Divergent regulation of KCNQ1/E1 by targeted recruitment of protein kinase A to distinct sites on the channel complex
Figures
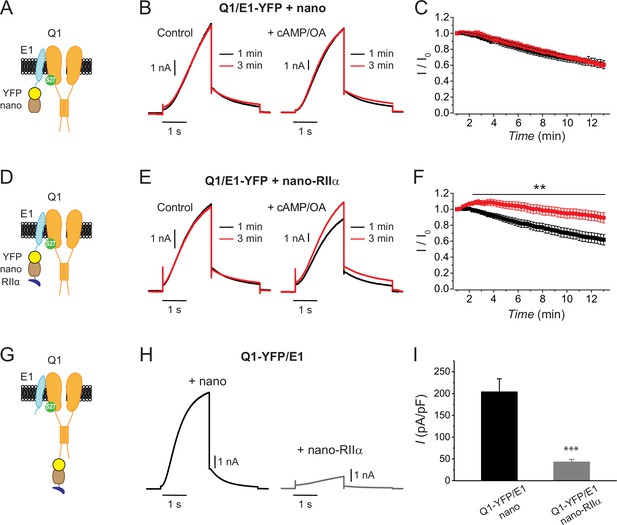
Differential functional effects of nanoRIIα targeted to either KCNE1 or KCNQ1 on IKs.
(A) Cartoon showing targeting GFP/YFP nanobody (nano) to Q1/E1 channel complex via a YFP tag on E1. (B) Exemplar IKs traces elicited by test pulses (+60 mV, –40 mV return) reconstituted in Chinese hamster ovary (CHO) cells expressing Q1/E1-YFP+nano at 1 min (black traces) or 3 min (red traces) after break-in to whole-cell configuration. Cells were dialyzed with internal solution either lacking (left) or including (right) 0.2 mM cAMP+0.2 μM okadaic acid (cAMP/OA). (C) Diary plot of population tail-current amplitudes (mean ± SEM) vs time with cAMP/OA either lacking (black symbols, n=10) or included (red symbols, n=11) in the patch pipette solution. (D–F) Cartoon, exemplar currents and population tail-current amplitude vs time for CHO cells expressing Q1/E1-YFP+nanoRIIα. Same format as (A–C). **p<0.01, two-tailed unpaired t-test. (G) Cartoon showing nanoRIIα targeting to Q1/E1 channel complex via YFP tag on Q1. (H) Exemplar IKs traces reconstituted in CHO cells expressing Q1-YFP/E1 with either nano (left) or nanoRIIα (right). (I) Population current densities (nano, n=26; nanoRIIα, n=17). ***p<0.001, two-tailed unpaired t-test.
-
Figure 1—source data 1
Differential functional effects of nanoRIIα targeted to either KCNE1 or KCNQ1 on IKs.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig1-data1-v2.zip
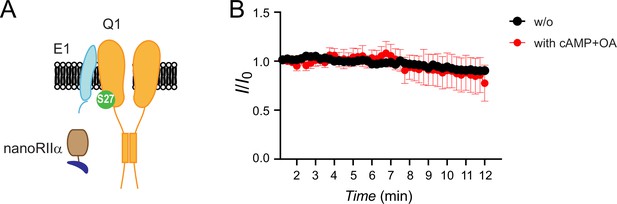
NanoRIIα does not reconstitute protein kinase A (PKA) regulation of IKs when co-expressed with untagged KCNQ1+KCNE1.
(A) Schematic showing nanoRIIα co-expressed with Q1/E1 channel complex. (B) Diary plot of population tail-current amplitudes (mean ± SEM) vs time with cAMP/OA either lacking (black symbols, n=5) or included (red symbols, n=5–9) in the patch pipette solution.
-
Figure 1—figure supplement 1—source data 1
NanoRIIα does not reconstitute protein kinase A (PKA) regulation of IKs when co-expressed with untagged KCNQ1+KCNE1.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig1-figsupp1-data1-v2.zip
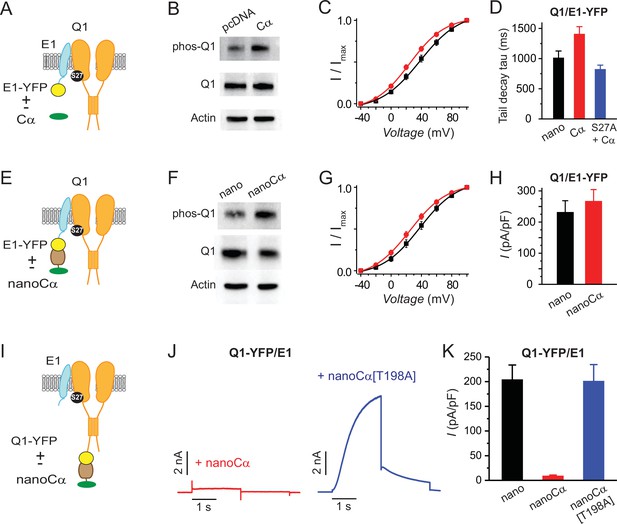
Differential functional effects of nano-Cα targeted to either Q1 or E1 on IKs.
