Systematic evaluation of intratumoral and peripheral BCR repertoires in three cancers
Figures
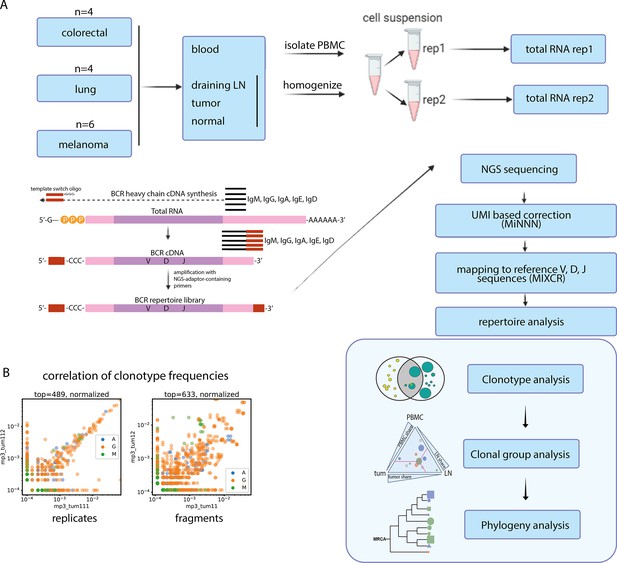
Experimental design.
(A) Overview of the experimental design and procedures. Cell suspensions from all samples were divided into 2 replicates and processed for BCR repertoire profiling. UMI-based correction was employed to compensate for PCR-based bias. MIXCR software was used for alignment to reference BCR V, D, and J genes. The following bioinformatic analysis was performed on several levels: individual clonotype level (clonotype overlap), clonal group level (clonal group sharing between tissues), and clonal phylogeny level. (B) Using technical replicates at the cell suspension level allows to account for sampling bias in the clonotype frequencies.
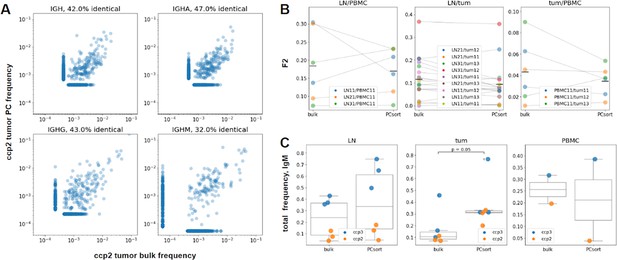
PC representation in bulk samples and repertoire similarities clonotype level, individual patient data from colorectal cancer patient ccp2 (panel A, B) or pooled patient data from colorectal cancer patients ccp2 and ccp3 (panel C).
(A) Correlation plots, comparing PC and bulk frequencies of top 400 clonotypes in total tumor repertoire and isotypes A, G and M separately for colon cancer patient ccp2. Up to 47% of identical amino acid sequences were found. (B) Comparison of F2-overlaps for three tissue repertoires’ pairs (LN and PBMC, LN and tumor, tumor and PBMC). No significant difference was found between bulk and PC-sorted samples (Wilcoxon signed-rank test). (C) Comparison between bulk and PC IgM proportions in lymph node, tumor and PBMC samples, the share of IgM in tumor was significantly higher in PC samples (p=0.05, Mann-Whitney test).
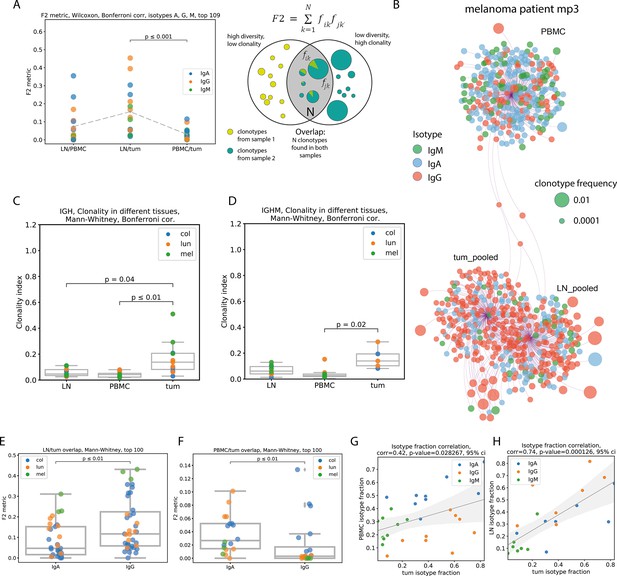
Repertoire overlap, clonality and isotype composition (clonotype level, pooled patient data (except B, data from melanoma patient 3)).
