Caspase-mediated nuclear pore complex trimming in cell differentiation and endoplasmic reticulum stress
Figures
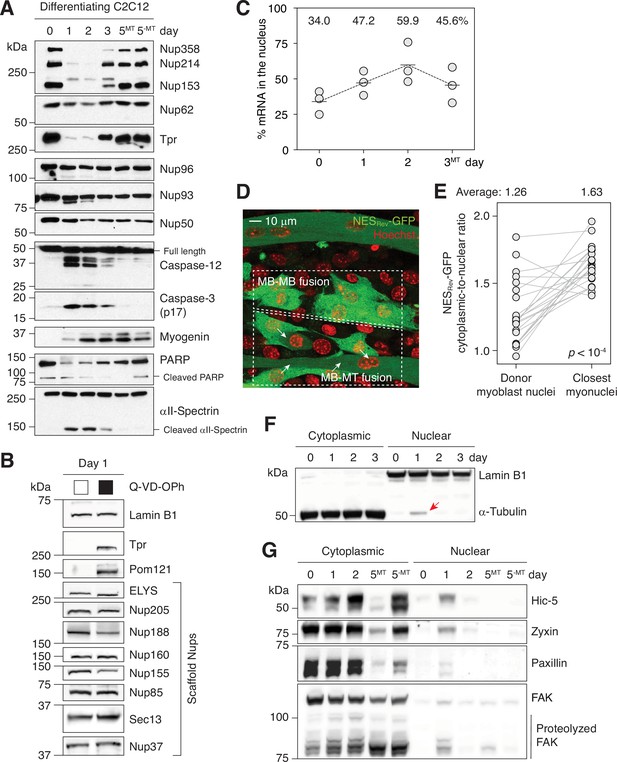
Caspases fully proteolyze 5 peripheral Nups during myogenesis.
(A) Immunoblots showing the degradation of Nups, PARP, and αII-spectrin, the activation of caspase-3/12 and the upregulation of myogenin in differentiating C2C12 cells. (B) Caspase-dependent proteolysis of Nups in differentiating C2C12 cells assessed by immunoblotting. Q-VD-OPh used at 30 μM. (C) Nuclear-to-total mRNA ratio in C2C12 cells undergoing myogenesis. The average value of three replicates shown for each time point. (D) A stable C2C12 cell line that expresses NESRev-GFP. In dashed boxes are cells undergoing MB-to-MB or MB-to-MT fusion. (E) Cytoplasmic-to-nuclear ratio of NESRev-GFP was quantified for 22 donor myoblast / closest myonuclei pairs. Grey lines link each pair. (F) Localization of lamin B1 and α-tubulin in differentiating C2C12 cells assessed by immunoblotting. Nuclear α-tubulin marked with an arrow. Thirty μg of protein loaded per lane. (G) Cytoplasmic and nuclear levels of focal adhesion proteins in differentiating C2C12 cells. Twenty-five μg of protein loaded per lane. MT: myotubes, MB: myoblasts.
-
Figure 1—source data 1
Unprocessed gel images.
- https://cdn.elifesciences.org/articles/89066/elife-89066-fig1-data1-v1.zip
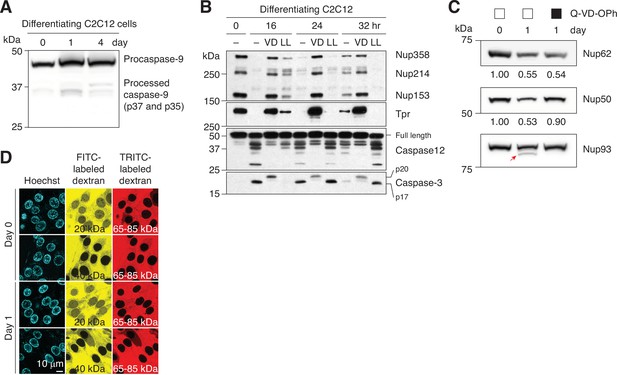
Caspase-mediated NPC trimming in differentiating C2C12 cells.
