Spermatogenesis: All eyes on FOXC2
Male fertility relies on the continuous production of sperm via a process known as spermatogenesis. This involves spermatogonial stem cells (SSCs) dividing to form undifferentiated spermatogonia (uSPGs), which then progress through the meiotic and haploid phases of spermatogenesis to form mature sperm (de Rooij, 1998). To ensure that the supply of sperm remains constant, SSCs must continuously provide new uSPGs while also self-renewing to maintain their stocks.
While the existence of SSCs in the adult testis is undisputed, their origin, identity and maintenance remain unclear. In fact, scientists still lack genetic markers that clearly allow them to distinguish these cells from the rest of the uSPG pool. So far, the hallmark feature of SSCs is their ability to re-establish full spermatogenesis when transplanted into testes devoid of germ cells (Kubota and Brinster, 2018; Lord and Oatley, 2018).
Previous work has identified three types of uSPGs – single, paired and aligned – which emerge during the first phase of the differentiation process. When a single uSPG divides, it can sometimes produce paired daughter cells that remain connected after mitosis. In turn, these paired uSPGs can expand to form chains of four to 32 aligned uSPGs, with some of these cells progressing through to the later stages of spermatogenesis to form mature sperm (de Rooij, 2017; Kubota and Brinster, 2018; de Rooij, 1998).
Transplantation experiments have revealed that most cells which can perform the hallmark feature of SSCs (that is, re-establishing full spermatogenesis in testes lacking germ cells) are found within the single uSPG population, but may also be present among paired and aligned progenitors (Kubota and Brinster, 2018). Meanwhile, genetic studies combined with lineage-tracing experiments have highlighted several genes predominantly expressed in single uSPGs that act as SSCs; however, these genes cannot represent strict SSC markers as they are also expressed in progenitors engaged in the differentiation process (Kubota and Brinster, 2018; Sharma et al., 2019). Now, in eLife, Wei Song and colleagues at the University of Dundee and the Peking Union Medical College – including Zhipeng Wang as first author – report findings which suggest that a transcription factor known as FOXC2 may represent a more precise marker of functional SSCs (Wang et al., 2023).
The team started by screening the expression profile of individual cells in a population of mouse uSPGs containing both SSCs and progenitors. Among the top ten genes preferentially enriched in these cells, Foxc2 was the only one to code for a protein exclusively present in the nucleus of uSPGs that also expressed ZBTB16, a protein important for SSCs to self-renew. A closer look showed that Foxc2 expression was most abundant in single uSPGs compared to paired or aligned uSPGs. Interestingly, FOXC2-producing uSPGs were mostly quiescent, with only 5% featuring markers associated with proliferation. This finding is consistent with the fact that many FOXC2-regulated genes are involved in cell cycle arrest.
To test whether FOXC2-producing uSPGs could underpin spermatogenesis, Wang et al. transplanted a population of uSPGs enriched in these cells into the testes of mice treated with busulfan, a toxic compound that kills endogenous germ cells. After two months, these animals had generated a much larger number of colonies of differentiating cells compared to control mice which had received a non-enriched uSPG population. Based on these results, Wang et al. set out to show that FOXC2-producing single uSPGs are in fact functional SSCs.
The first step was for the team to follow the fate of these cells for six weeks following transplantation. This revealed that this population could give rise to all subtypes of uSPGs, with some of the resulting progenitors differentiating into sperm that could fertilise eggs and generate offspring. However, FOXC2-producing uSPGs were also capable of self-renewal, forming cells which feature genetic markers associated with SSCs. More specifically, the lineage-tracing experiments showed that FOXC2-producing uSPGs could produce paired uSPGs that would then either divide to form two single uSPGs (including some that retained Foxc2 expression), or form chains of aligned uSPGs containing at most one FOXC2-producing cell (Figure 1A).
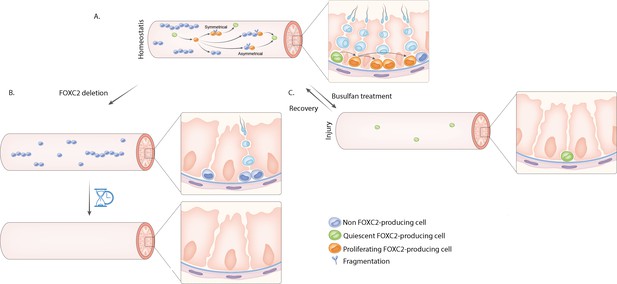
Undifferentiated spermatogonia which express Foxc2 may represent the entire population of spermatogonial stem cells in the adult testes of mice.
(A) Sperm is created inside seminiferous tubules (pink tubes) through spermatogenesis, a complex differentiation process that starts with the division of spermatogonial stem cells (SSCs). SSCs are difficult to distinguish from other types of undifferentiated spermatogonia (uSPGs) which also reside in the seminiferous tubules. Wang et al. propose that single uSPGs that express the gene encoding the transcription factor FOXC2 constitutes most, if not all, the SSC pool. About 95% of these cells are quiescent (green cells) and the remaining ~5% are proliferative (orange cells). Proliferating FOXC2-producing single uSPGs may divide asymmetrically to generate paired and aligned uSPGs that remain attached to each other after mitosis. These cells show differential expression of Foxc2. Some of the cells that do not produce FOXC2 (light blue cells with dark blue nucleus) will differentiate into progenitors (light blue cells) and ultimately become sperm. Others, which have retained Foxc2 expression, may split away from their sister cells through a fragmentation process (scissors) and return to a FOXC2-producing single uSPG state to contribute to the SSC pool. Self-renewal of SSCs may also be achieved by symmetrical division of a single FOXC2-producing SSC to form two single, FOXC2-producing daughter SSCs. (B) Deleting FOXC2-producing cells causes accelerated exhaustion of SSCs, and, in time (hourglass) leads to male infertility. (C) Quiescent FOXC2-producing single uSPGs are resistant to cytotoxins such as busulfan treatment; they can survive these environmental disruptions and replenish the pools of uSPGs, thereby maintaining SSC homeostasis.