(A) - repertoire overlap between pairs of tissues, by F2 metric, repertoires split by immunoglobulin isotype, (N=6); (B) network representation of Ig repertoires from PBMC, tumor-draining LN, and tumor of mp3 patient (melanoma); individual clonotypes of the same origin (PBMC, tumors, or draining LN) are shown as bubbles connected with the edges to one anchor node. Clonotypes shared between tissues were connected with two or three edges to the corresponding anchor nodes and were located between them. The size of the bubbles represents the relative frequency of clonotypes within a sample, and the color represents the isotype. The relative distance between anchor nodes corresponded to the similarity of repertoires (the number of shared clonotypes). (C, D) Clonality of Ig repertoires in PBMC, draining LNs, and tumors of 14 cancer patients. This reflects the presence of clonal expansion. Calculated as in Tumeh et al., 2014: [1-normalized Shannon-Wiener index]; (C) total IG repertoire; (D) IgM repertoire; (E) LN/tumor overlap for IgA and IgG repertoires (N=7); (F) PBMC/tumor overlap for IgA and IgG repertoires (N=9); (G, H) isotype fraction correlation between PBMC and tumor repertoires (G, N=9), or between LN and tumor repertoires (H, N=7).
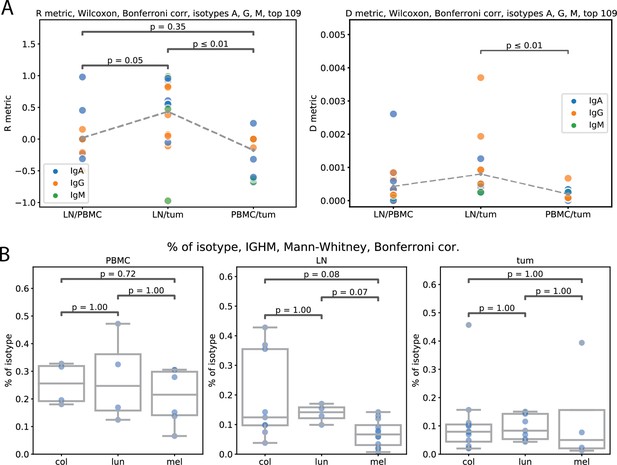
Repertoire similarity by R metric, repertoire overlap by D metric (A), isotype composition by tissue type and cancer (B).