(A) Activation of caspase-9 in differentiating C2C12 cells was assessed by immunoblotting. (B) Caspase-3 and –12 activation and peripheral Nup degradation during myogenesis were assessed in the absence or presence of a pan-caspase inhibitor (Q-VD-OPh [‘VD’], 30 μM) or a pan-calpain inhibitor (Z-LL-CHO [‘LL’], 50 μM) by western blotting. (C) C2C12 cells were differentiated in the absence or presence of 30 μM Q-VD-OPh. The levels of Nup62 and Nup50 were determined by densitometric analysis. Secondary band of Nup93 is marked with a red arrow. (D) NPC passive permeability barrier in C2C12 cells undergoing myogenic differentiation was assessed by dextran exclusion assay.
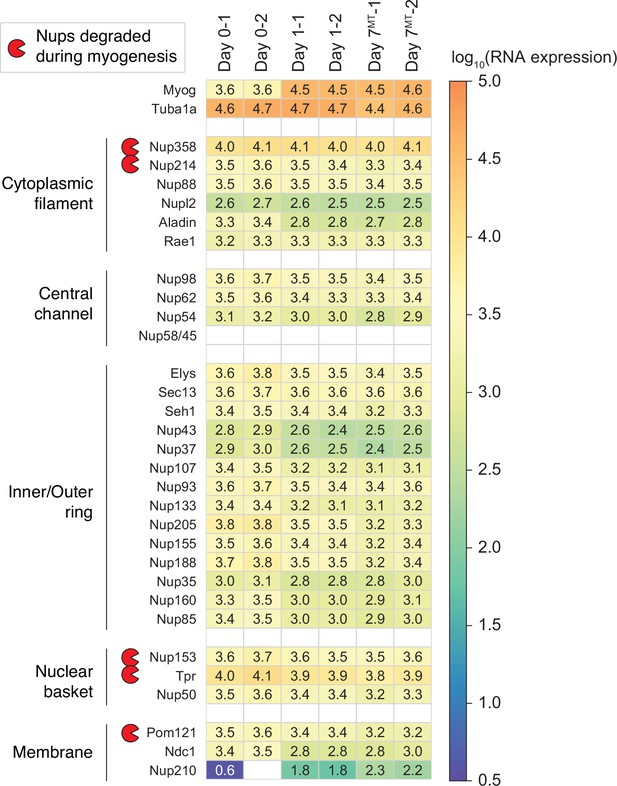
Transcript levels of NPC subunits in differentiating C2C12 cells.
Using RNA-seq, we monitored how the transcript levels of 30 Nups change over the course of myogenesis. Those of Myog and Tuba1a are also shown for comparison.
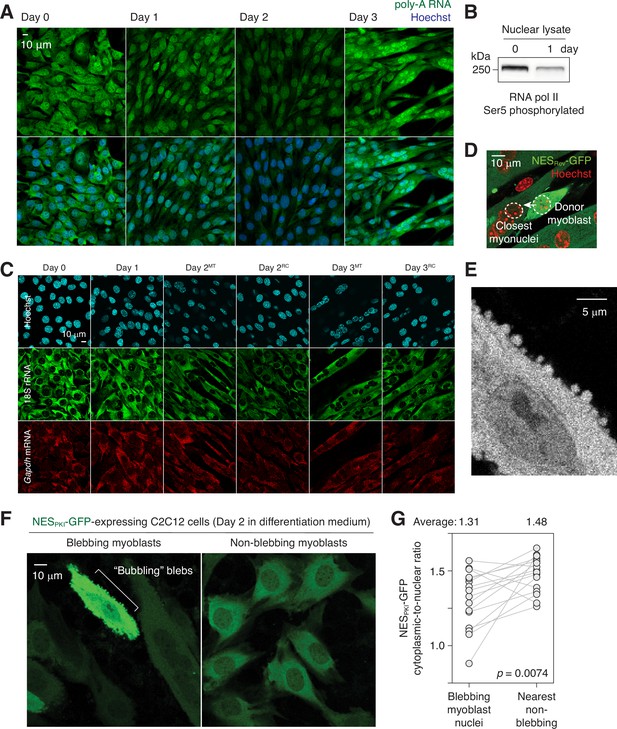
Nuclear export impairment in differentiating C2C12 cells.