(A) Cartoon showing Q1/E1-YFP complex co-expressed with or without free protein kinase A (PKA) Cα subunit. (B) Representative immunoblots of lysates from HEK293 cells co-expressing Q1/E1-YFP with either empty pcDNA3.1 vector or free Cα. Anti-pKCNQ1 (top) detects phosphorylated KCNQ1-S27, anti-KCNQ1 (middle) detects total KCNQ1, and anti-actin (bottom) detects total actin. N=1. (C) IKs activation curves in Chinese hamster ovary (CHO) cells co-expressing Q1, E1-YFP with either empty pcDNA3.1 vector (black symbols, n=13) or free PKA Cα (red symbols, n=13). (D) Tail-decay times for currents recorded from cells co-expressing Q1/E-YFP+yotiao and either nano or free PKA Cα, or cells co-expressing Q1[S27A]/E1-YFP+yotiao and free PKA Cα (p=0.0532, one-way ANOVA). (E–H) Cartoon, immunoblots, IKs activation curves, and population current densities of Q1/E1-YFP complex expressed with either nano (n=10) or nanoCα (n=10). (I) Cartoon showing targeting of nanoCα to Q1/E1 complex via YFP tag on Q1 C-terminus. (J) Exemplar IKs traces from CHO cells co-expressing Q1-YFP/E1 with either nanoCα (left) or catalytically inactive nanoCα [T198A] mutant (right). (K) Population current densities (nano, n=26; nanoCα, n=19; nanoCα[T198A], n=10).
-
Figure 2—source data 1
Differential functional effects of nano-Cα targeted to either Q1 or E1 on IKs.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig2-data1-v2.zip
-
Figure 2—source data 2
Differential functional effects of nano-Cα targeted to either Q1 or E1 on IKs.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig2-data2-v2.zip
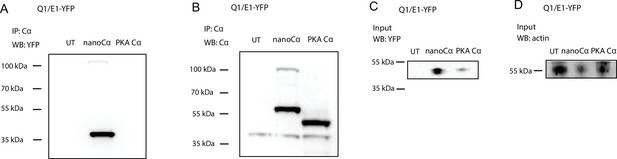
Evidence that nanoCα but not free protein kinase A (PKA) Cα is recruited to E1-YFP in the Q1/E1-YFP channel complex.
Representative immunoblots of lysates from untransfected HEK293 cells (UT) or co-expressing Q1/E1-YFP with either nanoCα or free Cα, immunoprecipitated with anti-PKA Cα and probed with (A) anti-YFP to detect E1-YFP, or (B) anti-PKA Cα to detect nanoCα and free Cα, respectively. (C) Input controls from same samples blotted with anti-YFP. (D) Anti-actin loading controls. N=2 for each blot.
-
Figure 2—figure supplement 1—source data 1
Full immunoblots of experiments showing nanoCα but not free PKA Cα is recruited to E1-YFP in the Q1/E1-YFP channel complex.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig2-figsupp1-data1-v2.zip
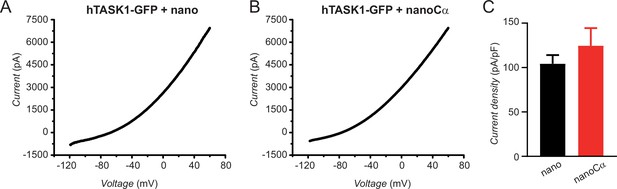
NanoCα targeted to the C-terminus of TASK1 via a GFP tag does not inhibit K+ current.
(A) Exemplar current-voltage relationship elicited by a ramp stimulus (–120 to +60 mV) in a Chinese hamster ovary (CHO) cell expressing hTASK1-GFP+nano. (B) Exemplar current-voltage relationship elicited by a ramp stimulus in a CHO cell expressing hTASK1-GFP+nanoCα. (C) Population current density at 0 mV for cells expressing hTASK1-GFP with either nano (black bar, n=7) or nanoCα (red bar, n=9).
