Extramacrochaetae regulates Notch signaling in the Drosophila eye through non-apoptotic caspase activity
Figures
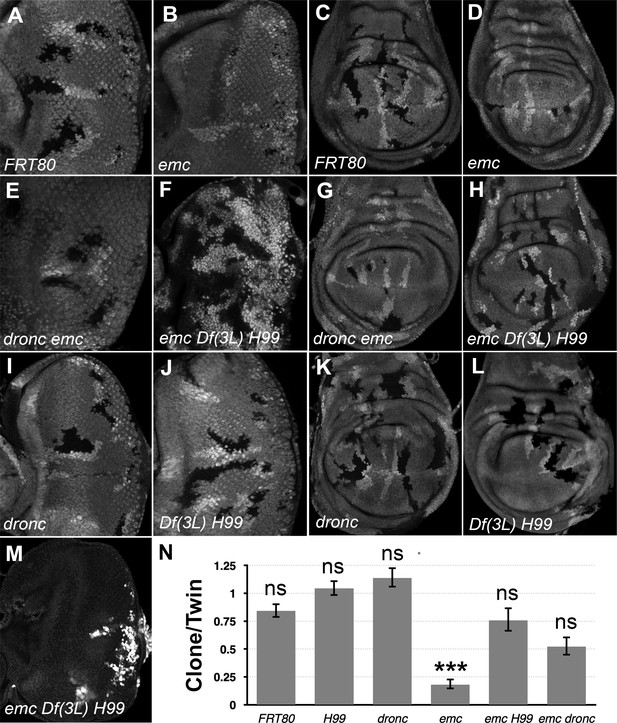
Df(3L)H99 recues growth defect of emc mutant clones.
(A–L) Control and mutant clones in eye and wing imaginal disc were induced at the end of first instar and are associated with sibling twin-spot clones marked by two copies of the GFP marker (brighter gray) that serves as internal control for growth. Clones are labeled by the absence of GFP. (M) emc H99 clones induced in the Minute background (N) Quantification of the ratio of clone size to twin-spot measured in wing imaginal discs. Geometrical means ± SEM are shown. After log-transformation of clone/twin-spot ratios to ensure normality, one-way ANOVA rejected the null hypothesis that these results are the same (p = 5.72 × 10−8). The Holm correction for multiple comparison was used to identify significant differences between all pairs of samples. *** denotes highly significant difference from the FRT80 control (p < 0.001), NS denotes no significant difference (p > 0.05). Whereas the clone/twin-spot ratio for emc homozygous clones was significantly different from the FRT80 control (p = 5.72 × 10−6), this was not true for any of the other genotypes (H99, p = 0.79; dronc, p = 0.906; emc H99, p = 0.92; emc dronc, p = 0.345). The clone/twin-spot ratio for emc homozygous clones was also significantly different from that for emc H99 or emc dronc (p = 4.12 × 10−6 and p = 0.0177, respectively), whereas emc H99 and emc dronc did not differ significantly from H99 or dronc clones (p = 1 and p = 0.136, respectively). Source data for (N) are provided in Figure 1—source data 1. Genotypes: (A, C) ywhsF;FRT80/[UbiGFP]FRT80, (B, D) ywhsF;emcAP6FRT80/[Ubi-GFP]FRT80, (E, G) ywhsF;dronci29emcAP6 FRT80/[UbiGFP]FRT80, (F, H) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP]FRT80, (I, K) ywhsF;dronci29 FRT80/[UbiGFP]FRT80, (J, L) ywhsF;;Df(3L)H99FRT80/[UbiGFP]FRT80, (M) ywhsF; emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80. N = 10 for each genotype.
-
Figure 1—source data 1
Clone size source data for Figure 1N.
- https://cdn.elifesciences.org/articles/91988/elife-91988-fig1-data1-v2.xlsx
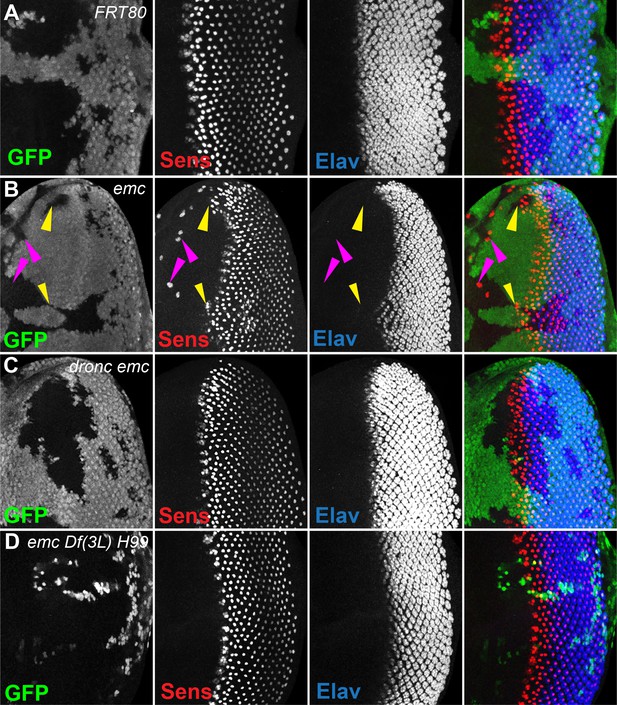
Morphogenetic furrow progression is affected by cell death pathways.