(A) poly-A RNAs were visualized in differentiating C2C12 cells by fluorescence in situ hybridization. (B) Ser5 phosphorylation level of RNA polymerase II in days 0 and 1 C2C12 cells. (C) 18 S rRNA and Gapdh mRNA fluorescence in situ images of differentiating C2C12 cells. MT: myotubes, RC: reserve cells. (D) Representative image showing how ‘donor myoblast’ and ‘closest myonuclei’ were determined. (E) Bubbling blebs on the plasma membrane of a fusion-competent myoblast. (F) Live imaging of C2C12 cells stably expressing NESPKI-GFP. Imaged 48 hours after switching from growth to differentiation medium. (G) Cytoplasmic-to-nuclear ratio of NESPKI-GFP was quantified for 16 pairs of blebbing and non-blebbing cells. Grey lines link each pair.
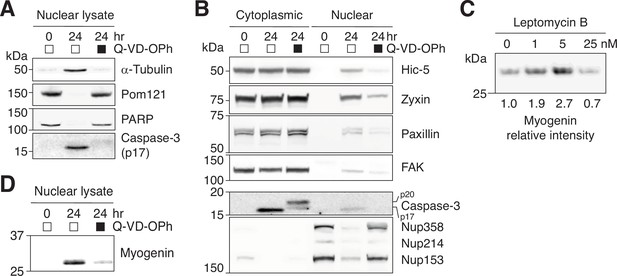
Nuclear retention of NES-containing proteins in differentiating C2C12 cells.
(A) Nuclear translocation of α-tubulin and (D) myogenin upregulation were assessed in C2C12 cells differentiated in the absence or presence of a pan-caspase inhibitor, Q-VD-OPh (30 μM). (B) C2C12 cells were differentiated in the absence or presence of Q-VD-OPh (30 μM) for 24 hr, and nuclear and cytoplasmic fractions were obtained. Nuclear accumulation of NES-containing focal adhesion proteins and caspase-mediated NPC trimming were examined by western blotting. A total of 17.5 μg of cytoplasmic or nuclear protein loaded per lane. (C) C2C12 cells were differentiated in the absence or presence of leptomycin b for 24 hr, and myogenin expression was determined by western blotting.
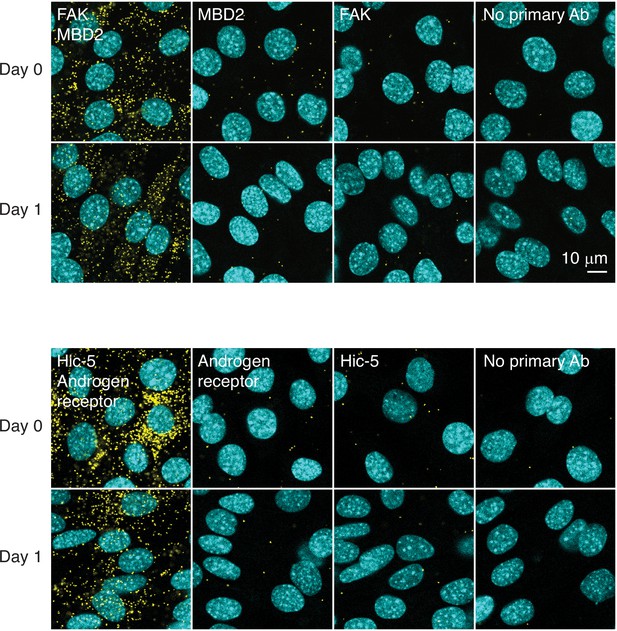
Proximity ligation assay.
Interaction between FAK and MBD2 and between Hic-5 and androgen receptor was assessed by proximity ligation assay (yellow puncta) in differentiating C2C12 cells.
