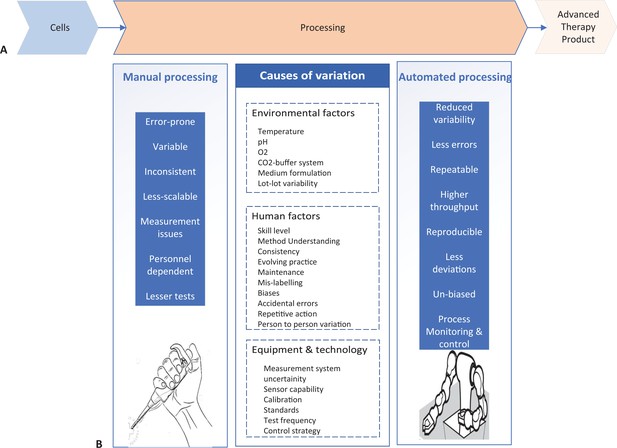
Comparison of manual and automated manufacturing of cell therapy products.
(A) Schematic of the development lifecycle of cell therapy products. Cells (left) undergo processing in the laboratory (center), where they are cultured under different conditions, often requiring several complex cell culture steps, to develop safe and efficacious therapy product (right). (B) The processing step of therapy product development can be done manually (left) or automatically (right). Manual processing has several factors (center left) that increase variation in the culture conditions. Causes of variation can be environmental factors, human factors or equipment and technology factors (center), and they lead to manufacturing being less consistent. Automated processing can correct the issues that manual processing introduces (center right).
In recent years, advanced imaging techniques and analysis have improved the reproducibility of the cell culture protocols used for the manufacture of cells in regenerative medicine (Imai et al., 2018). This suggests that further minimizing human error and variability could also greatly improve the manufacturing process. One way to do this would be to mechanize and automate the cell culture procedures. Now, in eLife, Yosuke Ozawa, Koichi Takahashi, Tohru Natsume, and colleagues (who are based at various institutions in Japan) – with Genki Kanda and Taku Tsuzuki as first authors – report on LabDroid Maholo, a robotics and artificial intelligence-based cell processing station that can reliably carry out complex manual cell culture procedures typically performed by trained scientists (Kanda et al., 2022).
Replacing a difficult and repetitive manual process with a machine is a critical step in reducing the variability that arises in cell culture protocols. Indeed, it has previously been shown that automating methods using robotics is an efficient way to reduce human error and operator-dependent variability in the laboratory (Archibald et al., 2016). Kanda et al. now demonstrate this principle in the context of cell culture by combining robotics and artificial intelligence to automate complex cell culture protocols.
The LabDroid Maholo System contains highly accurate and programmable robotic arms to manipulate the cell culture conditions, which allowed the robot to culture stem cells under different conditions. In this case, the LabDroid Maholo System was programmed to differentiate stem cells into retinal pigment epithelial cells. The resulting cells were imaged using the LabDroid Maholo’s integrated imaging system, and computer algorithms and statistical analyses were applied to analyze and quantify the level of cell differentiation against the given conditions using the captured images.
The data obtained from these images was then fed into a mathematical experimental design algorithm that uses a batch Bayesian optimization. This allowed the LabDroid Maholo System to determine which factors in the cell culture procedure needed to be optimized to produce more retinal pigment epithelial cells. Using this method allowed Kanda et al. to reduce millions of potential combinations of cell culture parameters to a more manageable number. Briefly, the batch Bayesian optimization used by Kanda et al. first compares the cell culture conditions against the obtained rate of differentiation, and then decides, with the help of artificial intelligence, whether to include further optimization steps. In this case, three successive optimization steps successfully identified the best cell culture conditions to repeatably differentiate stem cells into retinal pigment cells of adequate quality to be used in cell therapy research.
Kanda et al. provide one of the best examples of how to achieve process control and advanced optimization using a combined robotics and artificial intelligence system. The ability to characterize the cultured cells and establish an optimal cell culture protocol based on the data removes human intervention for these steps, reducing variability. Although the platform will need further improvements to provide end-to-end functionality, it is a step forward towards the reproducible manufacturing of cells and cell-derived products for regenerative treatments.
References
-
Comparability of automated human induced pluripotent stem cell culture: a pilot studyBioprocess and Biosystems Engineering 39:1847–1858.https://doi.org/10.1007/s00449-016-1659-9
-
Strategies for achieving measurement assurance for cell therapy productsStem Cells Translational Medicine 5:705–708.https://doi.org/10.5966/sctm.2015-0269
Article and author information
Author details
Publication history
Copyright
© 2022, Sebastian
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 4,287
- views
-
- 390
- downloads
-
- 3
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Computational and Systems Biology
Plasmid construction is central to life science research, and sequence verification is arguably its costliest step. Long-read sequencing has emerged as a competitor to Sanger sequencing, with the principal benefit that whole plasmids can be sequenced in a single run. Nevertheless, the current cost of nanopore sequencing is still prohibitive for routine sequencing during plasmid construction. We develop a computational approach termed Simple Algorithm for Very Efficient Multiplexing of Oxford Nanopore Experiments for You (SAVEMONEY) that guides researchers to mix multiple plasmids and subsequently computationally de-mixes the resultant sequences. SAVEMONEY defines optimal mixtures in a pre-survey step, and following sequencing, executes a post-analysis workflow involving sequence classification, alignment, and consensus determination. By using Bayesian analysis with prior probability of expected plasmid construction error rate, high-confidence sequences can be obtained for each plasmid in the mixture. Plasmids differing by as little as two bases can be mixed as a single sample for nanopore sequencing, and routine multiplexing of even six plasmids per 180 reads can still maintain high accuracy of consensus sequencing. SAVEMONEY should further democratize whole-plasmid sequencing by nanopore and related technologies, driving down the effective cost of whole-plasmid sequencing to lower than that of a single Sanger sequencing run.
-
- Biochemistry and Chemical Biology
- Computational and Systems Biology
Protein–protein interactions are fundamental to understanding the molecular functions and regulation of proteins. Despite the availability of extensive databases, many interactions remain uncharacterized due to the labor-intensive nature of experimental validation. In this study, we utilized the AlphaFold2 program to predict interactions among proteins localized in the nuage, a germline-specific non-membrane organelle essential for piRNA biogenesis in Drosophila. We screened 20 nuage proteins for 1:1 interactions and predicted dimer structures. Among these, five represented novel interaction candidates. Three pairs, including Spn-E_Squ, were verified by co-immunoprecipitation. Disruption of the salt bridges at the Spn-E_Squ interface confirmed their functional importance, underscoring the predictive model’s accuracy. We extended our analysis to include interactions between three representative nuage components—Vas, Squ, and Tej—and approximately 430 oogenesis-related proteins. Co-immunoprecipitation verified interactions for three pairs: Mei-W68_Squ, CSN3_Squ, and Pka-C1_Tej. Furthermore, we screened the majority of Drosophila proteins (~12,000) for potential interaction with the Piwi protein, a central player in the piRNA pathway, identifying 164 pairs as potential binding partners. This in silico approach not only efficiently identifies potential interaction partners but also significantly bridges the gap by facilitating the integration of bioinformatics and experimental biology.






