The ER folding sensor UGGT1 acts on TAPBPR-chaperoned peptide-free MHC I
Figures
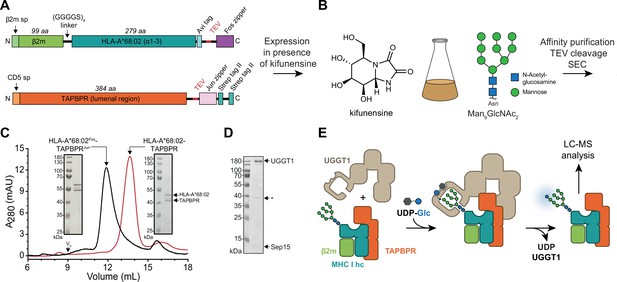
In-vitro system of UGGT1-catalyzed quality control and reglucosylation of MHC I.
(A) Construct design to obtain peptide-receptive HLA-A*68:02-TAPBPR complexes by transient co-transfection of HEK293-F cells. (B) Co-expression of HLA-A*68:02 and TAPBPR was performed in the presence of the α-mannosidase I inhibitor kifunensine to generate a defined Man9GlcNAc2 glycan tree on the MHC I that can be recognized by UGGT1. (C) The secreted leucine zippered complex was isolated from the cell culture supernatant by Strep-Tactin affinity purification. Leucine zippers and the Twin-Strep-tag of TAPBPR were removed by protease treatment. The MHC I-chaperone complexes were further analyzed and purified by size exclusion chromatography (SEC) on a Superdex 200 (Increase 10/300) column. (D) SDS-PAGE analysis of immobilized metal-affinity chromatography (IMAC)-purified human wildtype (wt) UGGT1 co-expressed with Sep15 and secreted from insect cells. The asterisk (*) indicates a degradation product of UGGT1. (E) The purified UGGT1-Sep15 complex was employed in the reglucosylation assay with the peptide-receptive HLA-A*68:02-TAPBPR complex of (C) harboring the high-mannose glycan on MHC I. Abbreviations: A280: absorption at 280 nm; aa: amino acids; kDa: kilodalton; mAU: milli-absorption units; MHC I hc: MHC I heavy chain; sp: signal peptide; UDP-Glc: UDP-glucose; V0: void volume.
-
Figure 1—source data 1
Original SDS-PAGE gel of HLA-A*68:02Fos-TAPBPRJun, Figure 1C.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig1-data1-v2.zip
-
Figure 1—source data 2
Original SDS-PAGE gel of HLA-A*68:02-TAPBPR, Figure 1C.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig1-data2-v2.zip
-
Figure 1—source data 3
Original SDS-PAGE gel of UGGT1-Sep15, Figure 1D.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig1-data3-v2.zip
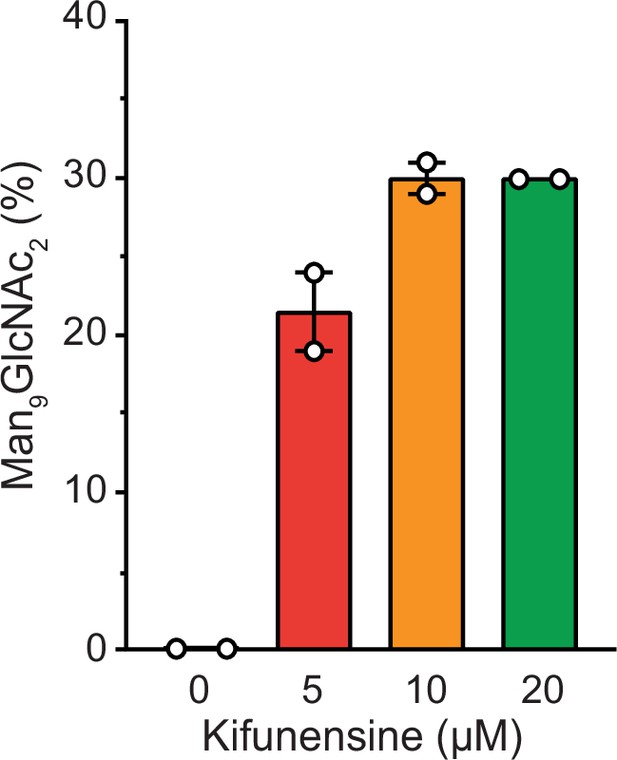
Influence of kifunensine concentration on the proportion of Man9GlcNAc2 among the Man7-9GlcNAc2 MHC I glycan species when co-expressed with TAPBPR in HEK293-F cells.
