The exocyst complex controls multiple events in the pathway of regulated exocytosis
Figures
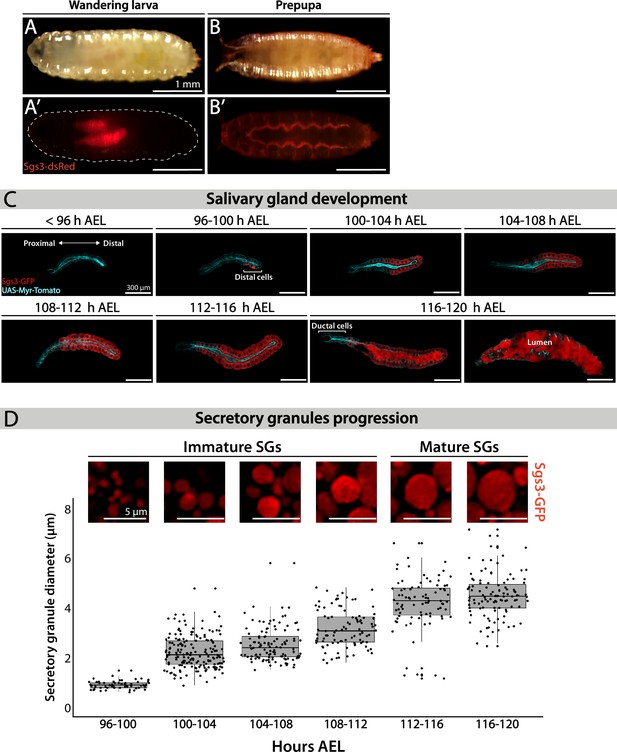
Larval salivary gland as a model for regulated exocytosis.
Images of a wandering larva (A, A’) and a prepupa (B, B’) expressing Sgs3-dsRed and visualized under white light (A, B) or by epifluorescence (A’, B’). Sgs3-dsRed localized in salivary glands of wandering larvae (A’) and outside the puparium in prepupae (B’). Scale bar 1 mm. (C) Confocal images of unfixed salivary glands dissected from larvae at the indicated time intervals (h AEL = hours after egg laying). Sgs3-GFP is shown in red and the plasma membrane labeled with myr-Tomato (sgs3-GFP, fkh-Gal4/UAS-myr-Tomato) is shown in cyan. At 96 h AEL, Sgs3 synthesis could be detected in the distal cells of the gland. Thereafter, Sgs3 expression gradually expanded proximally, and at 116 h AEL all salivary gland cells expressed Sgs3, with the exception of ductal cells. Exocytosis of secretory granules (SGs) began by this time point, and Sgs3 could be detected in the gland lumen. At 120 h AEL concerted exocytosis of SGs has ended. Scale bar 300 μm. (D) Confocal images of SGs labeled with Sgs3-GFP; SG diameter distribution at each time interval is displayed below. Based on its diameter, SGs are classified as immature (diameter <3 μm) or mature (diameter ≥3 μm). Only SGs from distal cells were used for diameter determination. Data points for this graph are shown in Table 1. For all time intervals analyzed n = 3, except for the 108–112 h AEL interval for which n = 4. ‘n’ represents the number of salivary glands analyzed. Scale bar 5 μm.
-
Figure 1—source data 1
Raw data used to generate Figure 1D.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig1-data1-v1.xlsx
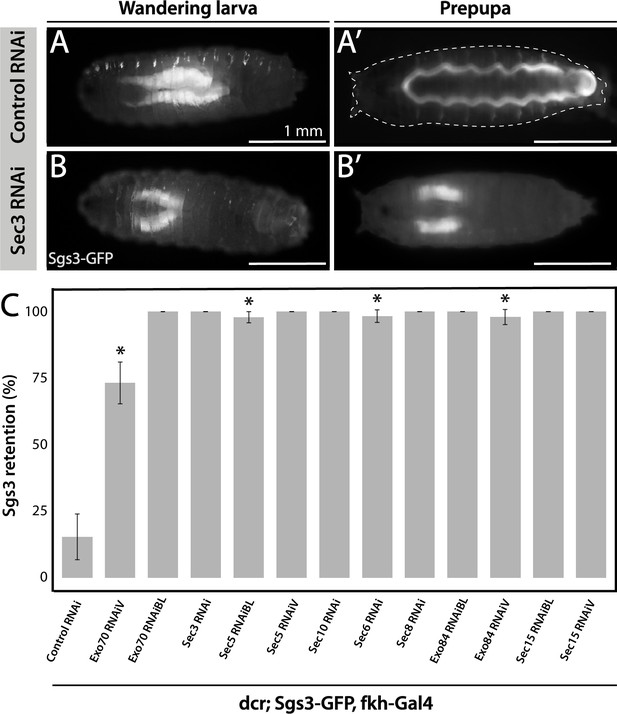
The exocyst is required for Sgs3 secretion.