-
Figure 2—figure supplement 2—source data 1
NanoCα targeted to the C-terminus of TASK1 via a GFP tag does not inhibit K+ current.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig2-figsupp2-data1-v2.zip
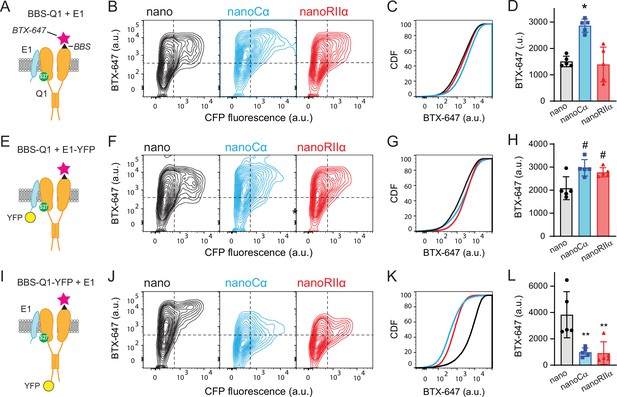
Tethering Cα and RIIα to either E1 or Q1 yields differential effects on channel surface density.
(A) Cartoon showing strategy for surface labeling of BBS-Q1/E1 using BTX-647. (B) Flow cytometry contour plots showing surface channels (BTX-647 fluorescence) and nano expression (CFP fluorescence) in cells expressing BBS-Q1/E1 with nano (left), nanoCα (middle), or nanoRIIα (right). (C) Corresponding cumulative distribution (CDF) histograms of BTX-647 fluorescence. Plot generated from population of CFP-positive cells. (D) Channel surface density (mean BTX-647 fluorescence in CFP-positive cells). *p=0.0003, one-way ANOVA and Tukey HSD post hoc test. (E–H) Cartoon, contour plots, CDF, and average surface labeling of BBS-Q1 in cells expressing BBS-Q1/E1-YFP with nano, nanoCα, or nanoRIIα, same format as A–D. #p<0.05, one-way ANOVA and Tukey HSD post hoc test. (I–L) Cartoon, contour plots, CDF, and normalized average surface labeling of BBS-Q1-YFP in cells expressing BBS-Q1-YFP/E1 with nano, nanoCα, or nanoRIIα, same format as A–D. **p<0.05, one-way ANOVA and Tukey HSD post hoc test.
-
Figure 3—source data 1
Tethering Cα and RIIα to either E1 or Q1 yields differential effects on channel surface density.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig3-data1-v2.zip
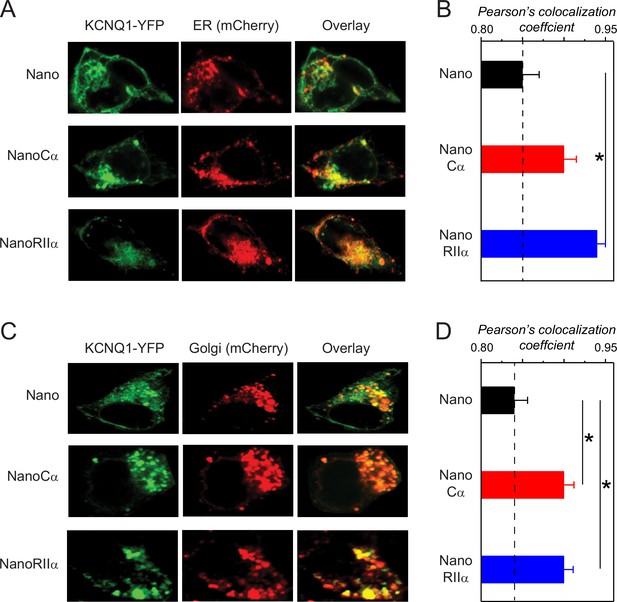
Subcellular localization of KCNQ1 tethered to nano, nanoCα, or nanoRIIα.
(A) Representative confocal images of HEK293 cells expressing Q1-YFP/E1 and ER-mCherry marker with nano, nanoCα, or nanoRIIα. (B) Co-localization of Q1-YFP with ER-mCherry assessed by Pearson’s co-localization coefficient; n=9 for nano, n=7 for nanoCα and n=8 for nanoRIIα. (C) Representative confocal images of HEK293 cells expressing Q1-YFP/E1 and Golgi-mCherry marker with nano, nanoCα, or nanoRIIα. (D) Co-localization of Q1-YFP with ER-mCherry assessed by Pearson’s co-localization coefficient; n=9 for nano, n=8 for nanoCα, and n=6 for nanoRIIα. *p<0.05, one-way ANOVA and Tukey HSD post hoc test.