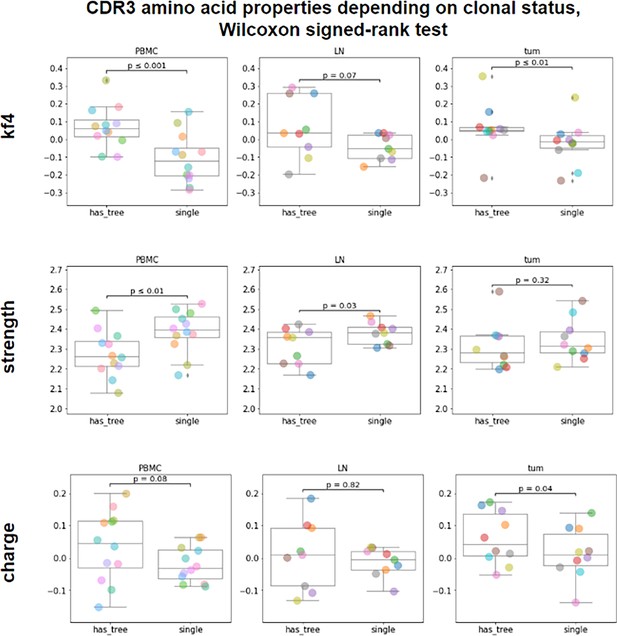
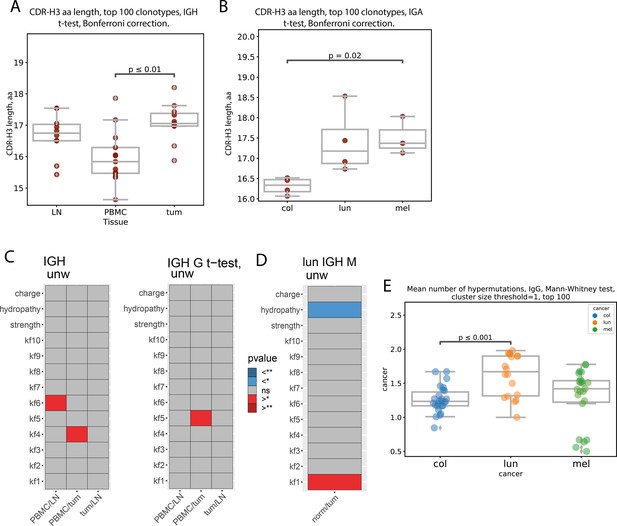
CDR-H3 amino acid properties (clonotype level, pooled patient data).
(A) Mean amino CDR-H3 length of top 100 most frequent clonotypes from tumor, lymph node and PBMC tissues irrespective of isotype CDR-H3 are on average significantly longer in tumor than PBMC for total repertoire (N=14, p<0.01, two-sided t-test, Bonferroni correction); (B) Comparison of mean amino acid CDR-H3 length of 100 most frequent clonotypes for colon, lung and melanoma cancer samples from, tumor. CDR-H3s of tumor-infiltrating clonotypes were shorter for colorectal cancer patients compared to melanoma in IgA repertoires (N=11, p=0.02); (C) Comparison of amino acid properties in the central region of CDR-H3, for total repertoire (C-left) or IGHG repertoire (C-right), all cancers (significantly increased - red, significantly decreased - blue) two-sided t-test, Bonferroni-Holm correction; (D) Comparison of amino acid properties in the central region of CDR-H3, for IGHM repertoire, lung cancer (significantly increased - red, significantly decreased - blue); (p<0.01, two-sided t-test, Bonferroni-Holm correction); (E) Average number of mutations relative to germline for tumor samples from different types of cancers, N=14.
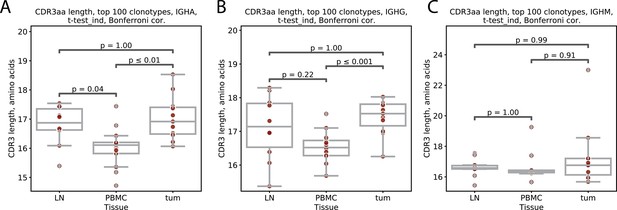
Mean CDR3 length for IGHA, IGHG and IGHM repertoires from lymph node, PBMC and tumor.
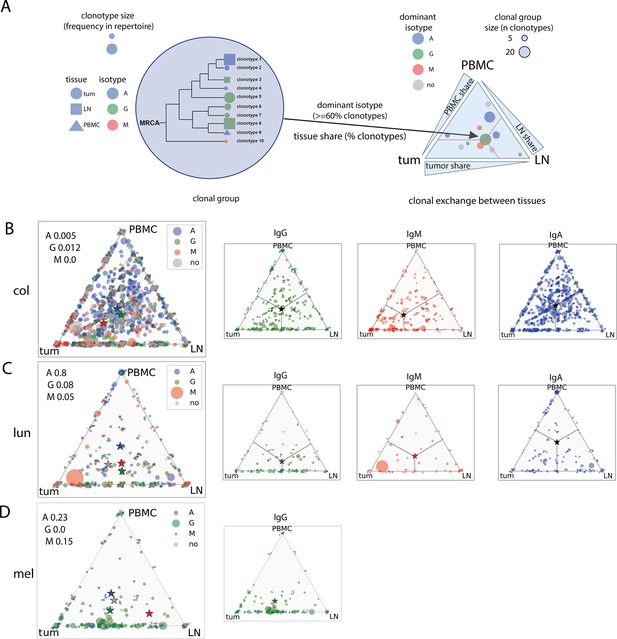
Immunoglobulin hypermutation analysis across tissues and isotypes (clonal group level, pooled patient data).
