Large-scale analysis of the integration of enhancer-enhancer signals by promoters
Figures
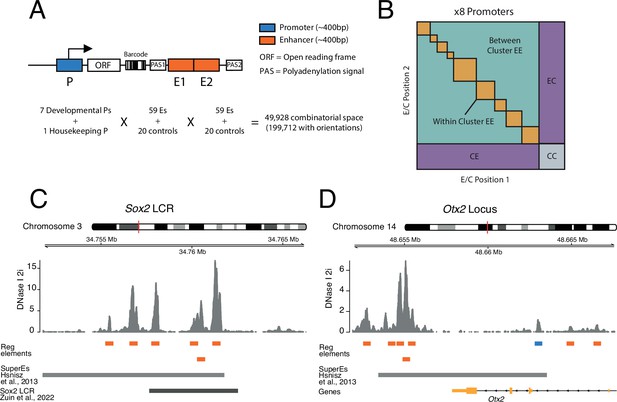
Schematic of three-way combinatorial approach.
(A) Three-way combinatorial massively parallel reporter assay (MPRA) design to test enhancer-enhancer-promoter combinations. Eight barcoded reporter assay libraries, one per promoter, were constructed. Pairs of DNA elements (enhancers and scrambled control sequences) were inserted after barcoded reporter. The enhancers and controls can be placed in both orientations in either the enhancer position 1 (E1) or enhancer position 2 (E2). (B) The design of the library yields eight matrices that contain control-control combinations (CC), enhancer-control combinations (EC and CE), and enhancer-enhancer combinations (EE). (C and D) Two example loci, Sox2 LCR (locus control region) (C) and Otx2 (D) from where we selected enhancers to test in the reporter libraries. Enhancers (orange) were defined around DNAse I hypersensitivty sites from mouse embryonic stem cells (mESC). Promoters (blue) were chosen according to transcription start site (TSS) annotation and mESC DNAse I hypersensitivty sites. DNAse I data is from Joshi et al., 2015.
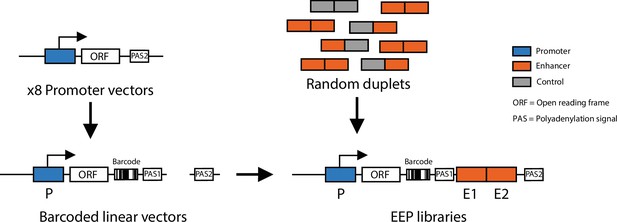
Schematic of cloning strategy to generate enhancer-enhancer-promoter libraries.
First the eight different promoters were cloned into a reporter vector. Each of the eight vectors was barcoded and linearised, random enhancer-enhancer, enhancer-control, and control-control combinations were cloned downstream of the barcoded reporter.
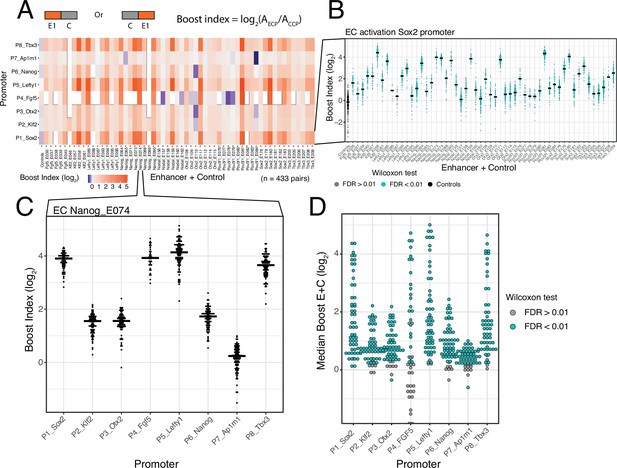
Effects of single enhancers across promoters.