The glycan status of TAPBPR-bound HLA-A*68:02 was analyzed by LC-MS after expression in presence of various concentrations of kifunensine. Proportion of Man9GlcNAc2 was calculated based on the total glycan intensity (Man7-9GlcNAc2). Data represent mean ± SEM (n=2) from two independent protein expressions and purifications.
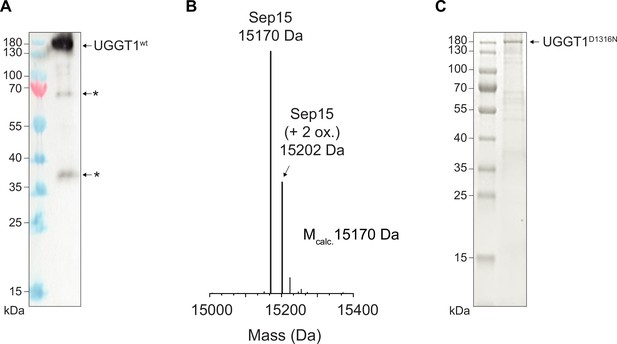
Analysis of purified human UGGT1.
(A) Anti-UGGT1 immunoblot analysis after immobilized metal-affinity chromatography (IMAC) purification of wildtype (wt) human UGGT1 co-expressed with Sep15 and secreted from insect cells. The asterisks (*) indicate degradation products of UGGT1. (B) Compositional analysis of purified UGGT1wt-Sep15 by liquid chromatography-mass spectrometry (LC-MS) (deconvoluted spectrum). The voltages and desolvation temperature were adjusted for optimal ionization of the 15 kDa protein Sep15, precluding simultaneous efficient ionization of the 175 kDa protein UGGT1 within the same measurement. (C) Non-reducing SDS-PAGE analysis of the catalytically inactive UGGT1D1316N mutant expressed in HEK293-F cells and purified by IMAC.
-
Figure 1—figure supplement 2—source data 1
Original anti-UGGT1 immunoblot, Figure 1—figure supplement 2A.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig1-figsupp2-data1-v2.zip
-
Figure 1—figure supplement 2—source data 2
Original SDS-PAGE gel of UGGT1D1316N, Figure 1—figure supplement 2C.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig1-figsupp2-data2-v2.zip
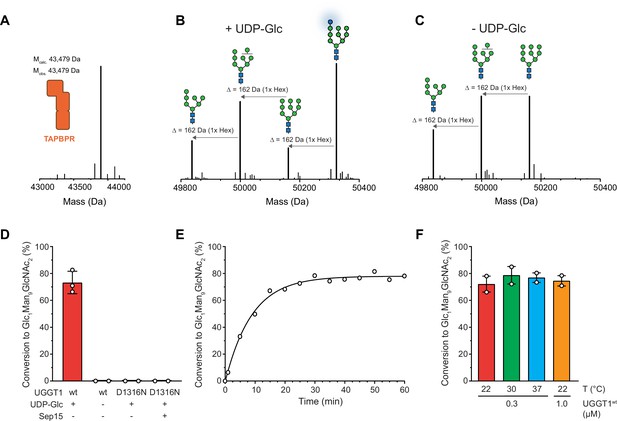
UGGT1 catalyzes reglucosylation of TAPBPR-bound HLA-A*68:02.