In all panels, the differentiating neurons are marked by Elav (in blue), and mutant cells are identified by the absence of GFP expression (in green). (A) The wave of retinal differentiation from posterior to anterior (right to left), marked here by Senseless expression in R8 photoreceptor cells (red) is normal in FRT80 control clones. (B) emc null clones lacking GFP show acceleration of retinal differentiation illustrated by yellow arrows (premature differentiation can also continue into wild-type regions ahead of such clones). In addition, ectopic neural differentiation also occurs sporadically anterior to the morphogenetic furrow, and unassociated with it (magenta arrows). (C) In contrast, retinal differentiation proceeds at the same pace in emc dronc double mutant clones as in nearby wild-type regions in 75% of the eye discs. (D) emc H99 double mutants show a stronger suppression of the acceleration of retinal differentiation (compare panel B). Genotypes: (A) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (B) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (C) ywhsF;dronci29emcAP6 FRT80/[UbiGFP] M(3)67C FRT80 (n = 12), (D) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80. N = 8 for each genotype.
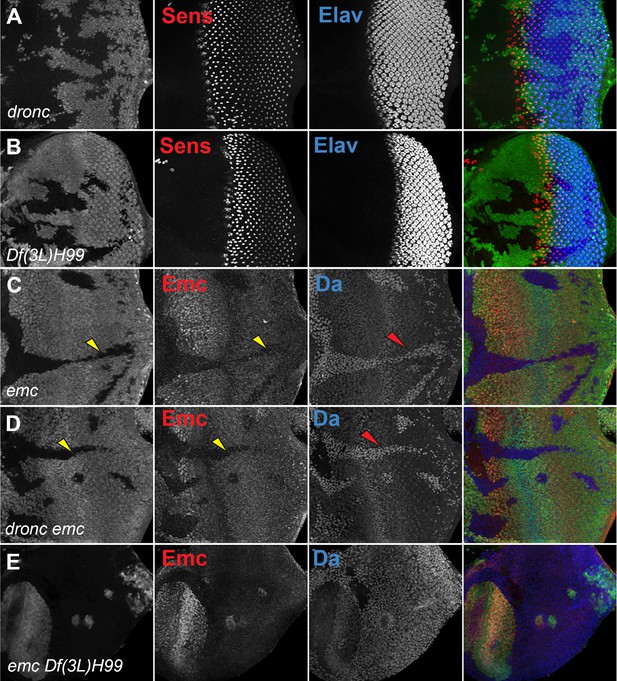
Morphogenetic furrow progression in emc mutant cells.
In eye imaginal discs, normal furrow progression was observed in (A) dronc−/− and (B) Df(3L)H99 homozygous clones, indicated by Senseless and Elav staining. Homozygous mutant clones of emc−/− (C) lacking GFP expression have no Emc staining (yellow arrowhead) and higher Da (red arrowhead). At the morphogenetic furrow as shown by yellow arrow, Emc expression goes down and Da goes up. Similar results were observed in (D) dronc−/− emc−/− clones and (E) emc−/− Df(3L) H99−/− clones confirm that these are emc null clones. Genotypes: (A) ywhsF;dronci29FRT80/[UbiGFP] M(3)67C FRT80, (B) ywhsF; Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80, (C) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (D) ywhsF;dronci29emcAP6 FRT80/[UbiGFP] M(3)67C FRT80, (E) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80. N = 4 for each genotype.
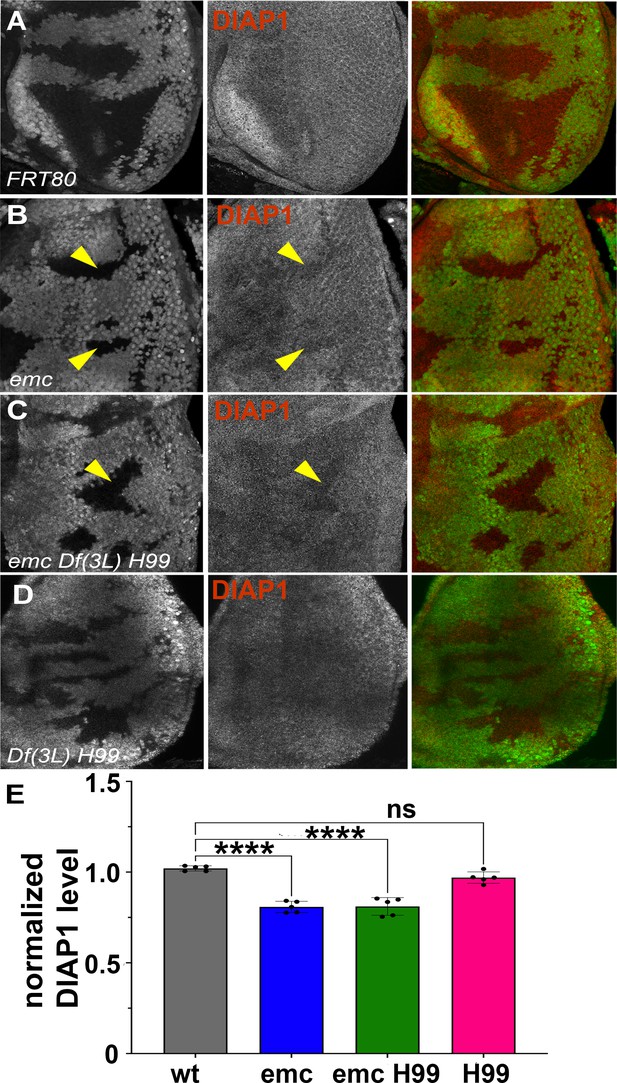
Diap1 expression in emc clones.