(A) Enhancer-promoter median boost index matrix of single enhancers. For each single enhancer a median boost index (see figure and Methods, log2(Activity_Combination/Activity_control-control_baseline)) was calculated across all enhancer-control combinations for that particular enhancer in any position and orientation, the baseline is the median activity of the promoter across all control-control combinations. For each control-control combination a boost index was calculated as the activity of the control-control combination over the median activity of all controls. A median control-control boost index was calculated based on this. Colour coding of the matrix corresponds to the median boost indexes, white spaces are missing data. (B) Distribution of boost indices for all enhancer+control combinations for the Sox2 promoter. Leftmost column corresponds to the boost index distribution for all control-control combinations for the Sox2 promoter. Each dot represents one enhancer+control combination. Horizontal lines correspond to the median of each distribution (same median as represented in the matrix in A). (C) Distribution of boost indices for all enhancer+control combinations for the enhancer Nanog_E074 across all promoters. Each dot represents one enhancer+control combination. Horizontal lines correspond to the median of each distribution. (D) Distribution of median boost indices for each single enhancer across promoters. Each dot represents the median of an enhancer across all enhancer+control combinations for that enhancers (same median as represented in the matrix in A). Colouring in B and D represents the significance at a 1% false discovery rate (FDR) for a Wilcoxon test comparing boost index distributions between enhancer+control combinations and the controls for each enhancer.
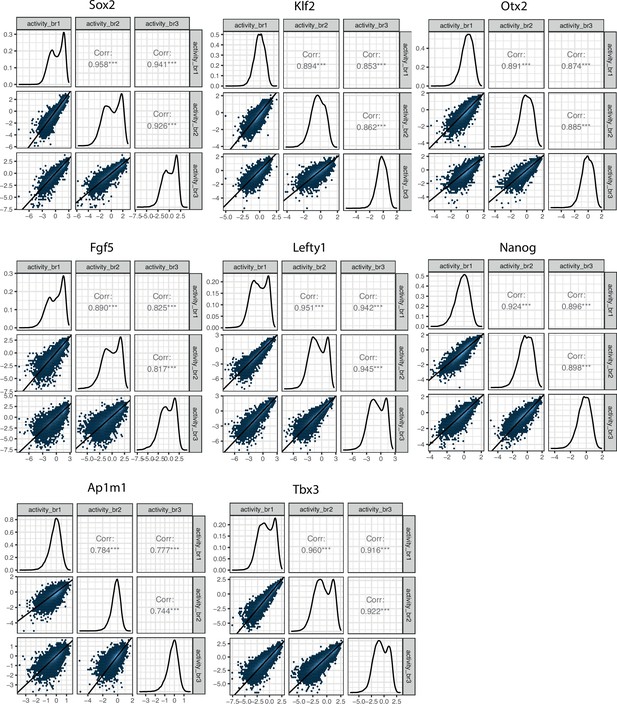
Reproducibility of experimental data matrix of replicate correlations for all eight enhancer-enhancer-promoter (EEP) libraries.
Lower left panels represent the two-dimensional density plots of replicate-replicate activity correlations. Middle panels represent the one-dimensional density plot of each replicate. Upper right panels are the Pearson’s correlation coefficients of the mirror lower left panels.
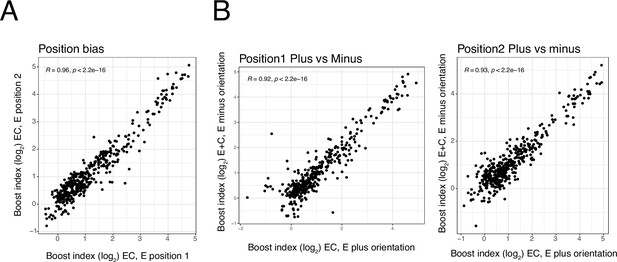
Position and orientation bias.
(A) Relationship between single enhancer boost indices across all enhancer-control (EC) combinations in position 1 versus in position 2 (see Figure 1). (B) Relationship between single enhancer boost indices across all EC combinations plus versus minus orientation, in position 1 (left) and in position 2 (right). For both panels each dot is one enhancer with one promoter. R is Pearson’s correlation.
