A genome-wide nucleosome-resolution map of promoter-centered interactions in human cells corroborates the enhancer-promoter looping model
Figures
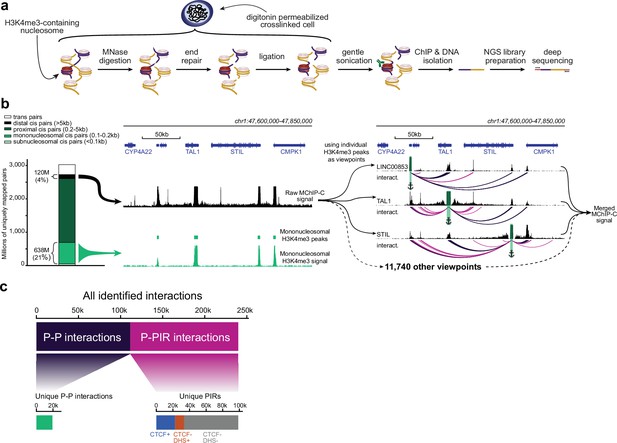
MChIP-C experimental and computational workflow.
(a) An overview of the MChIP-C experimental procedure. (b) General MChIP-C analysis pipeline. A 250 kb genomic region surrounding the TAL1 gene is shown with H3K4me3 MChIP-C profiles in K562 cells. Positions of individual viewpoints are highlighted by green rectangles and anchors. Identified MChIP-C interactions are shown as magenta (P-PIR) and dark violet (P-P) arcs. (c) Summary statistics for all 241,073 promoter-centered MChIP-C interactions identified in K562 cells.
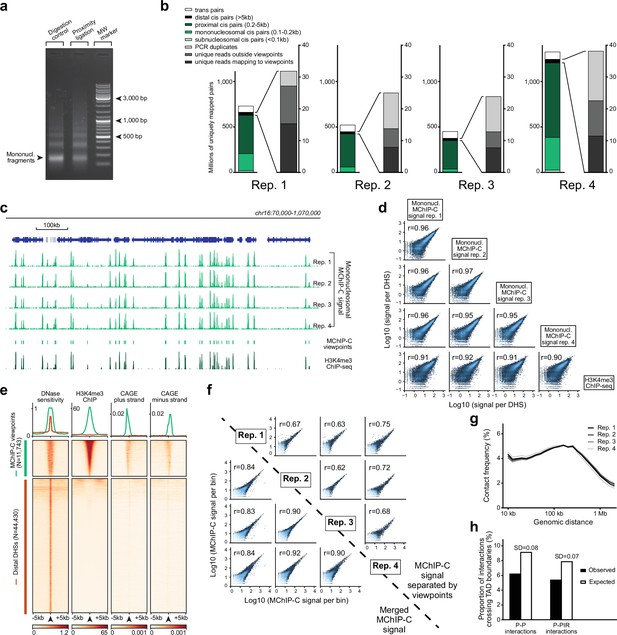
H3K4me3 MChIP-C experiment technical assessment.
(a) 1.5% agarose gel electrophoresis of MChIP-C MNase digestion and ligation controls. (b) Mapping and filtering statistics of four biological replicates of H3K4me3 MChIP-C experiments. (c) 1 Mb region on chromosome 16 with mononucleosomal MChIP-C profiles of 4 MChIP-C replicates reflecting H3K4me3 occupancy, positions of consensus MChIP-C mononucleosomal peaks, viewpoints and conventional H3K4me3 ChIP-seq profile. (d) Hexagonal heatmaps representing pairwise comparison of mononucleosomal MChIP-C profiles in four replicates and a conventional H3K4me3 ChIP-seq profile. Signals in K562 DHS sites are used for correlation and plotting. Pearson’s correlation coefficient (r) for each pair is shown. (e) Heatmaps and average profile plots of DNase sensitivity, H3K4me3 ChIP and CAGE signal in 10 kb windows centered on either MChIP-C viewpoints or distal DHS sites (not overlapping MChIP-C viewpoints). (f) Hexagonal heatmaps representing pairwise comparison of distal MChIP-C profiles in four replicates (either merged or separated by viewpoints). Pearson’s correlation coefficient (r) for each pair is shown. (g). Distance-dependent decay of distal MChIP-C signal in four replicates. Signal is calculated in 30 distance bins of equal size on a log10 scale between 3.5 (3,162 bp) and 6.5 (3,162 kbp). (h) Observed and expected numbers of cross-TAD boundary MChIP-C interactions. SDs of simulated expected numbers are indicated.
