Model Organisms: The holy grail of longevity research
Aging is a major risk factor for numerous chronic diseases, such as dementia, metabolic syndromes, and cancers, (Hou et al., 2019; Kennedy et al., 2014; Niccoli and Partridge, 2012), and age-related declines in health are poised to become significant economic and clinical challenges (European Commission, 2014). As a consequence, researchers, governments and drug companies have been trying to identify how aging is influenced by lifestyle choices and by biological, environmental and socio-economic factors (Crane et al., 2022). A key challenge is to develop innovative approaches that can help us to better understand the biology of aging and to accurately quantify age-dependent changes in physiology and cognition. The latter is necessary to evaluate the costs and benefits of potential interventions. Now, in eLife, Anne Brunnet (Stanford University) and colleagues – including Andrew McKay, Emma Costa and Jingxun Chen as joint first authors – report a fresh dimension to this quest (McKay et al., 2022).
Finding appropriate animal models is crucial in biomedicine, but it is rare for a single species (such as mice) to have all the characteristics required and also capture all the aspects of a target species (such as humans). The observation that “all models are wrong, but some are useful” captures this concept succinctly (even if it was first made about statistical models, not animal models; Box, 1976). However, it is possible to overcome this limitation by having a diverse pool of animal models that can help uncover fundamentally conserved phenomena and fuel innovative thinking (Mathuru et al., 2020). This is particularly relevant for aging studies, where longevity can be affected by species-specific adaptations and dramatically divergent evolutionary trajectories.
McKay et al. showcase new technologies and resources for the African turquoise killifish, Nothobranchius furzeri (Figure 1A). Compared to other vertebrate model organisms, killifish have an extremely short life cycle, during which they go through all the stages of life – from a larva to a senile adult – within a few weeks (Harel et al., 2015; Reichard and Polačik, 2019; Terzibasi Tozzini and Cellerino, 2020; Valenzano et al., 2015).
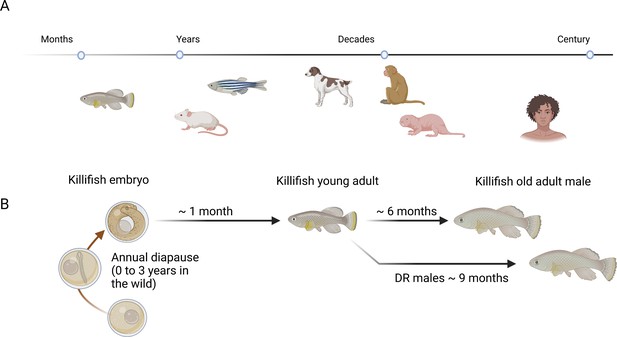
Killifish as a model organism to study aging.
(A) Of the various model organisms used to study aging and longevity in vertebrates, killifish (left) have the shortest lifespans. Other model organisms used to study aging and longevity in vertebrates include (in order of increasing lifespan) mice, zebrafish, dogs, macaques, naked mole rats and humans. (B) In the wild, killifish produce desiccation-resistant eggs and can pause egg development if environmental conditions are unfavorable. Once development resumes, killifish complete their lifecycle within six months. McKay et al. demonstrate that a restricted diet can extend the lifespan of male killifish (DR males) by up to three months.
Image created with BioRender.com.
For the experiments, adult fish were housed in individual transparent tanks. McKay et al. designed an automated feeding system, which has several advantages over the conventional, manual handling systems: it is less invasive; it is more precise and flexible; and it can be deployed across a large number of individual tanks. It is also designed to be open-source, easily transferable, and built from only 25 widely available components.
In a proof-of-concept study, McKay et al. used their design to compare the impact of a favorable diet and a restricted diet on aging. Their results suggest that males (but not females) raised on a restricted diet live longer, and that this change is accompanied by changes in the transcriptional profiles of liver cells (Figure 1B). Sex-specific effects of diet have also been seen in mammals, which suggest that this phenomenon may be widespread among vertebrates. The sex-specific gene expression and their potential connection to lifespan differences raises many interesting questions for future research.
A critical factor in longevity studies is to study the impact of interventions on cognition. Towards this goal, McKay et al. included a red LED light in the design of their automated feeding system: this light switched on a few seconds before the fish were fed, so they can learn to associate the red light with food delivery. After a few repetitions, if the fish learned this association, they would react to the red light switching on as if they expected to be fed. This assay allowed McKay et al. to test the learning abilities and memory retention of the killifish in their home tanks, and how these were affected by age. Even though the design of the associative learning paradigm was simple, McKay et al. were able to demonstrate that killifish rapidly learned such associations in five to eight repetitions.