(A, B) Images of a larva and a prepupa expressing Sgs3-GFP, and visualized with epifluorescence. Sgs3-GFP was inside the salivary glands in control wandering larvae (A), and outside the puparium (A’) in control prepupae (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi). Expression of sec3RNAi in salivary glands (UAS-sec3RNAi; sgs3-GFP, fkh-Gal4) did not affect expression of Sgs3-GFP in larvae (B), but blocked Sgs3-GFP release outside the puparium, so the protein was retained inside the salivary glands (B’). This phenotypic manifestation is referred as ‘retention phenotype’. Scale bar 1 mm. (C) Quantification of the penetrance of the retention phenotype in prepupae expressing the indicated RNAis. RNAis were expressed using fkh-Gal4 and larvae were cultured at 29°C. All RNAis tested against exocyst complex subunits displayed a retention phenotype, with a penetrance significantly different from the control RNAi (UAS-cherryRNAi) according to Likelihood ratio test followed by Tukey’s test (* = p-value <0.05). cherryRNAi n = 7; exo70RNAiV n = 11; exo70RNAiBL n = 8; sec3RNAi n = 6; sec5RNAiBL n = 17; sec5RNAiV n = 6; sec10RNAi n = 9; sec6RNAi n = 11; sec8RNAi n = 5; exo84RNAiBL n = 5; exo84RNAiV, n = 6; sec15RNAiBL n = 3; sec15RNAiV n = 4. ‘n’ represents the number of vials containing 20–30 prepupae per vial. For exocyst subunits with more than one RNAi line available, ‘BL’ indicates a Bloomington Stock Center allele and ‘V’ a Vienna Stock Center allele (see Table 2 for stock numbers).
-
Figure 2—source data 1
Raw data used to generate Figure 2C.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig2-data1-v1.xlsx
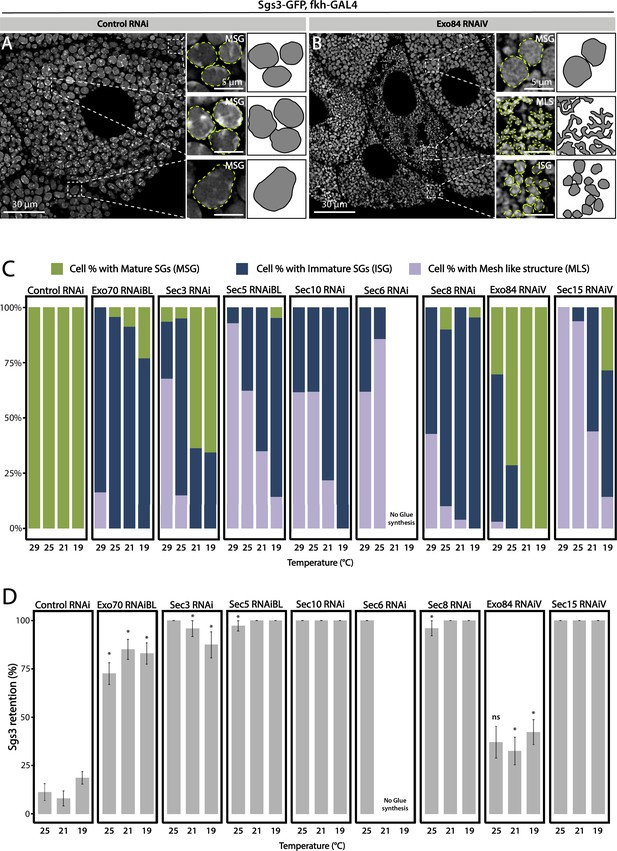
Phenotypic manifestations of exocyst subunits silencing.