-
Figure 1—figure supplement 1—source data 1
Uncropped image for gel shown in Figure 1—figure supplement 1a (original and with bands labelled).
- https://cdn.elifesciences.org/articles/91596/elife-91596-fig1-figsupp1-data1-v1.zip
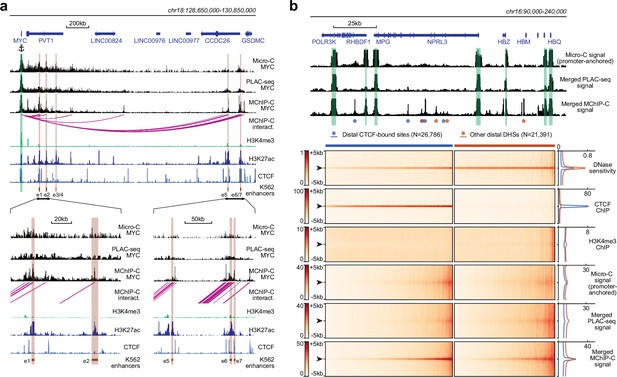
Comparison of MChIP-C with PLAC-seq and Micro-C.
(a) Top: MChIP-C, PLAC-seq and Micro-C interaction profiles of the MYC promoter in K562 cells. MChIP-C interactions of the MYC promoter are shown as magenta arcs. Positions of 7 (e1–e7) CRISPRi-verified K562 MYC enhancers are highlighted as orange rectangles. Bottom: zoom in on two enhancer clusters. (b) Systematic comparison of merged MChIP-C, merged PLAC-seq and merged promoter-anchored Micro-C signals in distal regulatory sites. Top: Merged MChIP-C, merged PLAC-seq and merged promoter-anchored Micro-C profiles in a 150 kb genomic region surrounding the α-globin gene domain. Viewpoints are highlighted as green rectangles. Positions of CTCF-bound and CTCF-less DNase hypersensitive sites outside viewpoints are depicted as blue and orange circles. Bottom: Heatmaps and averaged profiles of DNase sensitivity, CTCF ChIP, H3K4me3 ChIP, merged MChIP-C, merged PLAC-seq and merged promoter-anchored Micro-C signals centered on distal CTCF-bound and CTCF-less DNase hypersensitive sites.
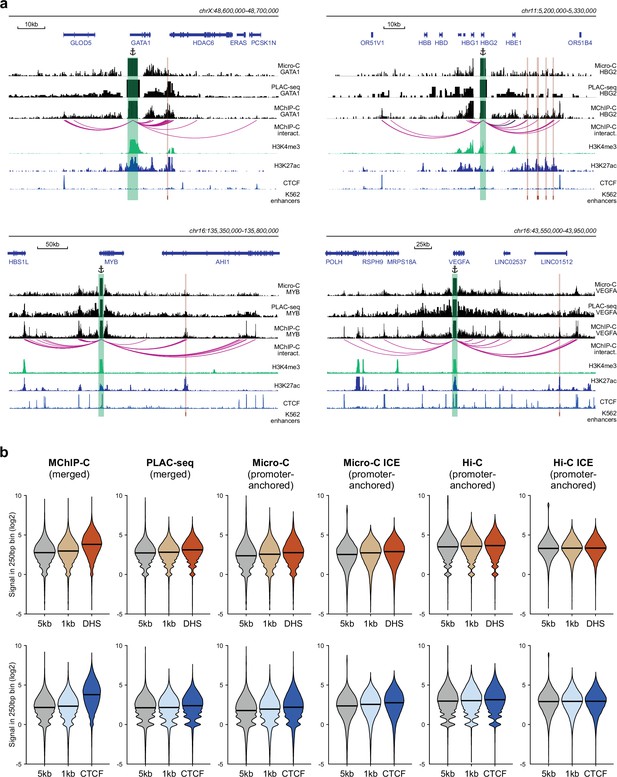
Comparison of MChIP-C with other C-methods.