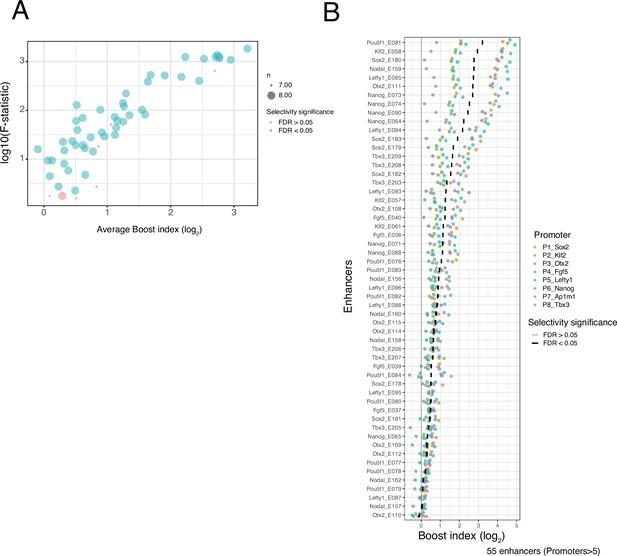
Selectivity of single enhancers.
(A) Relationship between average boost index of each single enhancer across all promoters and the F-statistic of a Welch F-test for that enhancer across all promoters. Colours indicate significance at a 5% false discovery rate (FDR). (B) Distributions of single enhancer boost indices for each enhancer across promoters. Each dot is one enhancer with one promoter and the boost index is the average boost index across all enhancer-control combinations. Colours of each point represents promoter identity. Vertical bars indicate the median boost index, and their grey or black shading indicates significance according to the Welch F-test at a 5% FDR.
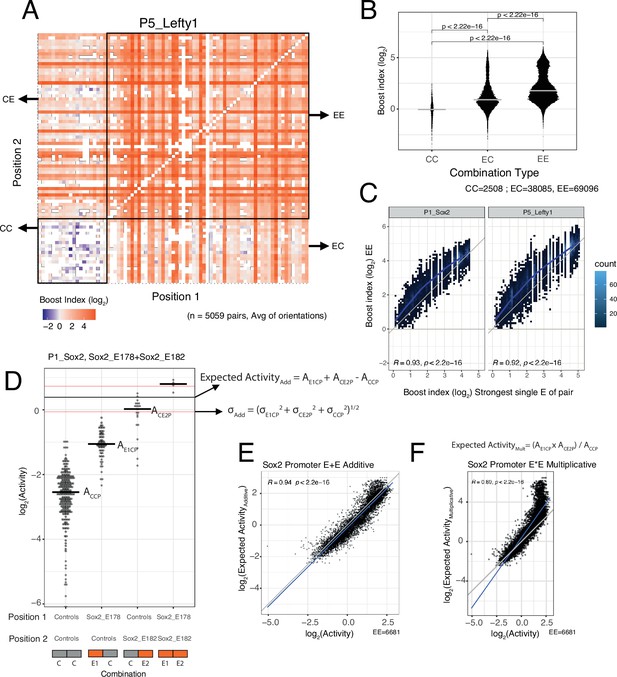
Effects of enhancer-enhancer (EE) combinations.
(A) Fragment-fragment combinatorial boost index matrix for the Lefty1 promoter. Each square represents one control-control (CC), enhancer-control (EC or CE), or EE combination. Colour coding corresponds to the average boost index for each combination across all orientations measured over the median control-control baseline. (B) Boost index distributions across all eight promoters for each combination type, control-control (CC), enhancer-control (EC regardless of position) or EE combinations. p-Values correspond to the result of a Wilcoxon test. (C) Relationship between observed boost index for each EE combination and the observed boost index of the strongest single enhancer of the pair for Sox2 and Lefty1 promoters. Blue lines represent the LOESS fit of the data. (D) Observed and expected additive activities for the Sox2_E178+Sox2_E182 combination with the Sox2 promoter and the individual activities of each of the elements. Each column represents the observed activities for the control-contol combinations, the enhancer-control combinations, and the EE combinations. The horizontal bars represent the median of each distribution. The horizontal black line represents the expected additive activity of the EE combination as calculated by the formula in the panel in the linear space. The horizontal red lines represent the propagated standard deviations of the expected additive activity of the EE combinations as calculated by the formula in the panel. (E and F) Relationship between observed and expected activities (additive in E, multiplicative in F) for all EE combinations for the Sox2 promoter. The blue lines represent the linear fit of the data. Grey diagonal line is the x=y identity line. In all panels R represents Pearson’s correlation coefficient. Expected activities are calculated in the linear space and then plotted in the log2 space.