At the end of larval development (116–120 h AEL) salivary gland cells of control individuals (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi) (A) were filled with mature secretory granules (SGs) (insets). In cells expressing exo84RNAiV (UAS-exo84RNAiV; sgs3-GFP, fkh-Gal4) (B), three different phenotypes could be visualized in a single salivary gland: cells with mature SGs (MSG), cells with immature SGs (ISG), and cells with no SG, in which Sgs3 was retained in a mesh (MLS). Scale bar 30 μm in main panels, and 5 μm in insets. For didactic purposes, MSG, ISG, and MLS were drawn on the right, next to the corresponding inset. (C) Quantification of the penetrance of each of the three phenotypes observed upon downregulation of each of the exocyst subunits. Larvae were grown at four different temperatures (29, 25, 21, or 19°C) to achieve different levels of RNAi expression. ‘n’ = number of salivary glands analyzed; controlRNAi (cherryRNAi) n = 4 (29°C), n = 5 (25, 21, and 19°C); exo70RNAiBL n = 11 (29°C), n = 7 (25 and 21°C), n = 6 (19°C); sec3RNAi n = 7 (29 and 25°C), n = 4 (21°C), n = 9 (19°C); sec5RNAiBL n = 4 (29°C), n = 12 (25°C), n = 9 (21°C), n = 6 (19°C); sec10RNAi n = 8 (29°C), n = 6 (25 and 19°C), n = 7 (21°C); sec6RNAi n = 9 (29°C), n = 4 (25°C); sec8RNAi n = 8 (29°C), n = 6 (25 and 19°C), n = 7 (21°C); exo84RNAiV n = 8 (29°C), n = 4 (25 and 19°C), n = 5 (21°C); sec15RNAiV n = 4 (29 and 25°C), n = 6 (21°C), n = 5 (19°C). Raw data used to generate this graph is shown in Table 3. (D) Quantification of the penetrance of the Sgs3-GFP retention phenotype in salivary glands of prepupae of the indicated genotypes. Only a few control individuals (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi) displayed the retention phenotype. Downregulation of exocyst subunits provoked significant retention of Sgs3 inside the salivary glands irrespective to the temperature (25, 21, or 19°C). Expression of sec6RNAi at 21 or 19°C resulted in no synthesis of Sgs3-GFP or Sgs3-dsRed, so the distribution of phenotypes was not assessed for this genotype at these temperatures. RNAis were expressed with fkh-Gal4. ‘n’ = number of vials with 20–30 larvae. controlRNAi (cherryRNAi) n = 7 (25°C), n = 9 (21°C), n = 19 (19°C); exo70RNAiBL n = 13 (25°C), n = 9 (21°C), n = 22 (19°C); sec3RNAi n = 9 (25°C), n = 6 (21 and 19°C); sec5RNAiBL n = 9 (25°C), n = 7 (21°C), n = 12 (19°C); sec10RNAi n = 10 (25°C), n = 5 (21°C), n = 4 (19°C); sec6RNAi n = 10 (25°C); sec8RNAi n = 5 (25°C), n = 8 (21°C), n = 22 (19°C); exo84RNAiV n = 8 (25 and 19°C), n = 6 (21°C); sec15RNAiV n = 5 (25°C), n = 9 (21°C), n = 8 (19°C). Statistical analysis was performed using a Likelihood ratio test followed by Tuckey´s test (*p-value <0.05). For those genotypes with 100% penetrance no statistical analysis was performed due to the lack of standard error. ns = not significant. Comparisons were made between RNAis for each of the temperatures analyzed.
-
Figure 3—source data 1
Raw data used to generate Figure 3C.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Raw data used to generate Figure 3D.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig3-data2-v1.xlsx
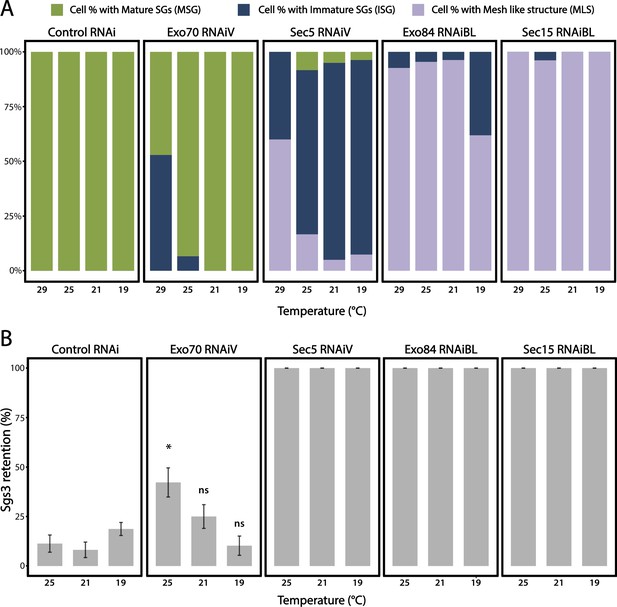
Penetrance of phenotypes observed after silencing subunits of the exocyst.