(a) MChIP-C, PLAC-seq and Micro-C interaction profiles for GATA1, HBG2, MYB and VEGFA genes. Viewpoints are highlighted by anchor symbols and green rectangles, K562 known enhancers are highlighted by orange rectangles (only enhancers localized within 5 kb-1Mb of the viewpoints are shown). (b) Aggregate ligation signal between all active promoters and CTCF-bound (bottom) and CTCF-less (top) distal DNase hypersensitive sites measured with various C-methods in K562 cells. Genomic bins separated by 1 and 5 kb from DHS center were used as background. ICE denotes iteratively balanced Micro-C and Hi-C datasets.
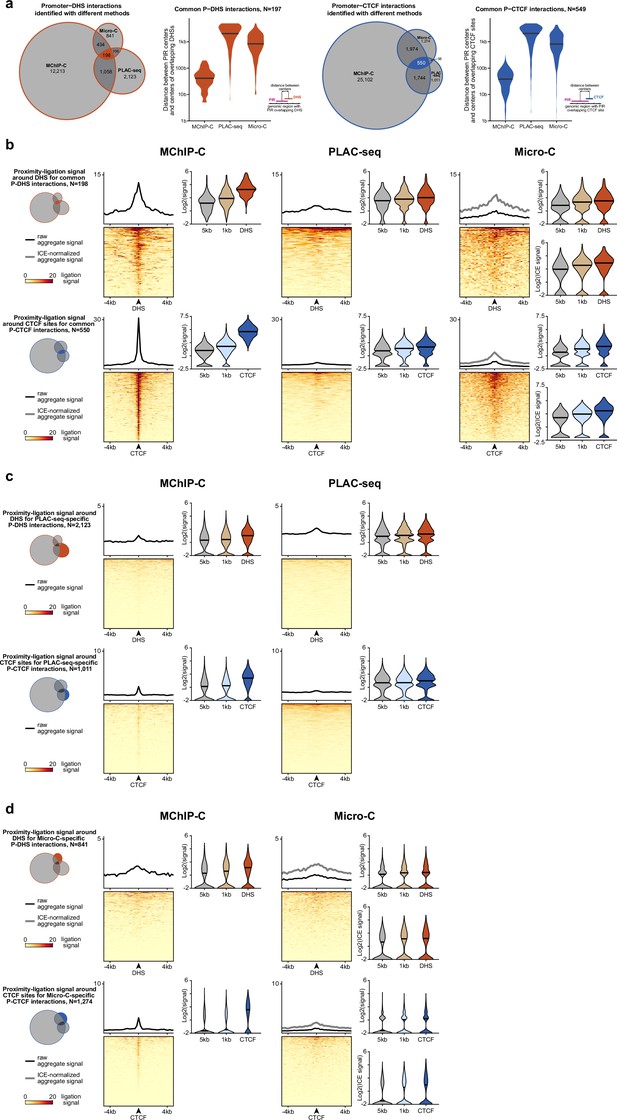
Consensus, PLAC-seq-specific and Micro-C-specific promoter-DHS and promoter-CTCF interactions.
(a) Quantity and overlap between promoter-DHS (P-DHS, left) and promoter-CTCF (P-CTCF, right) interactions identified by MChIP-C, PLAC-seq and Micro-C in K562 cells. Violin plots represent distances between centers of identified PIRs and centers of overlapping DHS/CTCF sites. (b) Heatmaps, aggregate profiles and violin plots reflecting ligation signal (to cognate promoter exclusively) around DHS (top) and CTCF (bottom) sites for consensus P-DHS and P-CTCF pairs measured with various C-methods. In violin plots genomic bins separated by 1 and 5 kb from DHS/CTCF center were used as background. ICE denotes usage of iteratively balanced Micro-C dataset. (c) The same as in b, but for PLAC-seq-specific interactions. (d) The same as in b, but for Micro-C-specific interactions.