In mosaic eye discs with (A) FRT80 clones, there is no difference in Diap1 levels within and outside the clone. However, both (B) emc mutant clones and (C) emc H99 clones show reduced Diap1 levels posterior to the morphogenetic furrow, compared to the heterozygous background. (D) H99 clones showed similar Diap1 levels, compared to the heterozygous background. (E) Quantification of Diap1 levels in different clone genotypes, compared to the background levels outside the clones. Means ± SEM are shown. Note that our experiments did not generate mosaics of wild-type and Df(3L)H99/+ cells for direct comparison of these genotypes. Statistical significance calculated by two-way ANOVA (****p ≤ 0.0001). Source data for (E) are provided in Figure 3—source data 1. Genotypes: (A) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (B) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (C) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80, (D) ywhsF; Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80. N = 6 for each genotype.
-
Figure 3—source data 1
Anti-Diap1 labeling source data for Figure 3E.
- https://cdn.elifesciences.org/articles/91988/elife-91988-fig3-data1-v2.xlsx
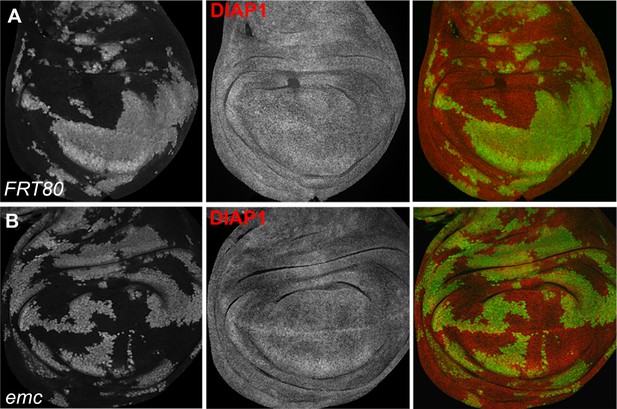
DIAP1 staining in wing disc clones.
In wing discs of emc clones (B) we did not detect changes in Diap1 levels in comparison to control clones (A). Genotypes: (A) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (B) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80.
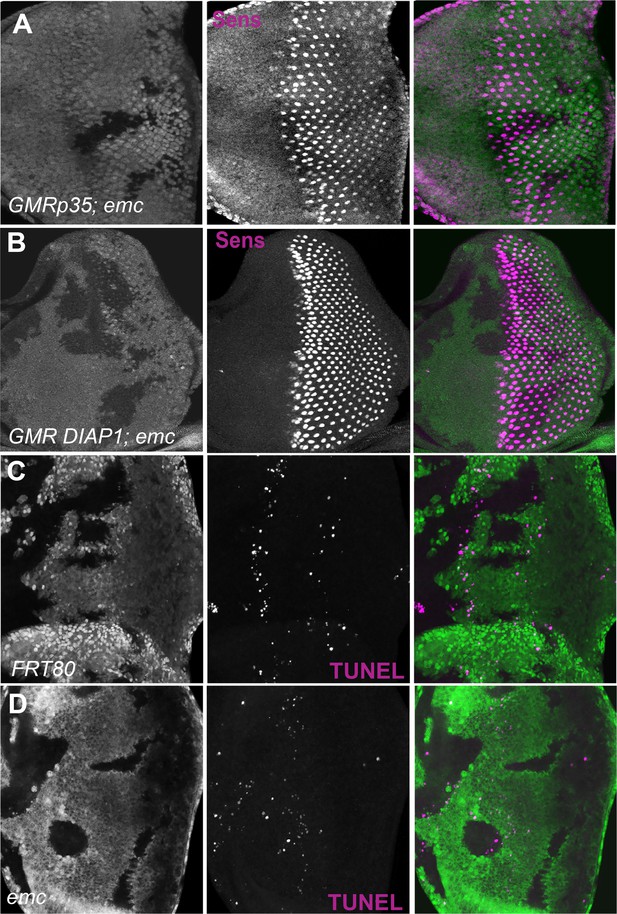
Apoptosis and furrow progression.
Representative images of eye imaginal discs stained for senseless. (A) In GMR-DIAP1 eye disc mutant for emc furrow progression is normal and is marked by Senseless staining. Similarly, in GMR-p35 eye disc mutant for emc also show normal furrow progression (B). Representative images of eye imaginal discs with terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining. (A) ywhsF/GMR-DIAP1;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (B) ywhsF/GMR-p35;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (C) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (D) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80. N = 4 for each genotype.