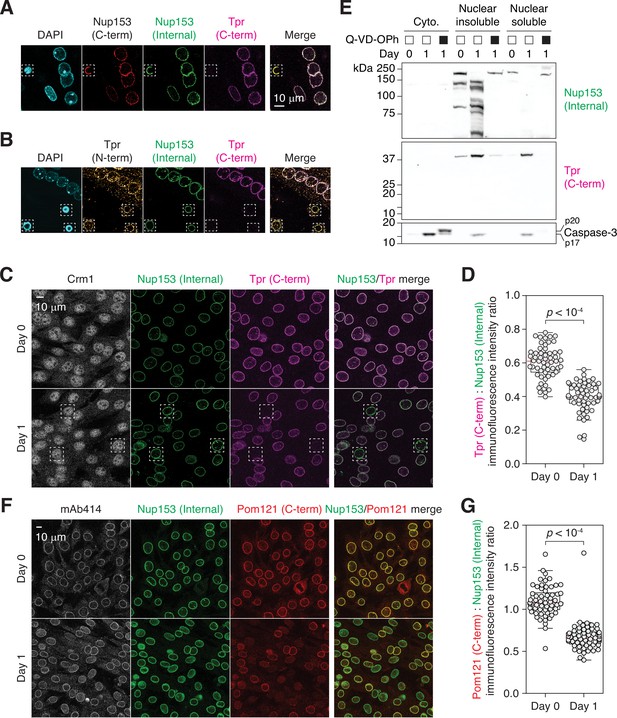
Tpr and Pom121 C-terminal fragments dissociate from the NPC during myogenesis.
(A and B) C2C12 cells immunostained using two Nup153 and two Tpr antibodies. In dashed boxes are apoptotic nuclei. (C) C2C12 cells undergoing myogenesis immunostained for Crm1, Nup153, and Tpr. In dashed boxes are TprC– cells. (D) Tpr C-term immunoreactivity (normalized against Nup153 internal domain immunoreactivity) determined for 50+nuclei. (E) Nup153, Tpr, and caspase-3 fragments in subcellular fractions of differentiating C2C12 cells visualized by immunoblotting. Q-VD-OPh used at 30 μM. (F) C2C12 cells undergoing myogenesis immunostained for mAb414, Nup153, and Pom121. (G) Pom121 C-term immunoreactivity (normalized against Nup153 internal domain immunoreactivity) determined for 50+nuclei.
-
Figure 2—source data 1
Unprocessed gel images.
- https://cdn.elifesciences.org/articles/89066/elife-89066-fig2-data1-v1.zip
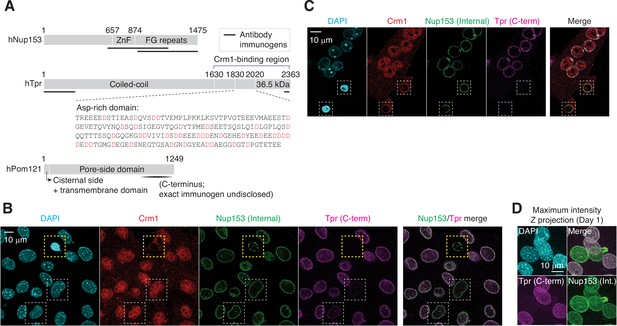
Immunofluorescence staining of apoptotic and differentiating C2C12 cells.
(A) Domain architecture of human Nup153, Tpr, and Pom121. Immunogenic fragments used to generate Nup153, Tpr, and Pom121 antibodies are mapped on each protein. Aspartates (D) in Tpr aspartate-rich region are colored in red. (B) C2C12 cells that have undergone myogenic differentiation for a day. In yellow dashed box is an apoptotic cell, and in white dashed boxes, differentiating TprC– cells. (C) C2C12 cells were immunostained for Crm1, Nup153, and Tpr. Apoptotic nuclei with condensed chromatin are shown in dashed boxes. (D) TprC– cells were identified by immunofluorescence. Maximum intensity Z projection micrographs were reconstructed from confocal imaging.
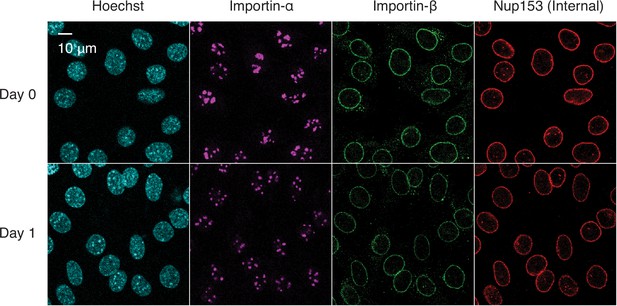
Immunofluorescence staining of differentiating C2C12 cells.