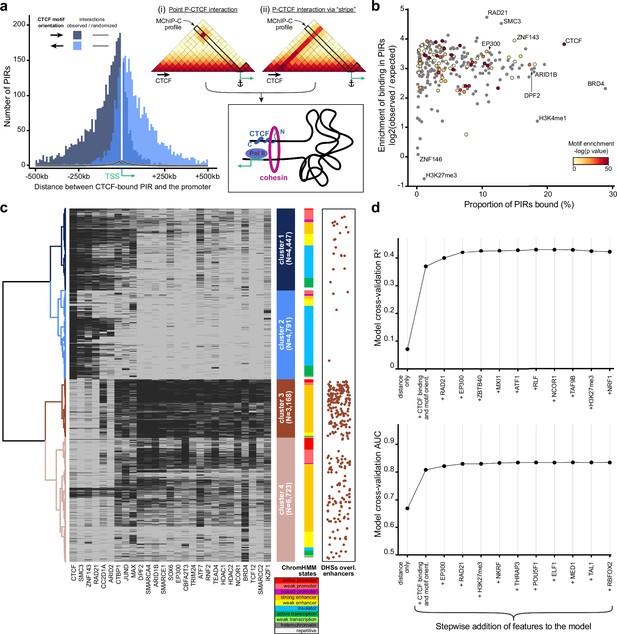
Analysis of protein factors underlying MChIP-C interactions.
(a) Left: CTCF-motif orientation bias in regions interacting with CTCF-less promoters. The majority (~79%) of CTCF motifs are oriented towards the interacting promoter. Right: Schematic of two hypothetical loop extrusion dependent mechanisms that can account for the observed pattern: promoter LE-barrier activity (i) or CTCF-originating interaction stripes (ii).(b) Enrichment of transcription-related factor (TRF) binding in MChIP-C PIRs. Y-axis represents enrichment (log2 observed/expected) of binding for 271 examined TRFs, x-axis – proportion of TRF-bound PIRs, color – enrichment of corresponding motifs in PIRs (grey color is assigned to TRFs lacking DNA-binding motif). (c) Hierarchical clustering of PIR-overlapping DHSs (N=19,129). The binding status of 164 TRFs highly enriched in PIRs are used as binary features. Binding of 27 selected TRFs (see Methods) in each PIR-overlapping DHS is shown as a heatmap. ChromHMM chromatin state distributions in each cluster are shown. DHSs overlapping CRISPRi-verified K562 enhancers (Fulco et al., 2019; Gasperini et al., 2019) are shown as orange dots. (d) Predictive performance (3-fold cross-validation R2/AUC) of random forest models predicting MChIP-C signal for DHS-promoter pairs. Starting with an initial model based on distance and CTCF, the most predictive TRF features are added incrementally to the model (left to right).
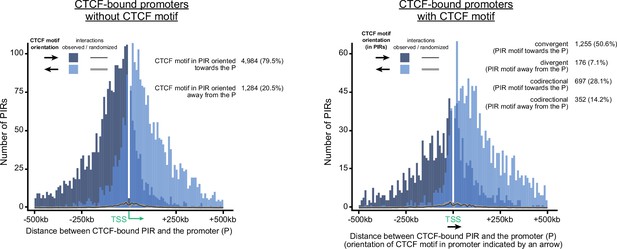
CTCF-orientation bias in PIRs interacting with CTCF-occupied promoters.
The graph demonstrates CTCF-motif orientation bias in regions interacting with CTCF-occupied promoters. A significant portion of these promoters does not contain canonical CTCF binding motifs. CTCF-bound promoters tend to interact with CTCF-motifs oriented towards them whether the CTCF-binding motif is present (right diagram) or absent (left diagram) in these promoters. Note that the bias holds even for codirectional CTCF motifs while one of them is localized within a promoter and the other within a PIR.
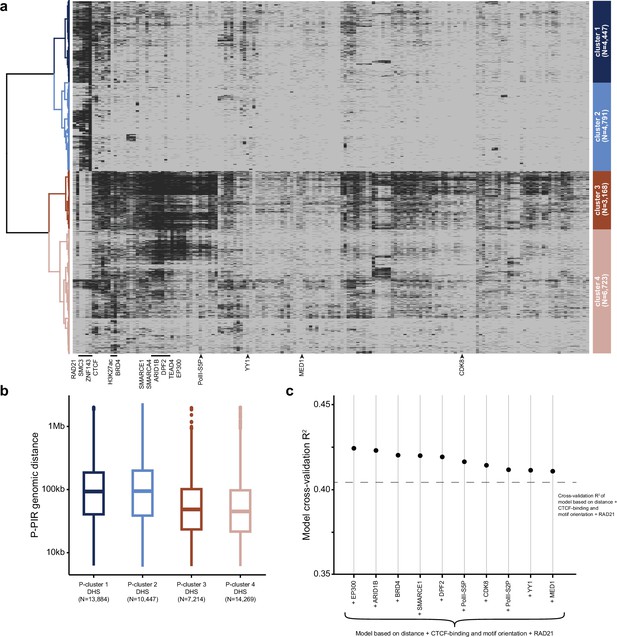
Extended characterization of protein factors underlying promoter-interacting DHSs.