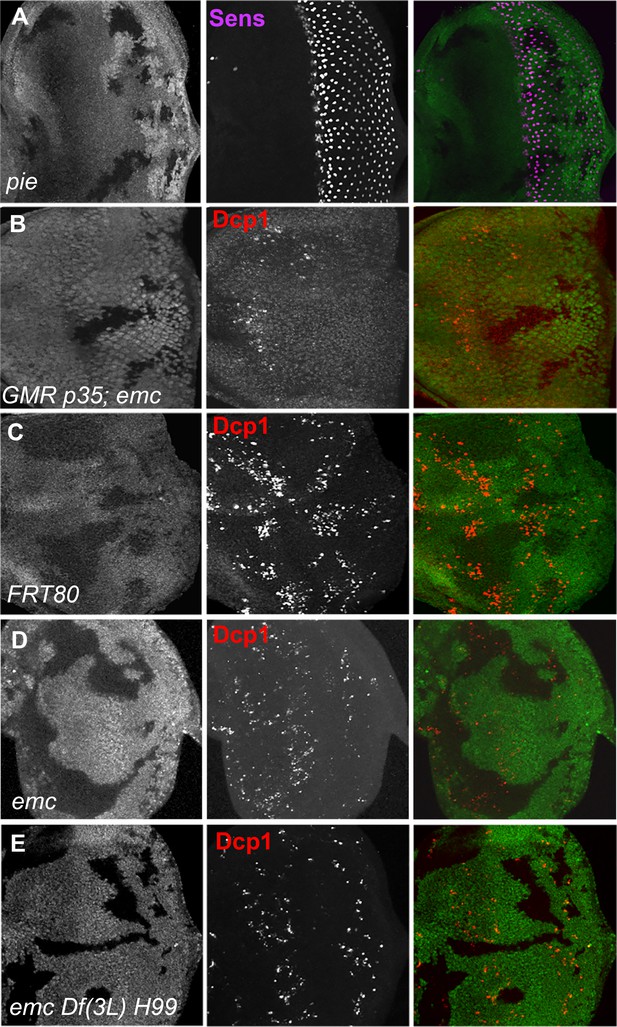
Emc clones show no caspase activity beyond the morphogenetic furrow.
In pieEB3 homozygous clones, furrow progression occurs at normal speed marked by Senseless staining in R8 photoreceptors (A). In GMR-p35 eye disc mutant for emc show lack of Dcp1staining (B). In emc mutant clones posterior to morphogenetic furrow show lack of Dcp1 staining (D) in comparison to FRT80 clones (C). Genotypes: (A) ywhsF/+; pieEB3FRT40/FRT40Alz, (B) ywhsF/GMR-p35;;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (C) ywhsF;;FRT80/[UbiGFP] M(3)67C FRT80, (D) ywhsF;;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (E) ywhsF;;emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80. N = 4 for each genotype.
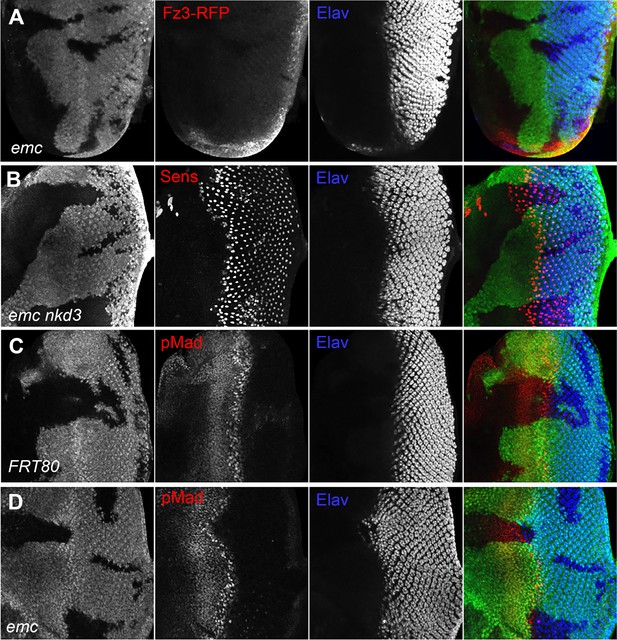
Wingless and Dpp signaling are not caspase targets.
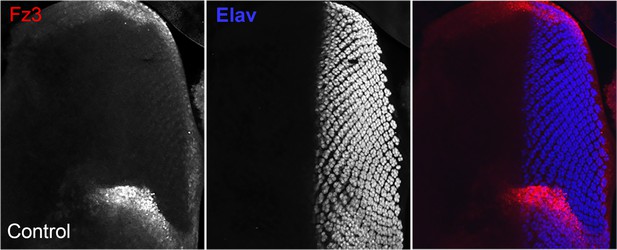
(A) No reduction in the Wg signaling reporter Fz3-RFP was detectable in emc clones. See Figure 4—figure supplement 1 for Fz3-RFP expression in the wild-type. (B) Retinal differentiation is accelerated in emc nkd3 double mutant clones like in emc clones. Senseless expression in R8 photoreceptor cells and Elav staining in differentiating photoreceptors are shown. (C) p-Mad accumulates around the morphogenetic furrow in eye discs containing control clones (Firth et al., 2010; Vrailas and Moses, 2006). (E) Except for the advanced progression, p-Mad levels were unchanged in emc clones. Genotypes: (A) Fz3-RFP/+; emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (B) ywhsF;emcAP6nkd3FRT80/[Ubi-GFP] M(3)67C FRT80, (C) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (D) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80. N = 8 for each genotype.