(A) - Schematic representation of analysis and triangle plot visualization of clonal group distribution between tissues; (B, C, D) clonal group distribution between tissues for colorectal (B), lung (C), and melanoma (D); stars represent non-weighted by size mean center of triangle coordinates.
Chi-2 test for goodness of fit was used to test whether each tissue equally contributed to clonal group formation.
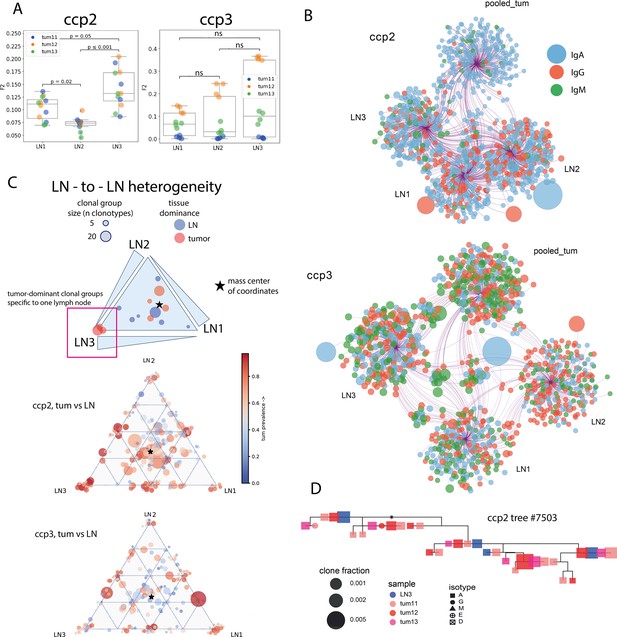
LN-to-LN difference of BCR repertoires in colorectal cancer (clonotype level - panels A, B; clonal group level - panels C, D; individual patient data).
(A) Repertoire overlap between tumor and three separate LNs from the draining lymph node pool; Mann-Whitney test, Bonferroni correction. (B) Network representation of Ig repertoires from tumor and three separate LNs, with circles representing individual CDR-H3 sequences, size of the circles corresponding to the clone frequency, and color corresponding to the isotype; (C) triangle plot visualization of clonal group distribution between three different LNs, with size corresponding to the number of individual CDR-H3 sequences (clonotypes) within a given clonal group, and color corresponding to the percentage of tumor-derived clonotypes–within the clonal group; (D) example of a clonal lineage consisting of CDR-H3 sequences derived from a lymph node (blue) and all three tumor fragments (shades of red) from patient ccp2, shapes representing isotypes and size representing frequency of a given sequence in a given sample.
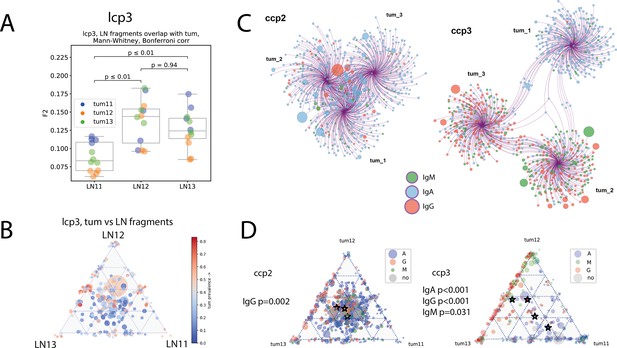
Intra-LN and intratumoral heterogeneity (clonotype level - panels A, C; clonal group level - panel B, D; individual patient data).