(a) Heatmap showing the binding of all 164 TRFs used for hierarchical clustering of promoter-interacting DHS. Columns corresponding to proteins associated with clusters 1 and 2 (RAD21, SMC3, ZNF143 and CTCF) as well as to enhancer-associated factors potentially involved in physical E-P interactions (BRD4, H3K27ac, SMARCE1, SMARCA4, ARID1B, DPF2, TEAD4, EP300, YY1, MED1, PolII-S5P, CDK8) are highlighted. (b) Distance distribution boxplots for MChIP-C P-PIR interactions anchored in DHSs from identified clusters. (c) Predictive power (3-fold cross-validation R2) of various enhancer-associated factors in the random forest model of MChIP-C signal strength. The predictive power of the initial model +RAD21 (6 features total) is shown as a dashed line. The predictive power of each factor is shown after adding the factor to the 6 features and retraining the model.

The majority of functionally-verified enhancers do physically interact with their target promoters.
(a) Bar plots representing the proportion of MChIP-C interacting pairs among nonfunctional DHS-P pairs and CRISPRi-verified E-P pairs. Heatmaps and average profiles of MChIP-C signal are shown for individual subsets. CRISPRi-verified enhancers shown in panel b are indicated by roman numerals. (b) Examples of MChIP-C, Micro-C and PLAC-seq profiles for promoters physically interacting with their functionally verified enhancers (i.e. MChIP-C interaction has been found) (i-iii) and not interacting with them (i.e. MChIP-C interaction has not been found) (iv-vi). Viewpoints are highlighted by anchor symbols and green rectangles, enhancers are highlighted by orange rectangles. (c) Distance distribution boxplots for verified E-P pairs with and without MChIP-C interactions.
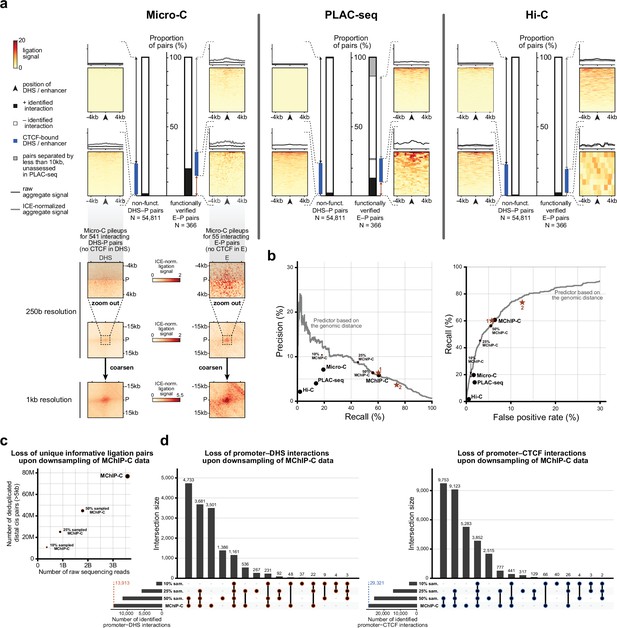
Recall (sensitivity), precision and false positive rate for predictions of functional enhancer-promoter pairs using different C-methods.