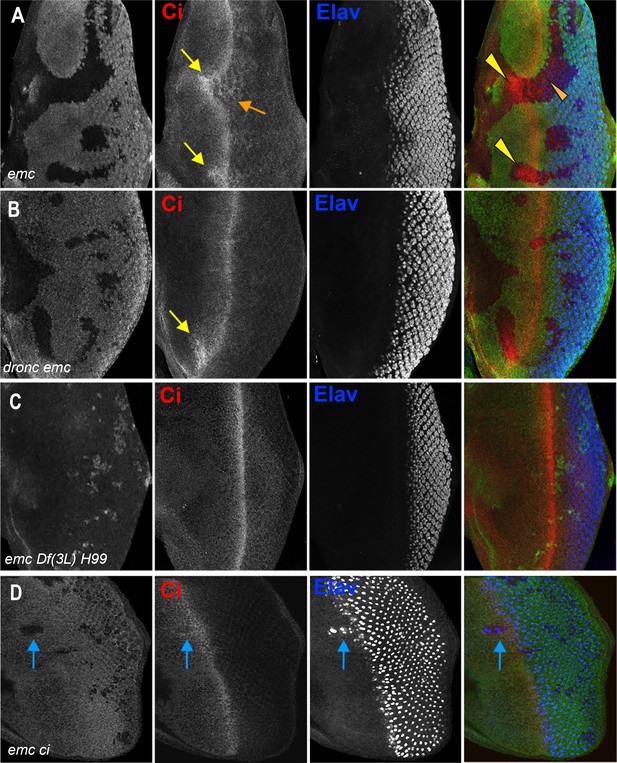
Ci expression and function in emc mutant clones.
(A) Ci is elevated within emc mutant cells (yellow arrows). This was also true posterior to the morphogenetic furrow (orange arrow). (B) Higher Ci was also seen in emc dronc mutant cells, even when the morphogenetic furrow progressed normally, as indicated by Elav staining. (C) Ci levels were completely normal in emc H99 clones. (D) emc and ci double mutant clones lacking GFP shows accelerated retinal differentiation (blue arrow). Genotypes: (A) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (B) ywhsF;dronci29emcAP6 FRT80/[UbiGFP] M(3)67C FRT80, (C) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80, (D) ywhsF;emcAP6FRT80/[Ubi-GFP] ci+ M(3)67C FRT80;ci[94]/ci[94]. N = 8 for each genotype except (D) which has n = 3.
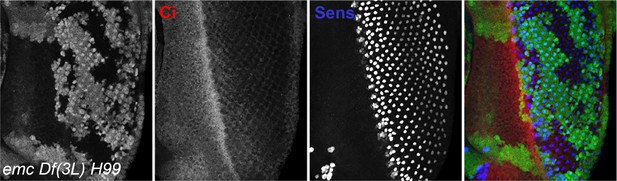
Ci155 levels when cell death pathways are blocked.
Representative image of eye imaginal disc in emc H99 clones in the non-Minute background, showing similar areas inside and outside clones. Genotype: ywhsF; emcAP6 Df(3L)H99 FRT80/[UbiGFP]FRT80. N = 4.
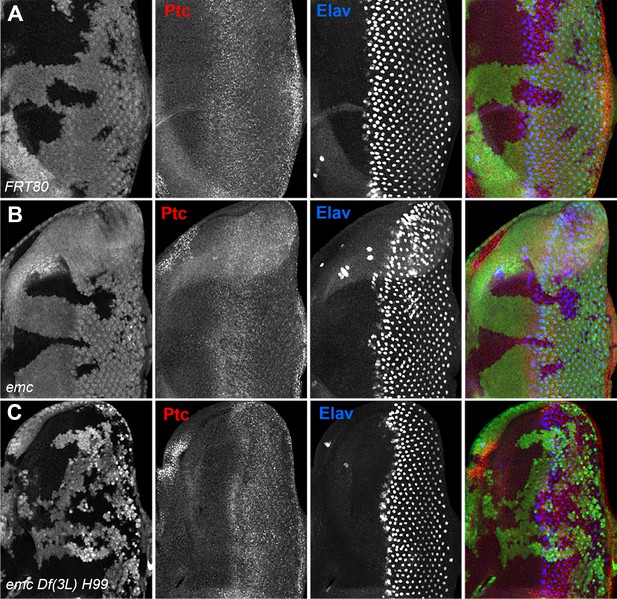
Patched staining in emc mutants.
(A) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (B) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (C) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT8.