(A) Repertoire overlap between tumor and lymph node fragments for colorectal cancer patient ccp6 and lung cancer patient lcp3; Mann-Whitney test, Bonferroni corr. (B) Triangle plot visualization of clonal group distribution between lymph node fragments, with size corresponding to the number of individual CDR-H3 sequences (clonotypes) within a given clonal group, and color corresponding to the percentage of tumor-derived clonotypes within the clonal group; (C) network representation of Ig repertoires from tumor fragments, with circles representing individual CDR-H3 sequences, size of the circles corresponding to the clone frequency, and color corresponding to the isotype, edges connect clonotypes to their fragment of origin; (D) triangle plot visualization of clonal group distribution between tumor fragments, with size corresponding to the number of individual CDR-H3 sequences (clonotypes) within a given clonal group, and color representing dominant clonotype; Stars represent non-weighted by size mean center of triangle coordinates. Chi-2 test for goodness of fit is used to test if each tumor segment equally contributes to clonal groups formation.
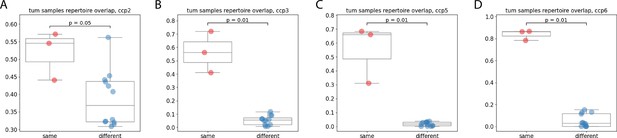
BCR repertoire overlap for separate fragments of tumor (different), and from cell suspension level replicates from the same fragment (same), patients ccp2 (A), ccp3 (B), ccp5 (C), ccp6 (D).
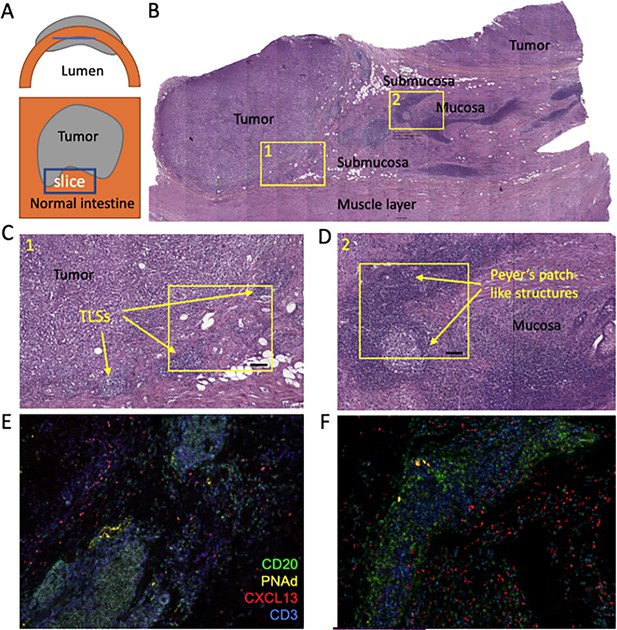
Histochemical analysis of lymphoid infiltration in ccp2 sample.
(A) Schematic representation of slice orientation relative to the intestine wall and tumor. (B) Tissue section stained with hematoxylin and eosin (H&E). Different layers of the intestine wall and tumor are marked. Regions 1 and 2 are magnified on the panels (C and D) respectively. (C, D. H&E) histology of the peritumoral region and mucosal/submucosal layers. Lymphoid structures in these regions were classified as TLS and Peyer’s patches, respectively and are marked with arrows. Regions on (C and D) orespond to the areas that are shown on fluorescent images of parallel slices in (E and F), respectively. CD20 fluorescence is shown in green, CD3 in blue, CXCL13 in red and PNAd in yellow. Scale bar is shown in black and corresponds to 100 µm.