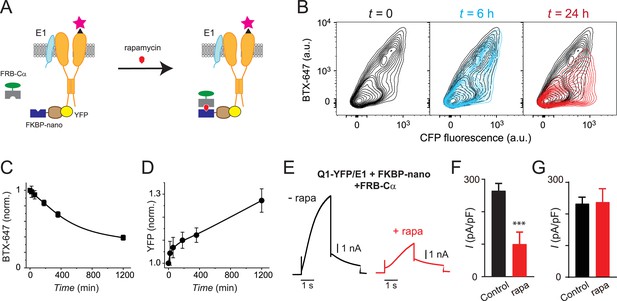
Slow temporal regulation of channel trafficking by targeted induced recruitment of nanoCα to Q1 C-terminus.
(A) Cartoon of FK506 binding protein (FKBP)/FKBP-rapamycin binding domain (FRB) heterodimerization strategy utilized for rapamycin-induced recruitment of engineered Cα to BBS-Q1-YFP/E1. (B) Exemplar flow cytometry contour plots showing surface expression (BTX-647 fluorescence) and CFP fluorescence in cells expressing BBS-Q1-YFP/E1 with FRB-Cα and FKBP-nano at times t=0 (left), t=6 hr (middle), and t=24 hr (right) after rapamycin addition. (C) Normalized mean Q1 surface density (BTX-647 fluorescence) plotted as a function of time after rapamycin induction. (D) Normalized mean Q1 total expression (YFP fluorescence) plotted as a function of time after rapamycin induction. (E) Exemplar IKs traces recorded in Chinese hamster ovary (CHO) cells co-expressing KCNQ1-YFP/KCNE1/nano-FKBP-FRB-Cα incubated 20 hr either without (left) or with (right) rapamycin. (F) Mean current densities in CHO cells co-expressing KCNQ1-YFP/KCNE1/nano-FKBP-FRB-Cα without rapamycin (black, n=10) or after 20 hr rapamycin incubation (red, n=14). ***p<0.001, paired t test. (G) Mean current densities in control cells co-expressing KCNQ1-YFP/KCNE1 without rapamycin (black, n=8) or after 20 hr rapamycin incubation (red, n=9).
-
Figure 5—source data 1
Slow temporal regulation of channel trafficking by targeted induced recruitment of nanoCα to Q1 C-terminus.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig5-data1-v2.zip
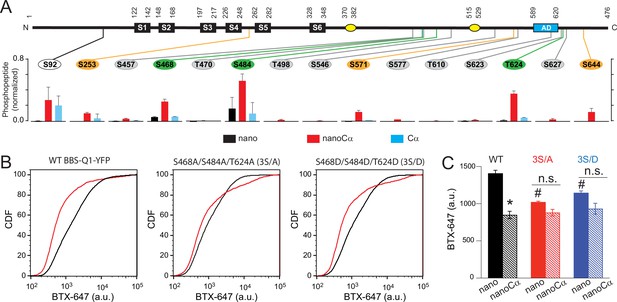
Potential phosphorylation sites involved in protein kinase A (PKA) modulation of KCNQ1 trafficking.
(A) Top, schematic of Q1 showing positions of Ser and Thr residues where phosphorylation was increased when nanoCα was targeted to Q1 C-terminus. Bottom, relative abundance of phosphorylated KCNQ1-YFP peptides identified using mass spectrometry in cells co-expressing nano (black), nanoCα (red), or free Cα (cyan). (B) Exemplar CDF plots showing channel surface density in cells expressing WT BBS-Q1-YFP (left), BBS-3S/A-YFP (middle), or BBS-3S/D-YFP (right) in the absence (black traces) or presence (red traces) of nanoCα. (C) Channel surface density (mean BTX-647 fluorescence in YFP-positive cells) in cells expressing WT BBS-Q1-YFP, BBS-3S/A-YFP, or BBS-3S/D-YFP in the presence of either nano or nanoCα. WT BBS-Q1-YFP (nano, N=4; nanoCα, N=4; *p<0.001, unpaired t-test). BBS-3S/A-YFP (nano, N=4; nanoCα, N=4; p=0.063, unpaired t-test). BBS-3S/D-YFP (nano, N=4; nanoCα, N=4; p=0.079, unpaired t-test). #p<0.001 compared to WT+nano, one-way ANOVA and Tukey HSD post hoc test.