Protein identities in the reconstituted glucosylation system and the glycan status of HLA-A*68:02 (Man7-9GlcNAc2 and Glc1Man9GlcNAc2) during the UGGT1-catalyzed reglucosylation reaction were determined by liquid chromatography-mass spectrometry (LC-MS). (A) Deconvoluted mass spectrum of TAPBPR. (B) HLA-A*68:02-TAPBPR (3 µM) complex was incubated with UGGT1 (1 µM) in the absence (-) and (C) presence (+) of UDP-glucose (UDP-Glc) for 1 h at room temperature. In the presence of UDP-Glc, UGGT1 catalyzed the glucose transfer, generating the mono-glucosylated glycan (Glc1Man9GlcNAc2), whereas the native glycan pattern remained unchanged in the absence of UDP-Glc. Exemplary deconvoluted mass spectra of glycosylated HLA-A*68:02 representing LC-MS analyses summarized in (D) are shown. (D) Comparison of glucosyltransferase activity, as measured by the amount of produced Glc1Man9GlcNAc2-HLA-A*68:02, upon incubating HLA-A*68:02-TAPBPR (3 µM) with UGGT1 proteins (1 µM) in the absence (-) and presence (+) of UDP-Glc and additional purified Sep15 (5 µM), respectively. Data represent mean ± SD (n=3) and (n=2), respectively. (E) Kinetics of reglucosylation of HLA-A*68:02-TAPBPR (3 µM) catalyzed by UGGT1 (1 µM). (F) Temperature dependence of UGGT1-catalyzed reglucosylation of HLA-A*68:02-TAPBPR (3 µM). Reactions were stopped after 60 min. Data represent mean ± SEM (n=2). Abbreviations: Da: dalton; Glc: glucose; GlcNAc: N-acetylglucosamine; Hex: hexose; Man: mannose; Mcalc: calculated mass; Mobs: observed mass.

UGGT1 concentration-dependent reglucoslyation kinetics of TAPBPR-bound HLA-A*68:02.
Dependence of Man9GlcNAc2-HLA-A*68:02-TAPBPR (3 µM) reglucosylation kinetics on UGGT1 concentration. Reactions were stopped after 10 min by adding EGTA (25 mM). Data represent mean ± SEM (n=2).
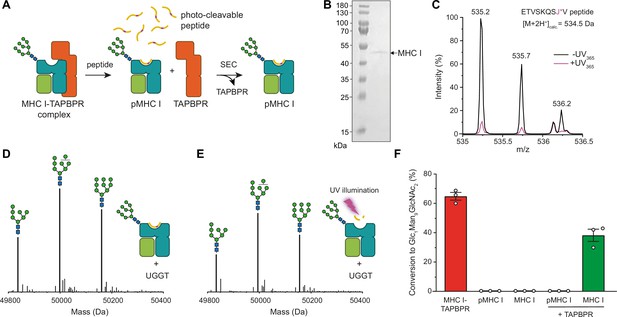
TAPBPR is indispensable for UGGT1-catalyzed reglucosylation of HLA-A*68:02.
(A) Schematic representation of loading a photo-cleavable peptide onto Man9GlcNAc2-HLA-A*68:02 via pre-assembled MHC I-TAPBPR complexes. Peptide binding results in TAPBPR dissociation, permitting isolation of photo-conditional pMHC I complexes. (B) Non-reducing SDS-PAGE analysis of SEC-isolated pMHC I. (C) Isotope-resolved raw mass spectrum of the double-charged photo-cleavable peptide ETVSKQSJ*V (J*, 3-amino-3-(2-nitrophenyl)-propanoic acid) bound to HLA-A*68:02 before (black) and after (violet) UV illumination at 365 nm. (D) Reglucosylation experiment of peptide-loaded and (E) peptide-free HLA-A*68:02 (1 µM) in the absence of TAPBPR. Man9GlcNAc2-HLA-A*68:02 devoid of peptide was produced by photo-cleavage and release of peptide fragments prior to incubation with UGGT1 (600 nM). Shown are representative LC-MS analyses of data depicted in F. (F) Reglucosylation activity of UGGT1 (600 nM) towards TAPBPR-bound HLA-A*68:02 (1 µM), peptide-bound HLA-A*68:02 (pMHC I, 1 µM), and UV-illuminated HLA-A*68:02 lacking peptide (1 µM). If TAPBPR (6 µM) is added back to peptide-deficient MHC I (1 µM), recognition of MHC I as a substrate by UGGT1 is restored. The amount of produced Glc1Man9GlcNAc2 glycan was determined after 60 min for all reactions. Data represent mean ± SEM (n=3). Abbreviations: calc.: calculated; Da: dalton; kDa: kilodalton; SEC: size-exclusion chromatography.