Additional RNA lines targeting exocyst subunits utilized to assess reduction-of-function phenotypes in the pathway of regulated exocytosis. (A) Quantification of the penetrance of each of the three phenotypes observed upon downregulation of each of the exocyst subunits. Larvae were grown at four different temperatures to achieve different levels of RNAi expression of exocyst subunits. n = number of salivary glands analyzed. controlRNAi (cherryRNAi) n = 4 (29°C), n = 5 (25, 21, and 19°C); exo70RNAiV n = 5 (29°C), n = 6 (25°C), n = 6 (21°C), n = 4 (19°C); sec15RNAiBL n = 5 (29°C), n = 6 (25°C), n = 5 (21°C), n = 5 (19°C); exo84RNAiBL n = 7 (29°C), n = 5 (25°C), n = 6 (21°C), n = 5 (19°C); sec5RNAiV n = 5 (29°C), n = 5 (25°C), n = 6 (21°C), n = 6 (19°C). (B) Quantification of the penetrance of the Sgs3-GFP retention phenotype in salivary glands of prepupae with the indicated RNAis at different temperatures. n = number of vials containing 20–30 larvae each. controlRNAi (cherryRNAi) n = 7 (25°C), n = 9 (21°C), n = 19 (19°C); exo70RNAiV n = 6 (25°C), n = 8 (21°C), n = 6 (19°C); sec5RNAiV, n = 4 (25°C), n = 6 (21°C), n = 5 (19°C); sec15RNAiBL n = 7 (25°C), n = 8 (21°C), n = 7 (19°C); exo84RNAiBL n = 5 (25°C), n = 4 (21°C), n = 8 (19°C). RNAis were expressed using fkh-Gal4. Comparisons of the retention phenotype were carried out between genotypes at each different temperature, and statistical analysis was performed using a Likelihood ratio test followed by Tukey’s test (*p value <0.05). For those RNAis with 100% penetrance no statistical analysis was performed. ns = not significant. This figure completes experiments of Figure 3C, D; all RNAis were expressed with controls at the same time, and therefore the controls are identical to those of Figure 3.
-
Figure 3—figure supplement 1—source data 1
Raw data used to generate Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig3-figsupp1-data1-v1.xlsx
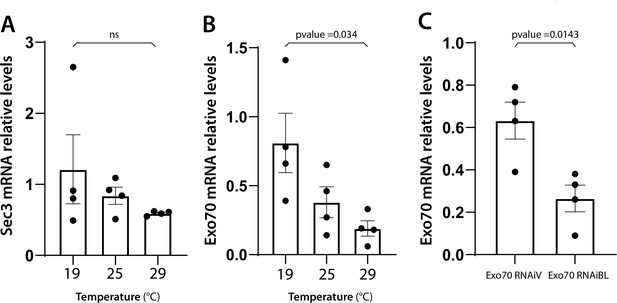
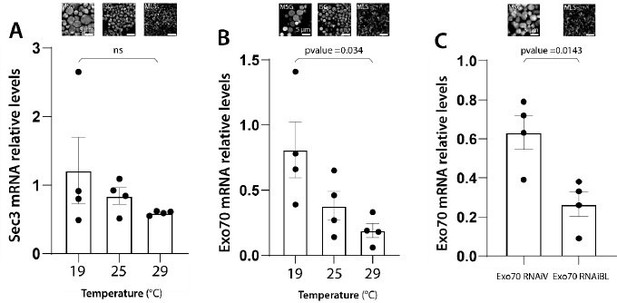
Remaining mRNA levels after RNAi-mediated knock-down of exocyst subunits correlate with the observed phenotypes.
mRNA levels of the indicated genes were measured by qRT-PCR in homogenates from salivary glands. sec3 (A) or exo70 (B) mRNA levels from larva grown at the indicated temperatures were determined in salivary glands expressing sec3RNAi (sgs3-GFP, fkh-Gal4/UAS-sec3RNAi)(A) or exo70RNAiBL (sgs3-GFP, fkh-Gal4/UAS-exo70RNAiBL) (B) relative to salivary glands expressing cherryRNAi (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi) from larvae grown at the same temperatures. (C) exo70 mRNA levels from larva grown at 29°C were determined in salivary glands expressing exo70RNAiV (sgs3-GFP, fkh-Gal4/UAS-exo70RNAiV) or exo70RNAiBL (sgs3-GFP, fkh-Gal4/UAS-exo70RNAiBL) relative to salivary glands expressing cherryRNAi (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi). n = 4 for all genotypes and conditions. Statistical analysis was performed by one-way analysis of variance (ANOVA) followed by Tukey’s test with a confidence interval higher than 95% (p < 0.05). ns = not significant.