C2C12 cells undergoing myogenesis were immunostained for importin-α, importin-β, and Nup153.
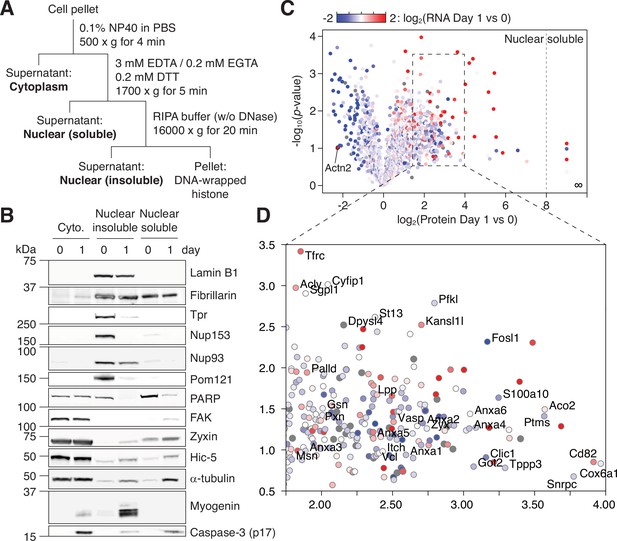
NPC trimming accompanies nuclear accumulation of cytoplasmic, plasma membrane, and mitochondrial proteins.
(A) Preparation of subcellular fractions for quantitative mass spectrometry. (B) Subcellular fractions were validated using multiple protein markers. (C) Volcano plot showing how protein levels in the ‘nuclear soluble’ fraction change during the first 24 hr of myogenic differentiation. Each data point colored to represent their transcriptional change during the same time frame. (D) Zoomed-in view of the dotted square in (C).
-
Figure 3—source data 1
Unprocessed gel images.
- https://cdn.elifesciences.org/articles/89066/elife-89066-fig3-data1-v1.zip
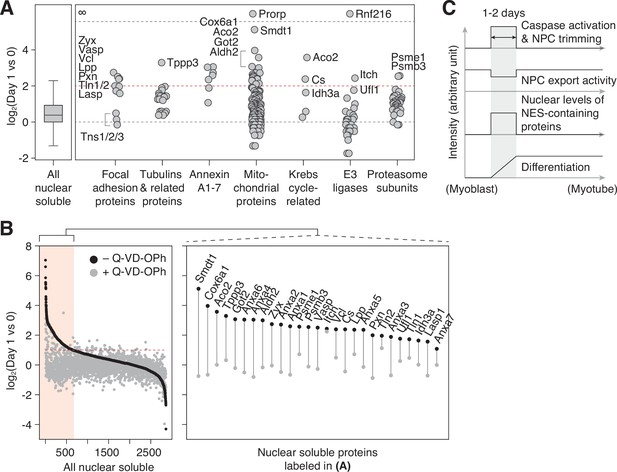
NPC trimming accompanies nuclear accumulation of cytoplasmic, plasma membrane, and mitochondrial proteins.
(A) Protein groups detected in the ‘nuclear soluble’ fraction. Red dotted line: y=2. (B) Protein level change in the ‘nuclear soluble’ fraction when C2C12 cells are differentiated with or without 30 μM Q-VD-OPh. Red dotted line: y=1. Data points for proteins labeled in (A) are shown in the right box. (C) Visual summary of multiple events that take place during myoblast-to-myotube transition.