-
Figure 3—source data 1
Original SDS-PAGE gel of SEC isolated HLA-A*68:02, Figure 3B.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig3-data1-v2.zip

SEC-MS analysis of photo-cleavable peptide bound to Man9GlcNAc2-HLA-A*68:02 before and after UV illumination.
This bar diagram quantifies the SEC-MS measurements in Figure 3C of bound peptide before (grey) and after UV exposure (purple). The N- and C-terminal peptide fragments generated by UV exposure are released from the MHC I peptide-binding groove. Data represent mean ± SEM (n=3).

Conformation-specific antibody W6/32 binds to peptide-loaded and peptide-receptive β2m-HLA-A*68:02.
The binding of the W6/32 antibody (2 µM) to (A) peptide-loaded HLA-A*68:02 (1 µM) and (B) peptide-free UV-illuminated HLA-A*68:02 (1 µM) as analyzed by UPLC-SEC. (C) UV-illuminated, peptide-free HLA-A*68:02 (1 µM) was incubated for 60 min at 21 °C prior to incubation with W6/32 (2 µM) and UPLC-SEC analysis. SEC profiles of 1 µM pMHC I and 2 µM W6/32, respectively, are shown as dashed lines for all UPLC-SEC runs.
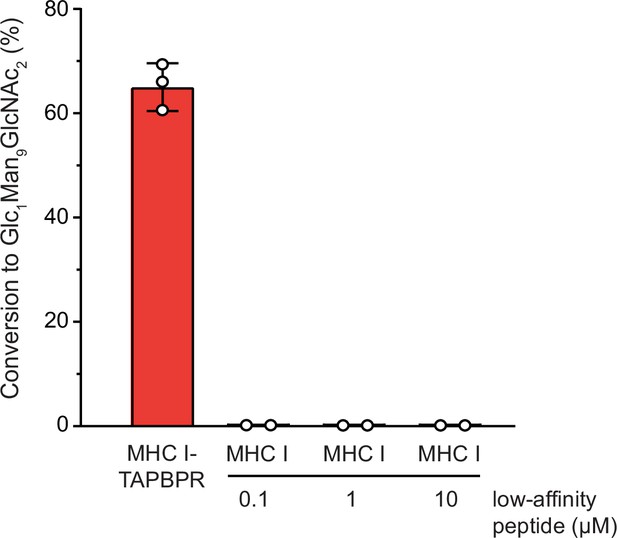
UGGT1-mediated reglucosylation is independent of the MHC I peptide-loading status.
Reglucosylation activity of UGGT1 (600 nM) towards TAPBPR-bound HLA-A*68:02 (1 μM) and MHC I (1 µM) loaded with low-affinity peptide. Pre-incubation of the UV-illuminated HLA-A*68:02 (1 μM) with various concentrations of low-affinity peptide could not restore the reglucosylation activity. The original data for HLA-A*68:02-TAPBPR are depicted in Figure 3 and are shown here for comparison. Data represent mean ± SEM (n=2/3).
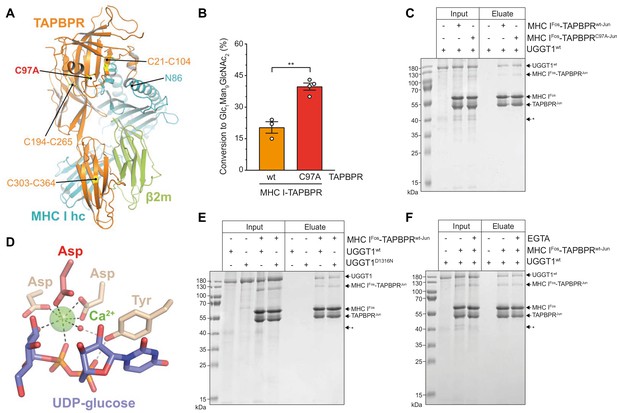
The interaction between UGGT1 and TAPBPR is independent of Cys97 and Ca2+.