Analysis of the downsampled MChIP-C datasets. (a) Bar plots representing the proportion of interacting pairs according to PLAC-seq, Micro-C, and Hi-C among nonfunctional DHS-P pairs and CRISPRi-verified E-P pairs. Heat maps and average profiles of proximity ligation signal are shown for separate subsets in each individual method. DHS-P and E-P pairs with interactions in the Micro-C dataset are also shown as pileups of Micro-C heatmap fragments. (b) Precision-Recall plot and ROC plot for various C-methods aiming to distinguish between nonfunctional DHS-P pairs and CRISPRi-verified E-P pairs in K562 cells. Stars denote the performance of distance-based predictors in which each analyzed DHS is assigned as an enhancer to the nearest active gene (1) or to the two nearest active genes (2). Curves represent the performance of the distance-based predictor using thresholds inversely proportional to the genomic distance between the DHS/enhancer and TSS of the target gene. Downsampled MChIP-C datasets are denoted as ‘50% MChIP-C’, ‘25% MChIP-C’ and ‘10% MChIP-C’. (c) Numbers of raw and deduplicated distal (>5 kb apart) read pairs upon MChIP-C data downsampling. (d) UpSet plots depicting gradual loss of identifiable promoter-DHS (left) and promoter-CTCF (right) interactions upon downsampling of the MChIP-C data.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | K562 | ATCC | ATCC:CCL-243 | |
| Antibody | anti-H3K4me3 (rabbit polyclonal) | Active Motif | Active Motif:39016 | (1:200) |
| Chemical compound, drug | Digitonin | Sigma-Aldrich | Sigma-Aldrich:D-5628 | |
| Chemical compound, drug | Protease Inhibitor Cocktail | Bimake | Bimake:B14001 | |
| Peptide, recombinant protein | Micrococcal Nuclease | Thermo Fisher Scientific | Thermo Fisher Scientific:EN0181 | |
| Peptide, recombinant protein | T4 Polynucleotide Kinase | New England Biolabs | New England Biolabs:M0201L | |
| Peptide, recombinant protein | DNA Polymerase I, Klenow Fragment | New England Biolabs | New England Biolabs:M0210L | |
| Peptide, recombinant protein | T4 DNA Ligase | Thermo Fisher Scientific | Thermo Fisher Scientific:EL0012 | |
| Commercial assay or kit | Protein A/G Magnetic Beads | Thermo Fisher Scientific | Thermo Fisher Scientific:88802 | |
| Commercial assay or kit | NEBNext Ultra II DNA Library Prep Kit | New England Biolabs | New England Biolabs:E7645 | |
| Commercial assay or kit | TruSeq DNA Single Indexes | Illumina | Illumina: 20015960 and Illumina:20015961 | |
| Commercial assay or kit | KAPA HiFi HotStart PCR Kit | Roche | Roche:07958897001 | |
| Software, algorithm | bwa, v.0.7.17 | Li, 2013 | RRID:SCR_010910 | https://github.com/lh3/bwa |
| Software, algorithm | Bowtie2, v.2.3.4 | Langmead and Salzberg, 2012 | RRID:SCR_016368 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Software, algorithm | pairtools, v.0.3.0 | Abdennur et al., 2023 | RRID:SCR_023038 | https://github.com/open2c/pairtools |
| Software, algorithm | samtools, v.1.15.1 | Danecek et al., 2021 | RRID:SCR_002105 | https://github.com/samtools/samtools |
| Software, algorithm | bedtools, v2.26.0 | Quinlan and Hall, 2010 | RRID:SCR_006646 | https://github.com/arq5x/bedtools2 |
| Software, algorithm | Python, v.3.7.12 | RRID:SCR_008394 | https://www.python.org | |
| Software, algorithm | numpy, v.1.21.6, | Harris et al., 2020 | RRID:SCR_008633 | https://github.com/numpy/numpy |
| Software, algorithm | pandas, v.1.3.5 | RRID:SCR_018214 | https://github.com/pandas-dev/pandas | |
| Software, algorithm | matplotlib, v.3.5.3 | Hunter, 2007 | RRID:SCR_008624 | https://github.com/matplotlib/matplotlib |