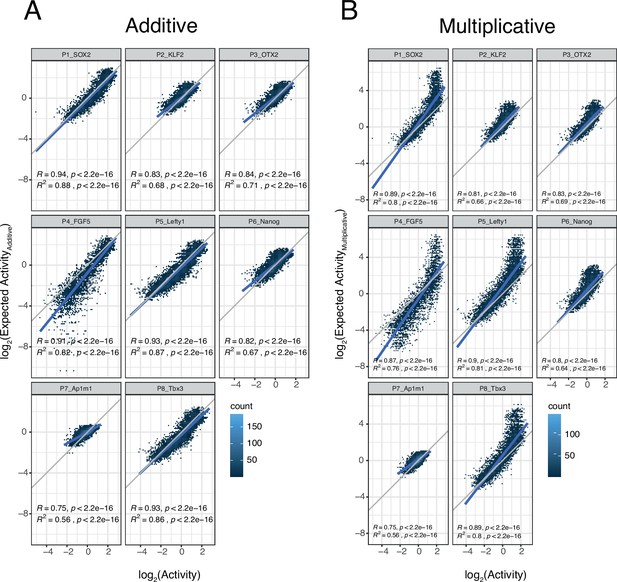
Additivity versus multiplicativity for all promoters.
Relationship between observed and expected activities (additive in A, multiplicative in B) for all enhancer-enhancer combinations for the all promoters. The blue lines represent the linear fit of the data. In all panels R and R2 are based on Pearson’s correlation.
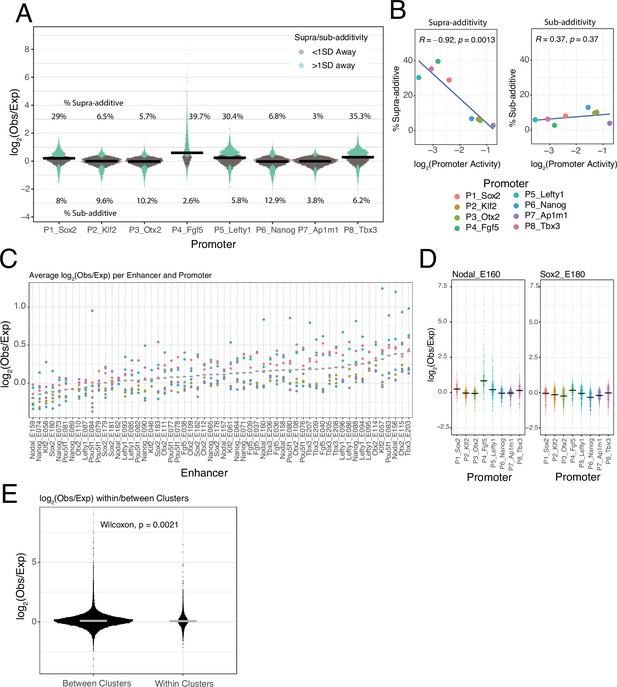
Supra- and sub-additive behaviours of enhancer combinations.