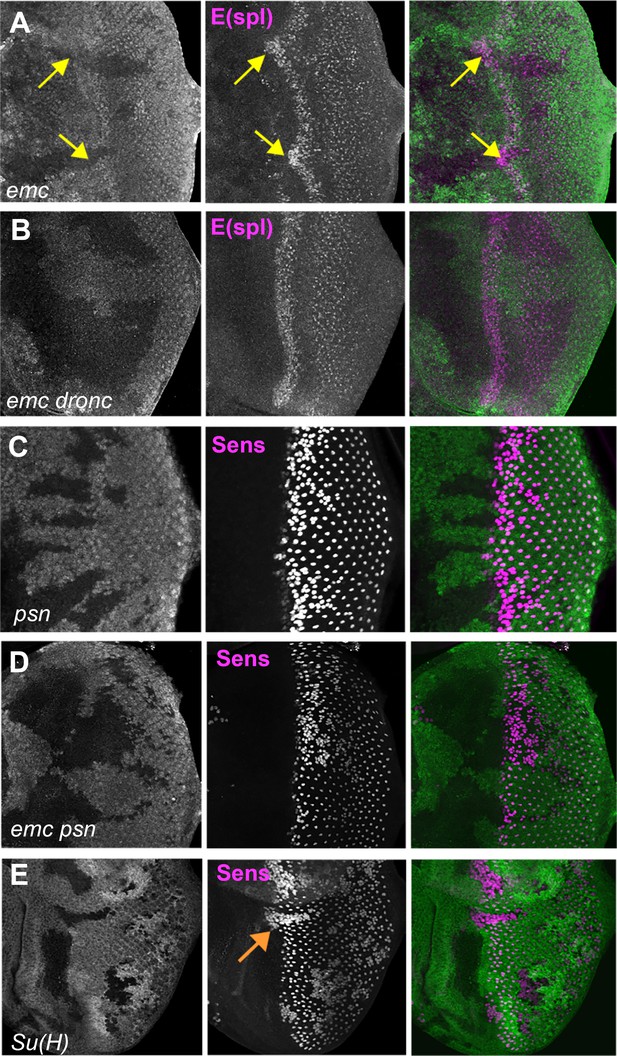
Notch activity and function in emc mutant clones.
(A) E(spl), an important N target for lateral inhibition, is elevated in the morphogenetic furrow region of emc clones (yellow arrows). (B) emc dronc clones have normal levels of E(spl) protein. (C) Senseless staining shows a neurogenic phenotype in psn mutant clones, due to reduced Notch signaling. Morphogenetic furrow progression is unaffected. (D) A neurogenic phenotype was also observed in emc psn clones, along with normal furrow progression. (E) Su(H) mutant clones identified by absence of GFP labeling show a strong neurogenic phenotype as well as advanced retinal differentiation (orange arrow) (Li and Baker, 2001). The cell-autonomous effect results in a discontinuity at the borders of Su(H) clones, where differentiation outside the clones lags that within clones (orange arrow) Genotypes: (A) ywhsF;emcAP6 FRT80/[Ubi-GFP] M(3)67C FRT80, (B) ywhsF; dronci29emcAP6 FRT80/[UbiGFP] M(3)67C FRT80, (C) ywhsF/+;psnV1 FRT80/[Ubi-GFP]M(3)67CFRT80, (D) ywhsF/+; emcAP6 psnV1 FRT80/[Ubi-GFP]M(3)67CFRT80, (E) ywhsF; Su(H)D47FRT40/FRT40[UbiGFP]. N = 6 for each genotype.
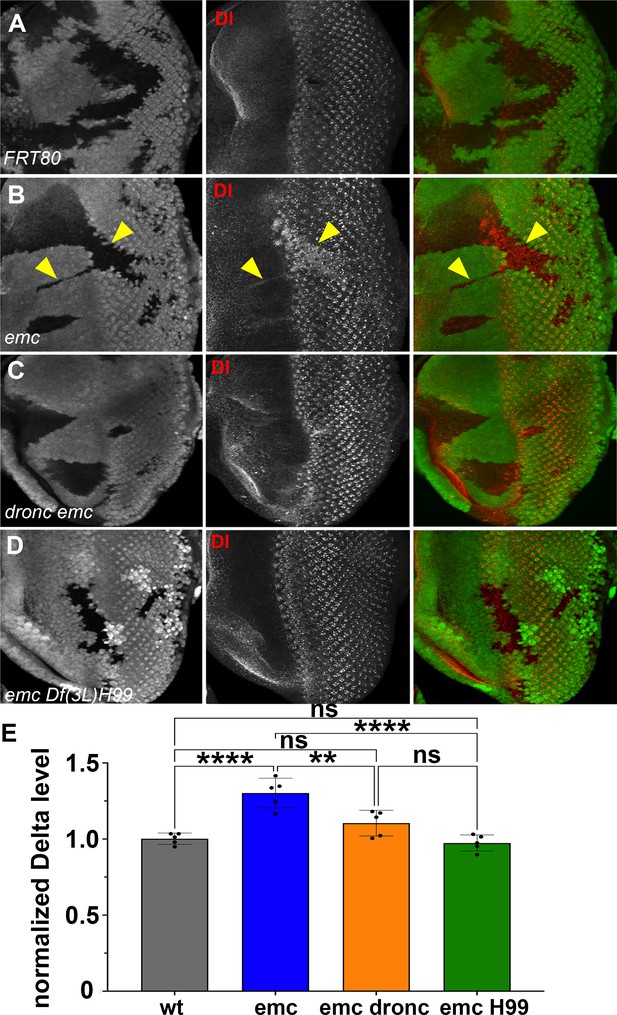
Caspases regulate Delta levels.