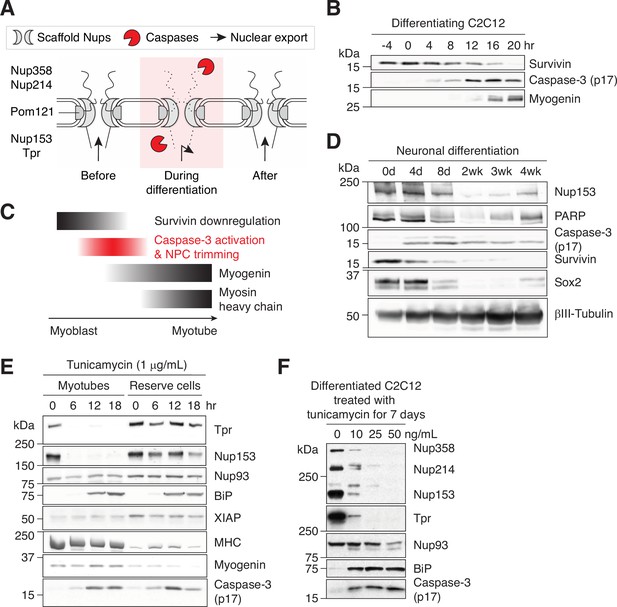
Caspase-mediated NPC trimming occurs during neurogenesis and ER stress.
(A) Schematic representation of caspase-mediated NPC trimming. (B) Down-/up-regulation of survivin, caspase-3, and myogenin in differentiating C2C12 cells. (C) Relative timing of key events in myogenesis. (D) Expression levels of Nup153, PARP, active caspase-3, survivin, and neuronal differentiation markers (Sox2 and βIII-tubulin) were determined in differentiating neurons. (E) Acute ER stress induced using 1 μg/mL tunicamycin in C2C12 myotubes and reserve cells. The levels of Nups, BiP, XIAP, myosin heavy chain (MHC), myogenin, and active caspase-3 levels were monitored by immunoblotting. (F) Chronic ER stress was induced in differentiated C2C12 cells for 7 days using low doses of tunicamycin. Nups, BiP, and active caspase-3 levels were assessed.
-
Figure 5—source data 1
Unprocessed gel images.
- https://cdn.elifesciences.org/articles/89066/elife-89066-fig5-data1-v1.zip
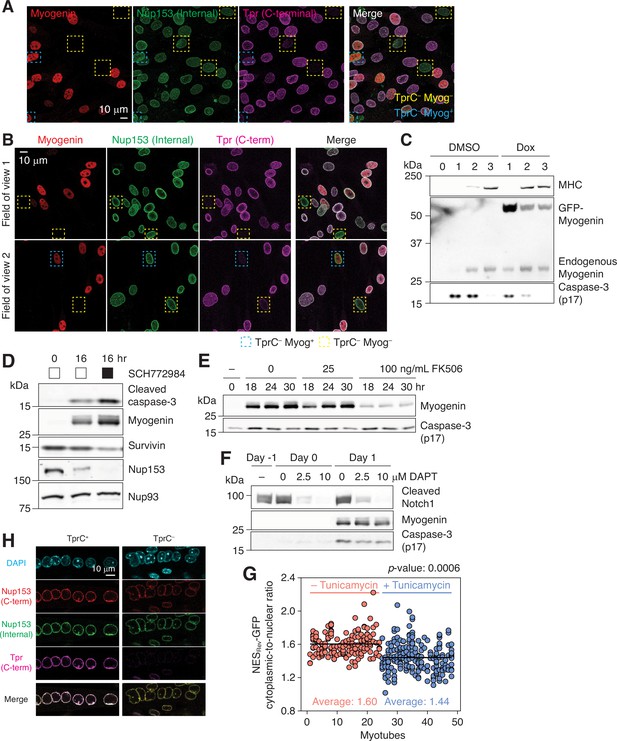
Activation of caspases and proteolysis of their substrates.