(A) Cartoon representation of the MHC I-TAPBPR complex (PDB ID: 5opi). Disulfide bridges in TAPBPR and C97, which was mutated to alanine, are shown as sticks and highlighted in yellow. The glycosylated N86 (depicted as sticks) of the MHC I heavy chain (hc) lies in the vicinity of C97. (B) UGGT1wt-catalyzed (1 µM) reglucosylation of Man9GlcNAc2-HLA-A*68:02 bound to TAPBPRwt (3 µM) or TAPBPRC97A (3 µM). Data represent mean ± SD (n=3 and 4). Statistical analysis was performed using unpaired t-test. Asterisks indicate the level of significance (p-values): **p≤0.01. (C) Pull-down experiment with UGGT1wt (3 µM, present in each lane) using Twin-Strep-tagged Man9GlcNAc2-HLA-A*68:02-TAPBPR complexes (5 µM) captured on Strep-Tactin Sepharose. (D) Ca2+ (green sphere) coordination in the active site of the Thermomyces dupontii UGGT glucosyltransferase domain (PDB ID: 5h18). The side chain of the corresponding aspartate residue that has been mutated to asparagine in human UGGT1D1316N (see panel (F)) is shown in red. The red sphere represents a water molecule that occupies one of the coordination sites. (E) Pull-down experiment with UGGT1wt (5 µM) and the catalytically inactive mutant UGGT1D1316N (5 µM) using Twin-Strep-tagged Man9GlcNAc2-HLA-A*68:02-TAPBPRwt complex (3 µM) in the presence of 5 mM CaCl2. Eluates of the pull-down experiments were analyzed by SDS-PAGE (12.5%, non-reducing, Coomassie-stained). (F) Ca2+ dependence of UGGT1wt (3 µM) binding to Man9GlcNAc2-HLA-A*68:02-TAPBPRwt (5 µM) was analyzed in presence (+) and absence (-) of EGTA. The asterisk (*) indicates a degradation product of UGGT1, representing the glycosyltransferase domain. The data are representative of two independent experiments. Abbreviation: kDa: kilodalton. The numbering of TAPBPR refers to the mature protein as defined by N-terminal sequencing (Zhang and Henzel, 2004).
-
Figure 4—source data 1
Original SDS-PAGE gel of pull-down experiment with UGGT1wt, Figure 4C.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig4-data1-v2.zip
-
Figure 4—source data 2
Original SDS-PAGE gel of pull-down experiment with UGGT1wt and UGGT1D1316N, Figure 4E.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig4-data2-v2.zip
-
Figure 4—source data 3
Original SDS-PAGE gel of pull-down experiment to test Ca2+ dependence of UGGT1wt, Figure 4F.
- https://cdn.elifesciences.org/articles/85432/elife-85432-fig4-data3-v2.zip
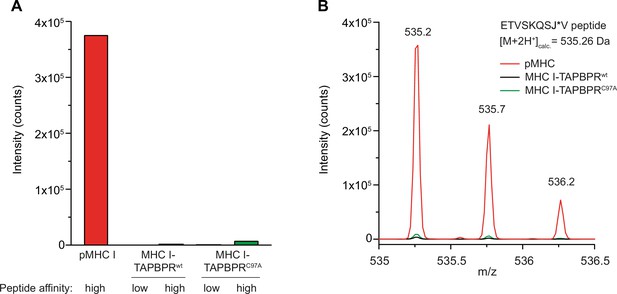
Tethered MHC IFos-TAPBPRJun complexes are not competent in peptide-binding.