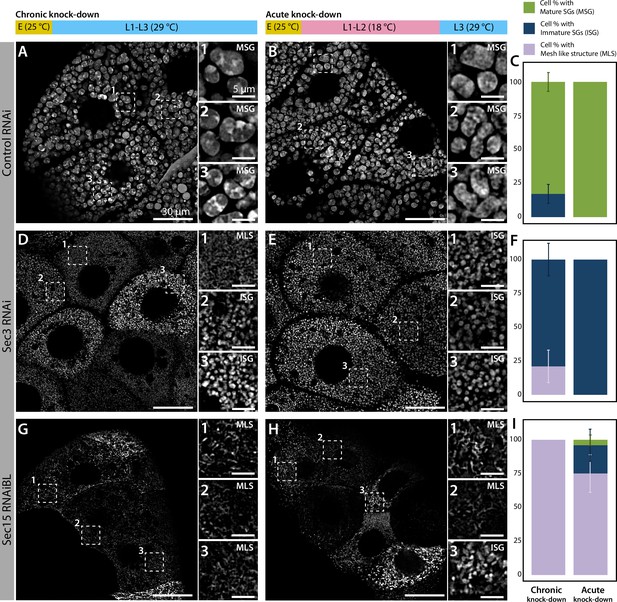
Chronic or acute knock-down of exocyst subunits generate comparable phenotypes.
Phenotypes of control (whiteRNAi) (A–C), sec3RNAi (D–F), or sec15RNAiBL (G–I) salivary glands. The thermo-sensitive Gal80 system (Gal80ts) was utilized to compare salivary glands after silencing the indicated genes throughout larval development ("chronic knock-down"): (A, D, G) or at the third larval instar only ("acute knock-down"): (B, E, H). Chronic RNAi expression was achieved by growing larvae at the restrictive temperature (29°C) from larval eclosion onwards (A, D, G). Acute RNAi expression was achieved by growing larvae at the permissive temperature (18°C) from larval eclosion until early L3 (larval third instar), (B, E, H) and then shifting larvae to the restrictive temperature (29°C) until analyzed. E: embryogenesis; L1: larval first instar; L2: second instar; and L3: third instar. The penetrance of each phenotype (mature secretory granules, immature secretory granules, or mesh-like structure) was quantified for each genotype and experimental condition (C, F, I). No substantial differences were detected between chronic and acute silencing of exocyst subunits. ‘n’ represents the total number of salivary glands analyzed. controlRNAi (whiteRNAi) (A) n = 5, (B) n = 3; sec3RNAi, (D) n = 6, (E) n = 7; sec15RNAiBL, (G, H) n = 4.
-
Figure 3—figure supplement 3—source data 1
Raw data used to generate Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig3-figsupp3-data1-v1.xlsx
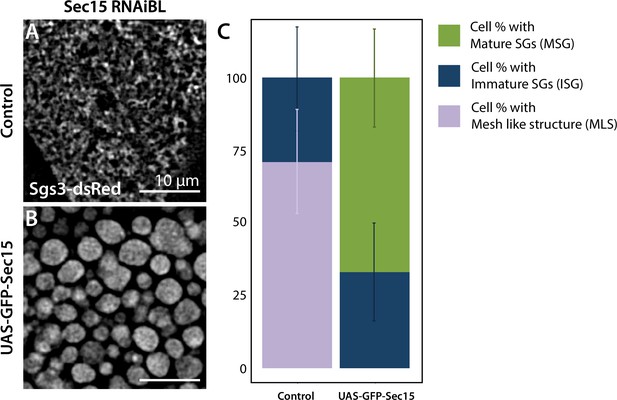
Expression of GFP-Sec15 can rescue secretory granule (SG) maturation after sec15 knock-down.