(A) Distributions of log2 observed activities over expected additive activities ratios of enhancer-enhancer combinations across promoters. Coloured in turquoise are supra- and sub-additive combinations for which the observed activity is more than one standard deviation away from the expected activity. Horizontal bars represent the median of each distribution. Numbers on the top part are the percentage of supra-additive combinations for each promoter. Numbers on the lower part are the percentage of sub-additive combinations for each promoter. (B) Relationship between the percentages of supra- and sub-additive enhancer-enhancer combinations and promoter control-control baselines. Blue lines are the linear fit of the data. R is Pearson’s correlation. (C) Average supra- or sub-additive behaviour of each single enhancer across enhancer-enhancer combinations for each promoter. Each dot represents the median log2 observed over expected for all enhancer-enhancer combinations of a single enhancer and a particular promoter. Grey bars represent the median of each distribution. (D) For two example enhancers, distribution of log2 observed over expected ratios for combinations of that enhancer with any other enhancer and promoter. Horizontal black bars represent the median of the distribution. (E) Distribution of log2 observed over expected ratios for enhancer-enhancer pairs from the same enhancer cluster (within clusters) or from different enhancer clusters (between clusters).The p-value results from comparing both distributions using a Wilcoxon test. Horizontal grey bars represent the median of each distribution. In all panels Obs/exp refers to observed activity over expected additive activity.
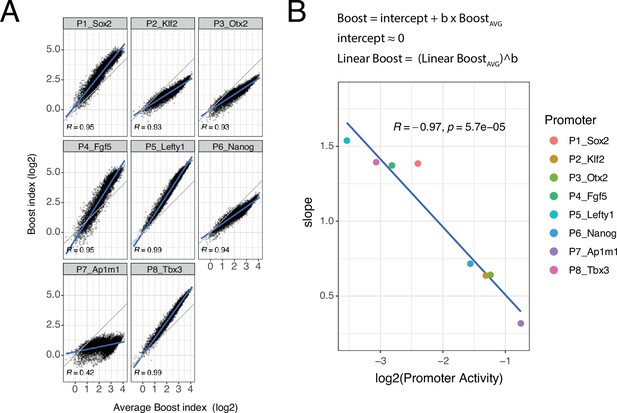
Non-linear responses of promoters to enhancer-enhancer combinations.
(A) Relationship between observed boost indices and average boost index across promoters for all shared enhancer-enhancer combinations. Blue lines represent the linear fit of the data. (B) Relationship between the slopes extracted from the linear fits in A and the baseline promoter activities derived from the control-control combinations. The formulae depict the relationship between the average boost indices and the observed boost indices of each promoter through the extracted slopes. For both panels R is Pearson’s correlation coefficient.
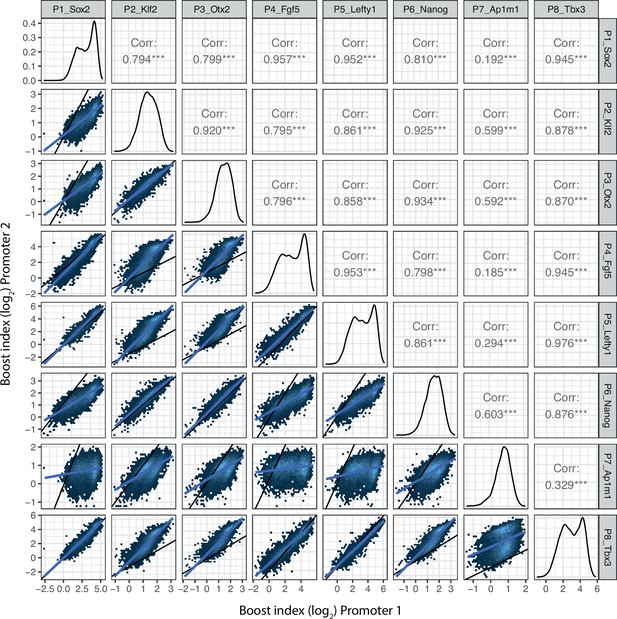
Promoter-promoter boost index correlations for all shared enhancer-enhancer combinations.
Lower left panels represent the two-dimensional density plots of promoter-promoter boost index correlations. Middle panels represent the one-dimensional density plot of each promoter. Upper right panels are the Pearson’s correlation of the mirror lower left panels. Blue lines are the linear fit of the data.