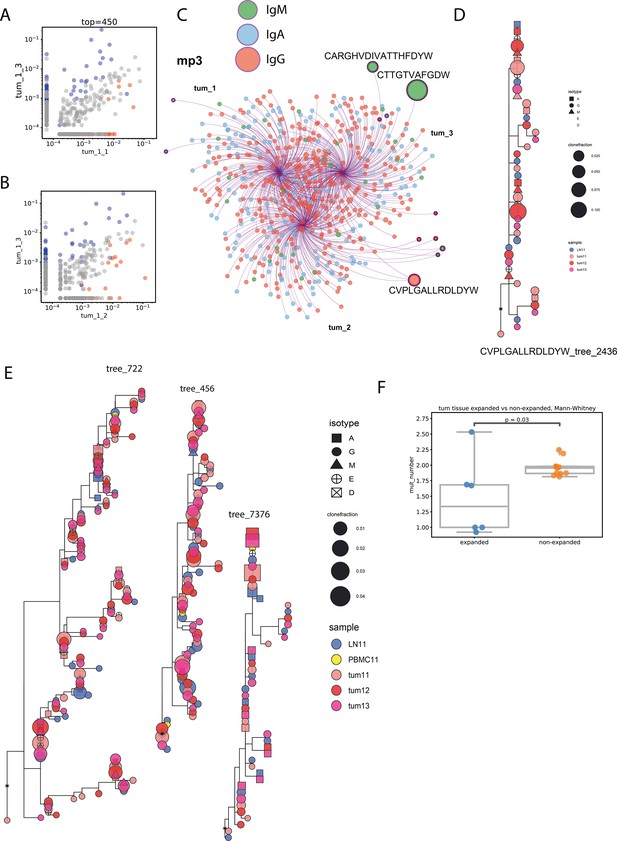
Expanded clonotypes and hypermutation analysis (clonotype level - A, B, C, F; clonotype group level - D, E; individual patient data).
(A, B) Visualization of expanded clonotypes on frequency correlation plots for pairs of tumor fragments of melanoma tumor from patient mp3; (C) Cytoscape network visualization of top 300 most frequent individual Ig CDR-H3 clonotypes, colored by isotype, with size of circles proportional to the frequency of a given clonotype; (D) example of a clonal lineage containing expanded CDR-H3 sequence; (E) examples of the biggest clonal lineages, none of which contain expanded CDR-H3 sequences; (F) average number of mutations in expanded and non-expanded clonotypes; Mann-Whitney test; N=11.
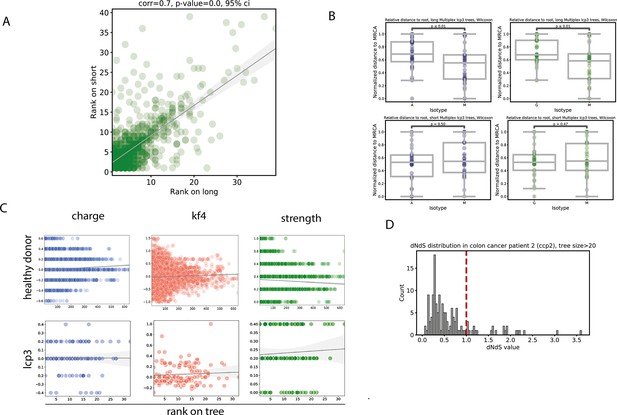
Short (CDR-H3-based) versus full-length IG trees phylogeny analysis.
(A) Distance to MRCA comparison for same clonal groups on short and long trees; (B) distance to MRCA comparison for IgM, IgA and IgG-dominated clonal groups on short and long trees; (C) distribution of CDR-H3 amino acid properties (charge, kf4 and strength) along short and long trees in healthy donor (top row) and cancer patients (bottom row); (D) dN/dS values for trees os size > 20.
Additional files
-
Supplementary file 1
Basic characteristics of sequencing output results.
- https://cdn.elifesciences.org/articles/89506/elife-89506-supp1-v1.xlsx
-
Supplementary file 2
Patient demographics and obtained samples.
- https://cdn.elifesciences.org/articles/89506/elife-89506-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89506/elife-89506-mdarchecklist1-v1.docx