(A) Normal levels of Delta protein are seen in FRT80 clones as compared to elevated Delta levels in (B) emc clones (arrowheads) whereas that phenotype is reversed in (D) emc H99 clones. However, as seen in (C), some dronc emc clones show intermediate Delta levels, although most resemble wild type. (E) Quantification of Delta levels in different clone genotypes, compared to the background levels outside the clones. Means ± SEM are shown. Significance was determined using one-way Anova with Tukey’s post hoc test. (**p ≤ 0.01, ****p ≤ 0.0001). Source data for (E) are provided in Figure 7—source data 1. Genotypes: (A) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (B) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (C) ywhsF;dronci29emcAP6 FRT80/[UbiGFP] M(3)67C FRT80, (D) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP]FRT80. N = 7 for each genotype.
-
Figure 7—source data 1
Anti-Dl labeling source data for Figure 7E.
- https://cdn.elifesciences.org/articles/91988/elife-91988-fig7-data1-v2.xlsx
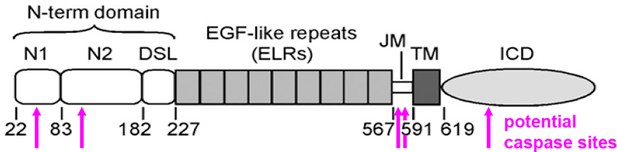
Potential caspase sites predicted in the Delta protein.
Like many proteins, Delta has multiple potential caspase cleavage sites, here predicted suing Cascleave 2.0 (Wang et al., 2014). None are high-confidence predictions and only one is in the intracellular domain where caspase access is plausible (position 675, predicted score 0.647). The intracellular domain is required for Delta signaling. Cleavage at position 675 would remove the main ubiquitylation site required for signaling, as well as the binding site for mindbomb, so is not anticipated to enhance signaling activity (Daskalaki et al., 2011).
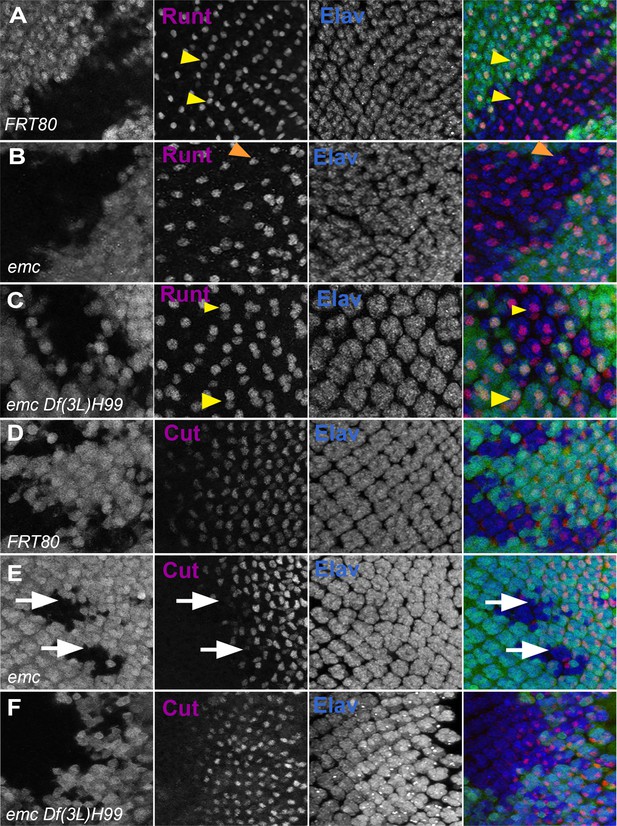
Caspases contribute to R7 photoreceptor and cone cell defects in emc mutants.
In all panels, emc mutant cells are marked by the absence of GFP expression (in green) and photoreceptor neurons are marked by Elav in blue. (A) Runt (in red) is expressed in R7 and R8 (yellow arrowhead) photoreceptor cells inside and outside the clone in FRT80 controls. (B) Inside emc clone, Runt expression is lost from R7 cells, while expression in R8 cells remains unaffected (orange arrowhead). (C) However, inside the emc H99 clones, Runt is expressed in both R7 and R8 cells. (D) Cut (in red) is expressed in cone cells in FRT80 controls. (E) Inside emc clones, cut is delayed. (F) However, inside the emc H99 clones, cut staining is not delayed. Genotypes: (A, D) ywhsF;FRT80/[UbiGFP] M(3)67C FRT80, (B, E) ywhsF;emcAP6FRT80/[Ubi-GFP] M(3)67C FRT80, (C, F) ywhsF;emcAP6 Df(3L)H99 FRT80/[UbiGFP] M(3)67C FRT80. N = 4 for each genotype.