-
Figure 6—source data 1
Potential phosphorylation sites involved in protein kinase A (PKA) modulation of KCNQ1 trafficking.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig6-data1-v2.zip
-
Figure 6—source data 2
Potential phosphorylation sites involved in protein kinase A (PKA) modulation of KCNQ1 trafficking.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig6-data2-v2.zip
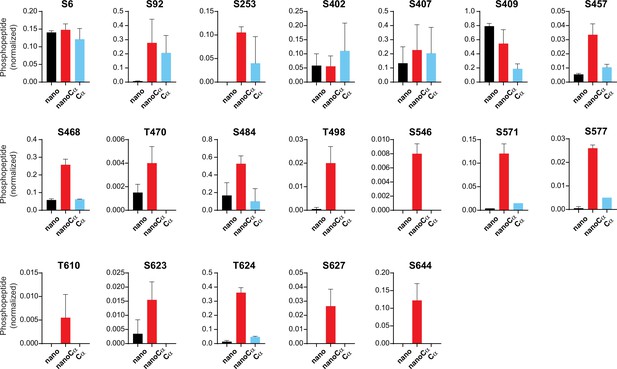
Relative abundance of phosphorylated KCNQ1-YFP peptides identified using mass spectrometry in cells co-expressing nano (black), nanoCα (red), or free Cα (cyan).
-
Figure 6—figure supplement 1—source data 1
Relative abundance of phosphorylated KCNQ1-YFP peptides identified using mass spectrometry in cells co-expressing nano, nanoCα, or free Cα.
- https://cdn.elifesciences.org/articles/83466/elife-83466-fig6-figsupp1-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293 | ATCC | RRID:CVCL_0045 | Laboratory of Dr. Robert Kass |
| Cell line (Homo sapiens) | CHO | ATCC | RRID:CVCL_0214 | CHO-K1, ATCC, CCL-61 |
| Antibody | Anti-Q1 (Rabbit polyclonal) | Alomone | RRID:AB_2040099 | IP (1:1000), WB (1:1000) |
| Antibody | Anti-PKA (Rabbit monoclonal) | Abcam | Cat# ab76238, RRID:AB_1523259 | IP(1:1000) WB (1:1000) |
| Antibody | Anti-actin (Rabbit polyclonal) | Abcam | Cat# ab197345 | WB (1:2000) |
| Antibody | Anti-pQ1 (Rabbit polyclonal) | PMID:12566567 | WB (1:250) | |
| Recombinant DNA reagent | BBS-Q1-YFP (plasmid) | PMID:25344363 | ||
| Recombinant DNA reagent | BBS-Q1 (plasmid) | PMID:25344363 | ||
| Recombinant DNA reagent | Q1-YFP (plasmid) | PMID:25344363 | ||
| Recombinant DNA reagent | Q1 (plasmid) | PMID:25344363 | From the lab of William Kobertz | |
| Recombinant DNA reagent | E1-YFP (plasmid) | PMID:25344363 | ||
| Recombinant DNA reagent | E1 (plasmid) | PMID:25344363 | From the lab of William Kobertz | |
| Recombinant DNA reagent | Yotiao (plasmid) | PMID:15528278 | ||
| Recombinant DNA reagent | Q1[S27A]-YFP | This paper | Made by site-directed mutagenesis; see Plasmid constructs and mutagenesis | |
| Recombinant DNA reagent | NanoCα-P2A-CFP (plasmid) | This paper | Made by gene synthesis (Genewiz) and cloning; see Plasmid constructs and mutagenesis | |
| Recombinant DNA reagent | NanoCα[T198A]-P2A-CFP (plasmid) | This paper | Made by site-directed mutagenesis; see Plasmid constructs and mutagenesis | |
| Recombinant DNA reagent | Cα-P2A-CFP (plasmid) | This paper | Made by gene synthesis (Genewiz) and cloning; see Plasmid constructs and mutagenesis | |
| Recombinant DNA reagent | NanoRIIα-P2A-CFP (plasmid) | This paper | Made by gene synthesis (Genewiz) and cloning; see Plasmid constructs and mutagenesis | |
| Recombinant DNA reagent | Nano-P2A-CFP (plasmid) | PMID:29256394 | ||
| Peptide, recombinant protein | Protein A/G Sepharose beads | Rockland | Cat# PAG50-00-0002 | |
| Peptide, recombinant protein | α-Bungarotoxin, Alexa Fluor 647 conjugate | Thermo Fisher scientific | Cat# B35450 | |
| Commercial assay or kit | Quik-Change Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200523 | |
| Chemical compound, drug | Rapamycin | Sigma | Cat# 553211-1MG | |
| Software, algorithm | FlowJo | FlowJo, LLC | RRID:SCR_008520 | |
| Software, algorithm | GraphPad Prism | GraphPad Software Inc | RRID:SCR_002798 | |
| Software, algorithm | Origin | OriginLab Corporation | RRID:SCR_014212 | |
| Software, algorithm | PulseFit | HEKA |