(A) The peptide content of tethered MHC IFos-TAPBPRJun complexes (3 µM) after pre-incubation with a 250-fold molar excess of low- or high-affinity peptides was analyzed by LC-MS. Neither high-affinity nor low-affinity peptides can bind to tethered MHC I-chaperone complexes containing wildtype (wt) or C97A TAPBPR. (B) Isotope-resolved raw spectrum of the double-charged mass of the high-affinity ETVSKQSJ*V peptide as shown in A.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (human) | TAPBPRwt-Jun | O’Rourke et al., 2019 | Lumenal region | |
| Gene (human) | TAPBPRC97A-Jun | This study | Lumenal region, TAPBPR version of TAPBPRwt-Jun (see material and methods, DNA constructs) | |
| Gene (human) | β2m-HLA-A*68:02 | O’Rourke et al., 2019 | Lumenal region | |
| Gene (human) | UGGT1wt-Sep15 | This study | See Materials and methods, DNA constructs | |
| Gene (human) | UGGT1D1316N | This study | UGGT version of UGGT1wt (Dr. Pietro Roversi, Zitzmann lab, University of Oxford) | |
| Gene (human) | Sep15 | This study | See Materials and methods, DNA constructs | |
| Strain, strain background (Escherichia coli) | BL21 (DE3) | Sigma-Aldrich | CMC0014 | Chemically competent |
| Cell line (Spodoptera Frugiperda) | Sf21 | Thermo Fisher Scientific | 11496015 | |
| Cell line (Human embryonic kidney) | HEK293-F | Thermo Fisher Scientific | R79007 | Routinely tested for mycoplasma contamination |
| Antibody | anti-human HLA-A/B/C (W6/32) (Mouse monoclonal) | BioLegend | Cat# 311402 RRID:AB_314871 | WB (1:3000) |
| Antibody | anti-UGGT/UGT1 (Rabbit monoclonal) | Abcam | Cat# ab124879 RRID:AB_10971344 | WB (1:3000) |
| Recombinant DNA reagent | pFastBacI-gp67 | PMID:2905996 | Transfer vector for Bac-to-Bac system | |
| Recombinant DNA reagent | pcDNA3.1(+) | Thermo Fisher Scientific | V79020 | Mammalian expression vector |
| Recombinant DNA reagent | pOPINGTTGneo | Dr. Pietro Roversi, Zitzmann lab, University of Oxford | Mammalian expression vector | |
| Peptide, recombinant protein | ETVSKQSJ*V | Dr. Ines Katharina Müller, Tampé lab | J* denotes photo-cleavable amino acid, see material and methods, peptide loading onto MHC I | |
| Peptide, recombinant protein | ILKCLEEPSV | This study | See Materials and methods, UGGT1-mediated reglucosylation | |
| Chemical compound, drug | UDP-Glc | Sigma-Aldrich | U4625 | |
| Chemical compound, drug | Kifunensine | BioMol | 10009437 | |
| Chemical compound, drug | VPA | Merck Millipore | P4543 | |
| Chemical compound, drug | Polyethyleneimine, linear | Sigma-Aldrich | 765090 | |
| Software, algorithm | OriginPro 2022b | |||
| Software, algorithm | Prism 10 | GraphPad Software | ||
| Software, algorithm | Unify 1.9.4.053 | Walters | ||
| Other | Superdex 200 Increase 10/300 | GE Healthcare | 28990944 | SEC column, see Materials and methods, Purification of MHC IFos-TAPBPRJun complexes |
| Other | Superdex 200 Increase 3.2/300 | GE Healthcare | 28990946 | SEC column, see Materials and methods, peptide loading onto MHC I |
| Other | UPLC Peptide BEH C18 Column | Walters | 186003555 | UPLC-column, see Materials and methods, LC-MS analysis |
| Other | UPLC Protein BEH C4 Column | Walters | 186008471 | UPLC-column, see Materials and methods, LC-MS analysis |
| Other | Äkta Purifier | Cytiva | Protein purification | |
| Other | Äkta Ettan | GE Healthcare | Protein purification | |
| Other | BioAccord LC-MS system | Walters | See Materials and methods, LC-MS analysis | |
| Other | Tabacco Etch Virus | In house production | TEV protease cleavage |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85432/elife-85432-mdarchecklist1-v2.pdf
-
Source data 1
This file contains all orignal gels, immunoblots, and data.
- https://cdn.elifesciences.org/articles/85432/elife-85432-data1-v2.xlsx