Overall, the study of McKay et al. opens up a number of exciting possibilities for future studies using killifish. For instance, experiments could focus on investigating how exactly dietary restriction, drug interventions, and sex-specific effects in gene expression intersect with cognitive fitness. The automated feeder should also be useful for studies looking at the effect of specific diet schedules, nutrients and circadian rhythms on longevity. Finally, as new technologies for killifish mature further (Platzer and Englert, 2016), comparative multi-species studies with other species – notably medaka and zebrafish – will become more realistic and offer the promise of even deeper insights into the biology of aging in vertebrates.
References
-
Science and statisticsJournal of the American Statistical Association 71:791–799.https://doi.org/10.1080/01621459.1976.10480949
-
Ageing as a risk factor for neurodegenerative diseaseNature Reviews. Neurology 15:565–581.https://doi.org/10.1038/s41582-019-0244-7
-
Why behavioral neuroscience still needs diversity?: A curious case of a persistent needNeuroscience and Biobehavioral Reviews 116:130–141.https://doi.org/10.1016/j.neubiorev.2020.06.021
-
Ageing as a risk factor for diseaseCurrent Biology 22:R741–R752.https://doi.org/10.1016/j.cub.2012.07.024
-
Nothobranchius furzeri: a model for aging research and moreTrends in Genetics 32:543–552.https://doi.org/10.1016/j.tig.2016.06.006
Article and author information
Author details
Publication history
Copyright
© 2022, Mathuru
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,606
- views
-
- 103
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Chromosomes and Gene Expression
- Genetics and Genomics
Models of nuclear genome organization often propose a binary division into active versus inactive compartments yet typically overlook nuclear bodies. Here, we integrated analysis of sequencing and image-based data to compare genome organization in four human cell types relative to three different nuclear locales: the nuclear lamina, nuclear speckles, and nucleoli. Although gene expression correlates mostly with nuclear speckle proximity, DNA replication timing correlates with proximity to multiple nuclear locales. Speckle attachment regions emerge as DNA replication initiation zones whose replication timing and gene composition vary with their attachment frequency. Most facultative LADs retain a partially repressed state as iLADs, despite their positioning in the nuclear interior. Knock out of two lamina proteins, Lamin A and LBR, causes a shift of H3K9me3-enriched LADs from lamina to nucleolus, and a reciprocal relocation of H3K27me3-enriched partially repressed iLADs from nucleolus to lamina. Thus, these partially repressed iLADs appear to compete with LADs for nuclear lamina attachment with consequences for replication timing. The nuclear organization in adherent cells is polarized with nuclear bodies and genomic regions segregating both radially and relative to the equatorial plane. Together, our results underscore the importance of considering genome organization relative to nuclear locales for a more complete understanding of the spatial and functional organization of the human genome.
-
- Chromosomes and Gene Expression
- Genetics and Genomics
Among the major classes of RNAs in the cell, tRNAs remain the most difficult to characterize via deep sequencing approaches, as tRNA structure and nucleotide modifications can each interfere with cDNA synthesis by commonly-used reverse transcriptases (RTs). Here, we benchmark a recently-developed RNA cloning protocol, termed Ordered Two-Template Relay (OTTR), to characterize intact tRNAs and tRNA fragments in budding yeast and in mouse tissues. We show that OTTR successfully captures both full-length tRNAs and tRNA fragments in budding yeast and in mouse reproductive tissues without any prior enzymatic treatment, and that tRNA cloning efficiency can be further enhanced via AlkB-mediated demethylation of modified nucleotides. As with other recent tRNA cloning protocols, we find that a subset of nucleotide modifications leave misincorporation signatures in OTTR datasets, enabling their detection without any additional protocol steps. Focusing on tRNA cleavage products, we compare OTTR with several standard small RNA-Seq protocols, finding that OTTR provides the most accurate picture of tRNA fragment levels by comparison to "ground truth" Northern blots. Applying this protocol to mature mouse spermatozoa, our data dramatically alter our understanding of the small RNA cargo of mature mammalian sperm, revealing a far more complex population of tRNA fragments - including both 5′ and 3′ tRNA halves derived from the majority of tRNAs – than previously appreciated. Taken together, our data confirm the superior performance of OTTR to commercial protocols in analysis of tRNA fragments, and force a reappraisal of potential epigenetic functions of the sperm small RNA payload.