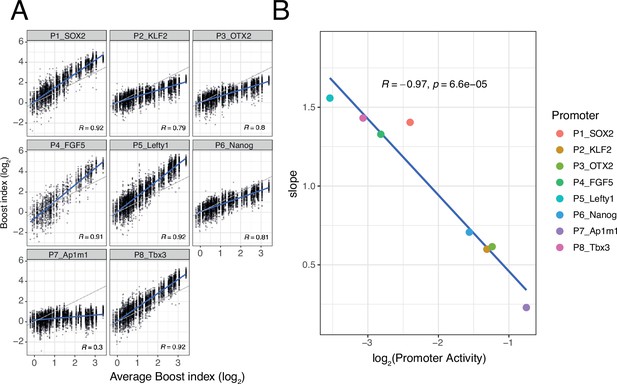
Non-linear responses of promoters to enhancer-control (single enhancers) combinations.
(A) Relationship between observed boost indices of single enhancers and average boost index across promoters for all shared single enhancer-control combinations. Blue lines represent the linear fit of the data. (B) Relationship between the slopes extracted from the linear fits in A and the baseline promoter activities derived from the control-control combinations. For both panels R is Pearson’s correlation coefficient.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Mus musculus) | E14tg2a mouse embryonic stem cell (mESC) | ATCC | CRL-1821 | |
| Gene (M. musculus) | Klf2 | GenBank | 16598 | Promoters and enhancers; coordinates in Supplementary file 1 |
| Gene (M. musculus) | Sox2 | GenBank | 20674 | Promoters and enhancers; coordinates in Supplementary file 1 |
| Gene (M. musculus) | Otx2 | GenBank | 18424 | Promoters and enhancers; coordinates in Supplementary file 1 |
| Gene (M. musculus) | Lefty1 | GenBank | 13590 | Promoters and enhancers; coordinates in Supplementary file 1 |
| Gene (M. musculus) | Fgf5 | GenBank | 14176 | Promoters and enhancers; coordinates in Supplementary file 1 |
| Gene (M. musculus) | Tbx3 | GenBank | 21386 | Promoters and enhancers; coordinates in Supplementary file 1 |
| Gene (M. musculus) | Nanog | GenBank | 71950 | Promoters and enhancers |
| Gene (M. musculus) | Ap1m1 | GenBank | 11767 | Promoters and enhancers |
| Recombinant DNA reagent | Downstream Assay vector (JvAp102) | Martinez-Ara et al., 2022 | JvAp102 | Downstream reporter assay plasmid; ‘see Methods |
| Sequence-based reagent | Enhancer and promoter oligos | This paper | Primers | Supplementary file 4 |
| Sequence-based reagent | Barcoding primers and sequencing primers | Martinez-Ara et al., 2022 | Barcoding reagents | Supplementary file 4 |
| Sequence-based reagent | Synthetic DNA negative controls | This paper | Negative controls | Supplementary file 2 |
| Peptide, recombinant protein | Gibson Assembly Master Mix | NEB | Cat# E2611S | |
| Peptide, recombinant protein | I-CeuI | NEB | #R0699S | |
| Peptide, recombinant protein | I-SceI | NEB | #R0694S | |
| Commercial assay or kit | Mouse Embryonic Stem Cell Nucleofector Kit | Lonza | #VPH-1001 | |
| Chemical compound, drug | TRIsure | Bioline | (#BIO-38032) | |
| Commercial assay or kit | GeneJET RNA extraction kit | Thermo Fisher | #K0732 | |
| Peptide, recombinant protein | DNase I | Roche | #04716728001 | |
| Peptide, recombinant protein | Maxima Reverse transcriptase | Thermo Fisher | #EP0743 | |
| Peptide, recombinant protein | MyTaq Red mix | Bioline | #BIO-25043 | |
| Strain, strain background (Escherichia coli) | e. cloni 10G supreme | Lucigen | #60081-1 | Electrocompetent cells |
| Peptide, recombinant protein | Takara ligation kit version 2.1 | Takara | #6022 | |
| Peptide, recombinant protein | Klenow HC 3′ ->5′ exo | NEB | #M0212L | |
| Commercial assay or kit | ISOLATE II PCR and Gel Kit | Bioline | BIO-52059 | |