| Software, algorithm | cooler, v.0.9.1 | Abdennur and Mirny, 2020 | RRID:SCR_024194 | https://github.com/open2c/cooler |
| Software, algorithm | cooltools, v.0.5.1 | Abdennur et al., 2024 | RRID:SCR_026118 | https://github.com/open2c/cooltools |
| Software, algorithm | R, v. 4.2.1 | RRID:SCR_001905 | https://cran.r-project.org | |
| software, algorithm | dplyr, v.1.0.9 | RRID:SCR_016708 | https://github.com/tidyverse/dplyr | |
| Software, algorithm | tidyr, v.1.2.0 | RRID:SCR_017102 | https://github.com/tidyverse/tidyr | |
| Software, algorithm | ggplot2, v.3.3.6 | RRID:SCR_014601 | https://github.com/tidyverse/ggplot2 | |
| Software, algorithm | gplots, v. 3.1.3 | RRID:SCR_025035 | https://github.com/talgalili/gplots | |
| Software, algorithm | data.table, v.1.14.8 | RRID:SCR_026117 | https://github.com/Rdatatable/data.table | |
| Software, algorithm | GenomicRanges, v.1.48.0 | Lawrence et al., 2013 | RRID:SCR_000025 | https://github.com/Bioconductor/GenomicRanges |
| Software, algorithm | reshape2, v.1.4.4 | RRID:SCR_022679 | https://github.com/cran/reshape2 | |
| Software, algorithm | fitdistrplus, v.1.1–8 | Delignette-Muller and Dutang, 2015 | RRID:SCR_024274 | https://github.com/lbbe-software/fitdistrplus |
| Software, algorithm | RColorBrewer, v.1.1–3 | RRID:SCR_016697 | https://github.com/cran/RColorBrewer | |
| Software, algorithm | dendextend, v.1.17.1 | Galili, 2015 | RRID:SCR_026116 | https://github.com/talgalili/dendextend |
| Software, algorithm | dendroextras, v.0.2.3 | RRID:SCR_026115 | https://github.com/jefferis/dendroextras | |
| Software, algorithm | GGally, v.2.1.2 | RRID:SCR_026114 | https://github.com/ggobi/ggally | |
| Software, algorithm | gridExtra, v.2.3 | RRID:SCR_025249 | https://github.com/baptiste/gridExtra | |
| Software, algorithm | eulerr, v.7.0.1 | Larsson and Gustafsson, 2018 | RRID:SCR_022753 | https://github.com/jolars/eulerr |
| Software, algorithm | ranger, v.0.16.0 | Wright and Ziegler, 2017 | RRID:SCR_022521 | https://github.com/imbs-hl/ranger |
| Software, algorithm | caret, v.6.0–93 | Kuhn, 2008 | RRID:SCR_022524 | https://github.com/topepo/caret |
| Software, algorithm | PRROC, v.1.3.1 | RRID:SCR_026113 | https://github.com/cran/PRROC | |
| Software, algorithm | UpSetR, v.1.4.0 | Conway et al., 2017 | RRID:SCR_026112 | https://github.com/hms-dbmi/UpSetR |
| Software, algorithm | HOMER, v.4.11.1 | Heinz et al., 2010 | RRID:SCR_010881 | http://homer.ucsd.edu/homer/motif/ |
| Software, algorithm | CrossMap, v.0.6.0 | Zhao et al., 2014 | RRID:SCR_001173 | https://github.com/liguowang/CrossMap |
| Software, algorithm | nextflow, v.22.10.4 | Di Tommaso et al., 2017 | RRID:SCR_024135 | https://github.com/nextflow-io/nextflow |
| Software, algorithm | ditiller-nf pipeline, v.0.3.4 | RRID:SCR_026111 | https://github.com/open2c/distiller-nf | |
| Software, algorithm | Mustache, v.1.3.2 | Roayaei Ardakany et al., 2020 | RRID:SCR_026110 | https://github.com/ay-lab/mustache |
| Software, algorithm | bedGraphToBigWig | http://hgdownload.soe.ucsc.edu/admin/exe/ | ||
| Software, algorithm | liftOver | http://hgdownload.soe.ucsc.edu/admin/exe/ | ||
| Software, algorithm | wigToBigWig | http://hgdownload.soe.ucsc.edu/admin/exe/ | ||
| Software, algorithm | Integrative Genomics Viewer, v.2.8.0 | Robinson et al., 2011 | https://igv.org/doc/desktop/ | |
| Software, algorithm | Adobe Illustrator, v.23.0.1 | https://www.adobe.com/products/illustrator.html |
Additional files
-
Supplementary file 1
MChIP-C processed datasets and statistics.
- https://cdn.elifesciences.org/articles/91596/elife-91596-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91596/elife-91596-mdarchecklist1-v1.docx