(A) C2C12 cells that have undergone myogenic differentiation for 1 day were immunostained for myogenin, Nup153, and Tpr. In yellow and blue dashed boxes are myogenin-negative and -positive TprC– cells, respectively. (B) Primary myoblasts were differentiated for 24 hr. Cells were immunostained for myogenin, Nup153, and Tpr. Two different fields of view are shown. Nuclei that lack Tpr C-terminal epitope are boxed in yellow (myogenin-negative) or blue (myogenin-positive). (C) C2C12 stable cell line that expresses GFP-myogenin in a doxycycline-dependent manner was differentiated in the absence or presence of 0.5 μM doxycycline (Dox). Myosin heavy chain (MHC), exogenous and endogenous myogenin, and active caspase-3 levels were determined by immunoblotting. (D) C2C12 cells were differentiated in the absence or presence of an ERK1/2 inhibitor (SCH772984, 1 μM). Expression levels of active caspase-3, myogenin, survivin, Nup153, and Nup93 were determined by immunoblotting. (E) Upregulation of myogenin and formation of caspase-3 p17 were evaluated in the presence of 0, 25, or 100 ng/mL FK506 by immunoblotting. (F) Immunoblots showing the expression levels of cleaved Notch1, myogenin, and active caspase-3 in C2C12 cells. DAPT was added at 0, 2.5, or 10 μM on day –1, and maintained throughout differentiation. (G) The cytoplasmic-to-nuclear ratio of NES-GFP was determined for myonuclei in 24 myotubes treated with DMSO (red) or tunicamycin (blue, 1 μg/mL for 24 hr). Each data point represents a myonucleus, and ones that share the same x values are from the same myotube. At least 3 myonuclei per myotube were quantified. (H) Myotubes were treated with 1 mg/mL tunicamycin for 24 hr, and immunostained for Nup153 and Tpr. Tpr C-terminal epitope was undetectable (TprC–) in 5–10% of the population.
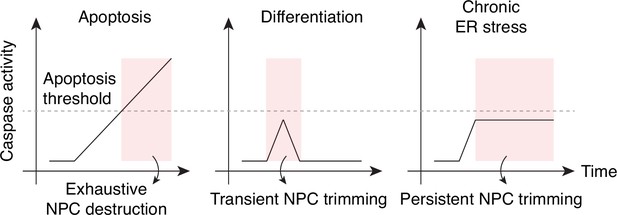
Caspase activation and NPC proteolysis patterns in apoptosis, cell differentiation, and chronic ER stress.
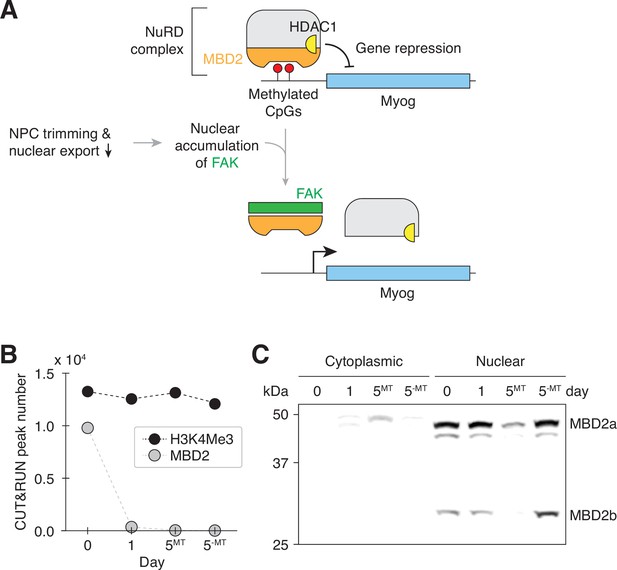
Transient nuclear accumulation of FAK resets MBD2-mediated genome regulation during myogenesis.
(A) NPC trimming is upstream of FAK-mediated MBD2 genome-binding reset event. (B) The number of MBD2 and H3K4Me3 CUT&RUN peaks in differentiating C2C12 cells. (C) Cytoplasmic and nuclear levels of MBD2 in differentiating C2C12 cells were determined by western blotting. Twenty μg of protein was loaded per lane.
Additional files
-
Supplementary file 1
Raw data for the mass spectrometry experiment described in Figures 3 and 4.
- https://cdn.elifesciences.org/articles/89066/elife-89066-supp1-v1.xlsx
-
Supplementary file 2
Antibodies, plasmids, RNA probes, and chemicals used in this study.
(a) Primary antibodies used in this study (b) Secondary antibodies used in this study (c) Plasmids used in this study (d) RNA FISH probes used in the study (e) Chemicals used in this study
- https://cdn.elifesciences.org/articles/89066/elife-89066-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89066/elife-89066-mdarchecklist1-v1.pdf
-
Source data 1
Ponceau S staining and loading control for immunoblots shown in this study.
- https://cdn.elifesciences.org/articles/89066/elife-89066-data1-v1.pdf