| Peptide, recombinant protein | Fast-link ligase | Lucigen | LK0750H | |
| Peptide, recombinant protein | End-It DNA End-Repair Kit | Epicentre | #ER0720 | |
| Commercial assay or kit | dsDNA High sensitivity Qubit kit | Invitrogen | #Q33231 | |
| Commercial assay or kit | CleanPCR magnetic beads | CleanNA | #CPCR-0050 | |
| Peptide, recombinant protein | XcmI | NEB | #R0533S | |
| Peptide, recombinant protein | T4 DNA ligase | Roche | #10799009001 | |
| Peptide, recombinant protein | AvrII | Thermo Fisher | #ER1561 | |
| Peptide, recombinant protein | NheI | NEB | #R0131S | |
| Strain, strain background (Escherichia coli) | 5-alpha Competent E. coli | NEB | #C2987 | Competent cells |
| Peptide, recombinant protein | LIF | Sigma-Aldrich | #ESG1107 | 2i+LIF media |
| Chemical compound, drug | Monothioglycerol | Sigma-Aldrich | #M6145-25ML | 2i+LIF media |
| Chemical compound, drug | CHIR-99021 | MedChemExpress | #HY-10182 | 2i+LIF media |
| Chemical compound, drug | PD0325901 | MedChemExpress | #HY-10254 | 2i+LIF media |
| Other | BSA | Gibco | #15260-037 | 2i+LIF media |
| Other | DMEM-F12medium | Gibco | #11320-033 | 2i+LIF media |
| Other | Neurobasal medium | Gibco | #21103-049 | 2i+LIF media |
| Chemical compound, drug | N27 | Gibco | #17504-044 | 2i+LIF media |
| Chemical compound, drug | B2 | Gibco | #17502-048 | 2i+LIF media |
| Commercial assay or kit | MycoAlert Mycoplasma Detection Kit | Lonza | #LT07-318 | |
| Software, algorithm | BatchPrimer3 version 1.0 | You et al., 2008 | version 1.0 | https://wheat.pw.usda.gov/demos/BatchPrimer3/ |
| Software , algorithm | Starcode | Zorita et al., 2015 | version 1.1 | https://github.com/gui11aume/starcode |
| Software, algorithm | Python | Rossum and Drake, 2009 | version 3.6 | https://www.python.org/downloads/release/python-362/ |
| Software, algorithm | Bowtie2 | Langmead and Salzberg, 2012 | version 2.3.4 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Software, algorithm | R | R Development Core Team, 2021 | version 4.0.5 | https://www.r-project.org/ |
| Software, algorithm | ggplot2 | Wickham, 2016 | ggplot2 | https://ggplot2.tidyverse.org/ |
| Software, algorithm | Snakemake | Köster and Rahmann, 2012 | version 4.4.0 | https://anaconda.org/bioconda/snakemake/files?version=4.4.0 |
Additional files
-
Supplementary file 1
mm10 genomic coordinates of the regulatory elements tested in this study.
- https://cdn.elifesciences.org/articles/91994/elife-91994-supp1-v1.zip
-
Supplementary file 2
Sequences of the synthetic controls used in this study.
- https://cdn.elifesciences.org/articles/91994/elife-91994-supp2-v1.zip
-
Supplementary file 3
Processed and normalised data for all the enhancer-enhancer-promoter (EEP) combinations tested in this study.
- https://cdn.elifesciences.org/articles/91994/elife-91994-supp3-v1.zip
-
Supplementary file 4
Sequences of the primers used in this study.
- https://cdn.elifesciences.org/articles/91994/elife-91994-supp4-v1.zip
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91994/elife-91994-mdarchecklist1-v1.docx