Wang et al. then inactivated Foxc2 in the germ cells of adult testes to better investigate FOXC2 function. This gradually exhausted the number of available uSPGs, leading to smaller testes and eventual infertility (Figure 1B). If Foxc2 was deleted in male germ cells before mice started to produce sperm, however, an initial wave of spermatogenesis was still able to occur but without subsequent, continuous sperm production. This is consistent with the fact that the first wave of sperm cell formation does not rely on SSCs, while subsequent spermatogenesis does.
Finally, Wang et al. tested whether FOXC2-producing uSPGs contribute to germline regeneration, an important property that allows sperm production to resume after being disrupted. They exposed adult mice to busulfan and found that the remaining population of uSPGs was primarily formed of quiescent FOXC2-producing cells; this aligns with previous findings showing that quiescence helps to protect stem cells from environmental insults (Murley et al., 2022; Tümpel and Rudolph, 2019). After a month, FOXC2-producing cells showed signs of higher levels of proliferation (yet the size of the population remained stable), and after four months spermatogenesis had been fully re-established (Figure 1C).
Together, these results suggest that single uSPGs which express Foxc2 could indeed constitute the reservoir of SSCs in the mammalian testis. According to these findings, FOXC2 may promote a reversible quiescent state through negative regulation of cell cycle progress. However, a small fraction of this population (~5%) undergoes active proliferation, creating a number of paired and then aligned uSPGs which may include a single cell that continues to express Foxc2. Such Foxc2-expressing cells may detach themselves from their sister cells in pairs or chains, returning to a single uSPG state and contributing to the renewal of the SSC pool. Meanwhile, other paired and aligned uSPGs that are not expressing Foxc2 progress through spermatogenesis to form sperm.
Overall, this work provides strong evidence that FOXC2 could mark functional SSCs more precisely while also actively shaping the fate of these cells. This transcription factor is highly conserved and, as Wang et al. show, it is expressed in a similar pattern in human and mouse testes (Wei et al., 2018). FOXC2 may therefore emerge as a useful marker and important regulator for investigating fertility issues in men.
References
-
Stem cells in the testisInternational Journal of Experimental Pathology 79:67–80.https://doi.org/10.1046/j.1365-2613.1998.00057.x
-
The nature and dynamics of spermatogonial stem cellsDevelopment 144:3022–3030.https://doi.org/10.1242/dev.146571
-
Quiescence: good and bad of stem cell agingTrends in Cell Biology 29:672–685.https://doi.org/10.1016/j.tcb.2019.05.002
-
The forkhead transcription factor FOXC2 is required for maintaining murine spermatogonial stem cellsStem Cells and Development 27:624–636.https://doi.org/10.1089/scd.2017.0233
Article and author information
Author details
Publication history
Copyright
© 2023, Yan and McCarrey
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 305
- views
-
- 50
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Developmental Biology
- Evolutionary Biology
Seahorses, pipefishes, and seadragons are fishes from the family Syngnathidae that have evolved extraordinary traits including male pregnancy, elongated snouts, loss of teeth, and dermal bony armor. The developmental genetic and cellular changes that led to the evolution of these traits are largely unknown. Recent syngnathid genome assemblies revealed suggestive gene content differences and provided the opportunity for detailed genetic analyses. We created a single-cell RNA sequencing atlas of Gulf pipefish embryos to understand the developmental basis of four traits: derived head shape, toothlessness, dermal armor, and male pregnancy. We completed marker gene analyses, built genetic networks, and examined the spatial expression of select genes. We identified osteochondrogenic mesenchymal cells in the elongating face that express regulatory genes bmp4, sfrp1a, and prdm16. We found no evidence for tooth primordia cells, and we observed re-deployment of osteoblast genetic networks in developing dermal armor. Finally, we found that epidermal cells expressed nutrient processing and environmental sensing genes, potentially relevant for the brooding environment. The examined pipefish evolutionary innovations are composed of recognizable cell types, suggesting that derived features originate from changes within existing gene networks. Future work addressing syngnathid gene networks across multiple stages and species is essential for understanding how the novelties of these fish evolved.
-
- Developmental Biology
- Neuroscience
We established a volumetric trans-scale imaging system with an ultra-large field-of-view (FOV) that enables simultaneous observation of millions of cellular dynamics in centimeter-wide three-dimensional (3D) tissues and embryos. Using a custom-made giant lens system with a magnification of ×2 and a numerical aperture (NA) of 0.25, and a CMOS camera with more than 100 megapixels, we built a trans-scale scope AMATERAS-2, and realized fluorescence imaging with a transverse spatial resolution of approximately 1.1 µm across an FOV of approximately 1.5×1.0 cm2. The 3D resolving capability was realized through a combination of optical and computational sectioning techniques tailored for our low-power imaging system. We applied the imaging technique to 1.2 cm-wide section of mouse brain, and successfully observed various regions of the brain with sub-cellular resolution in a single FOV. We also performed time-lapse imaging of a 1-cm-wide vascular network during quail embryo development for over 24 hr, visualizing the movement of over 4.0×105 vascular endothelial cells and quantitatively analyzing their dynamics. Our results demonstrate the potential of this technique in accelerating production of comprehensive reference maps of all cells in organisms and tissues, which contributes to understanding developmental processes, brain functions, and pathogenesis of disease, as well as high-throughput quality check of tissues used for transplantation medicine.