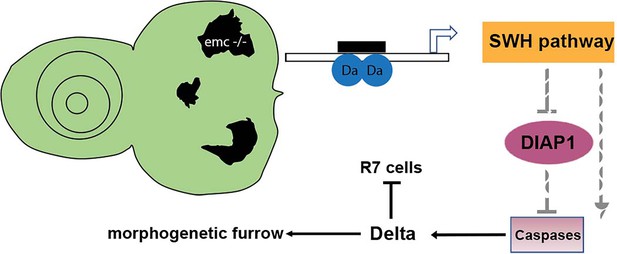
Model of emc effects on Drosophila eye development.
Loss of emc allows Da protein to form homodimers and activate ex transcription, increasing Salvador–Warts–Hippo (SWH) pathway activity. SWH activity reduces DIAP1 expression, thereby derepressing caspase activity. In the eye, non-apoptotic caspase activity increases expression of the Notch ligand Delta. Elevated Delta expression accelerates morphogenetic furrow progression, while cis-inhibiting Notch signaling posterior to the morphogenetic furrow, inhibiting R7 cell specification and cone cell differentiation. In wild-type cells, most Da is likely heterodimerized with either a proneural protein or with Emc protein, and there is no role of caspases in Dl expression.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | emc | GenBank | FBgn0000575 | |
| Genetic reagent (D. melanogaster) | emc [AP6] | PMID:7947322 | FBal0051626 | |
| Genetic reagent (D. melanogaster) | dronc [i29] | PMID:15800001 | FBal0190283 | |
| Genetic reagent (D. melanogaster) | Df(3L)H99 | PMID:8171319 | FBab0022359 | |
| Genetic reagent (D. melanogaster) | nkd[3] | PMID:2081466 | FBal0013025 | |
| Genetic reagent (D. melanogaster) | PBac{y+ w + ci+}VK33 | This paper | ||
| Genetic reagent (D. melanogaster) | psn[v1] | Bloomington Drosophila Stock Center | FBal0316340 BDSC: 63237 | |
| Genetic reagent (D. melanogaster) | Fz3-RFP | PMID:21869817 | ||
| Genetic reagent (D. melanogaster) | pie[eB3] | PMID:1634999 | FBal0032439 | |
| Genetic reagent (D. melanogaster) | Su(H)Δ47 | PMID:1617730 | FBal0103950 | |
| Antibody | Emc (Rabbit polyclonal) | Y.N. Jan | (1:8000) | |
| Antibody | Da (Mouse monoclonal) | PMID:3802198 | (1:200) | |
| Antibody | GFP (Rat monoclonal) | Nacalai Tesque | Cat #GF090R RRID:AB_2314545 | (1:50) |
| Antibody | GFP (Rabbit polyclonal) | Invitrogen | Cat #A-11122 RRID:AB_221569 | (1:500) |
| Antibody | β-Gal (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat #40-1a RRID:AB_528100 | (1:100) |
| Antibody | β-Gal (Rabbit polyclonal) | Cappel (MP Biomedicals) | Cat #55976 RRID:AB_2313707 | (1:100) |
| Antibody | Runt (Guinea pig polyclonal) | PMID:9683745 | (1:500) | |
| Antibody | E(spl)bHLH mAb323 (Mouse monoclonal) | PMID:1618155 | (1:50) | |
| Antibody | Senseless (Guinea pig polyclonal) | PMID:10975525 | (1:500) | |
| Antibody | Cleaved Drosophila Dcp-1 (Rabbit polyclonal) | Cell Signaling Technology | Cat #9578 RRID:AB_2721060 | (1:100) |
| Antibody | DIAP1 (Rabbit polyclonal) | PMID:12021769 | (1:50) | |
| Antibody | phospho-Smad1/5 (Rabbit monoclonal) | Cell Signaling Technology | Cat #9516 RRID:AB_491015 | (1:100) |
| Antibody | Delta (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat #C594.9B RRID:AB_528194 | (1:2000) |
| Antibody | Cut (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat #2B10 RRID:AB_528186 | (1:50) |
| Antibody | Ptc (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat #Apa 1 RRID:AB_528441 | (1:40) |
| Antibody | Elav (Rat monoclonal) | Developmental Studies Hybridoma Bank | Cat #7E8A10 RRID:AB_528218 | (1:50) |
| Antibody | Elav (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat #9F8A9 RRID:AB_528217 | (1:100) |
| Antibody | Ci (Rat monoclonal) | Developmental Studies Hybridoma Bank | Cat #2A1 RRID:AB_2109711 | (1:10) |
| Antibody | Cy2, Cy3, and Cy5 | Jackson ImmunoResearch | (1:200) | |
| Antibody | Alexa 555 (Guinea pig polyclonal) | Invitrogen | Cat #A-21435 RRID:AB_2535856 | (1:500) |
| Recombinant DNA reagent | genomic Ci | PMID:33084577 | ||
| Commercial assay or kit | ApopTag Red in situ apoptosis detection kit | Millipore Sigma | Cat #S7165 | |
| Software | Cascleave 2.0 | PMID:24149049 |