Expression of sec15RNAiBL at 18°C (sgs3-dsRed; UAS-PLCγPH-GFP, fkh-Gal4/UAS-sec15RNAiBL) (A) generates salivary glands that do not form mature SGs (MSGs) in any of the cells analyzed. Simultaneous expression of GFP-Sec15 (UAS-GFP-sec15/Sgs3-dsRed; UAS-sec15RNAiBL/fkh-Gal4) (B) generates salivary glands with MSGs in 67% of the cells. (C) Quantification of each of the three possible phenotypes mature SGs (MSGs), immature SGs (ISGs), or mesh-like structure (MLS) in control larvae (sgs3-dsRed; UAS-PLCγPH-GFP, fkh-Gal4/UAS-sec15RNAiBL) or after expression of the rescue construct (UAS-GFP-sec15/sgs3-dsRed; UAS-sec15RNAiBL/fkh-Gal4). ‘n’ represents the total number of salivary glands analyzed. Control genotype (UAS-PLCγPH-GFP) n = 7; rescue genotype (UAS-GFP-sec15) n = 5.
-
Figure 3—figure supplement 4—source data 1
Raw data used to generate Figure 3—figure supplement 4.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig3-figsupp4-data1-v1.xlsx
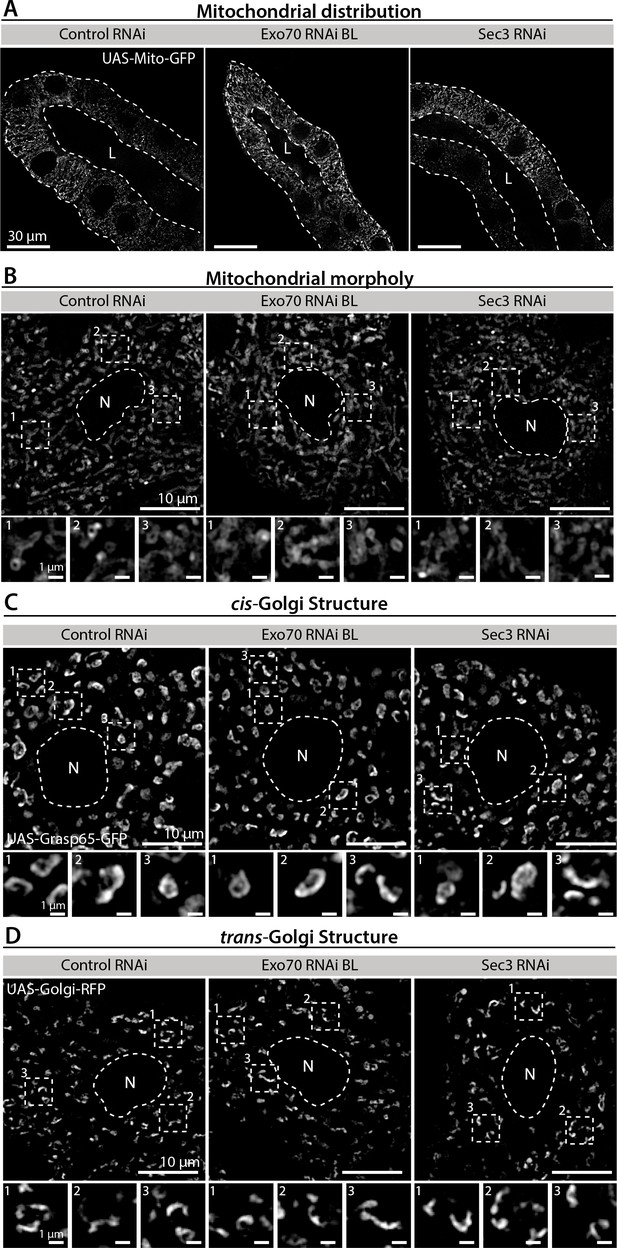
Exocyst down-regulation does not affect general cellular health or homeostasis.
Mitochondrial distribution and morphology (A, B), cis- and trans-Golgi complex distribution and morphology (C, D), nuclear size (E), and autophagy (F) were analyzed in salivary glands of control (UAS-whiteRNAi) and exocyst knock-down salivary glands. Mitochondria were labeled with mito-GFP (UAS-mito-GFP), cis-Golgi with GRASP65 (UAS-GRASP65-GFP), trans-Golgi with RFP-Golgi (UAS-RFP-Golgi), nuclei with EGFP (UAS-EGFP), and autophagy with mCh-Atg8 (UAS-GFP-mCh-atg8). Larvae were grown at 25°C to achieve a condition at which knock-down of the exocyst subunits mostly generate cells with immature SGs. RNAis were expressed using fkh-Gal4.
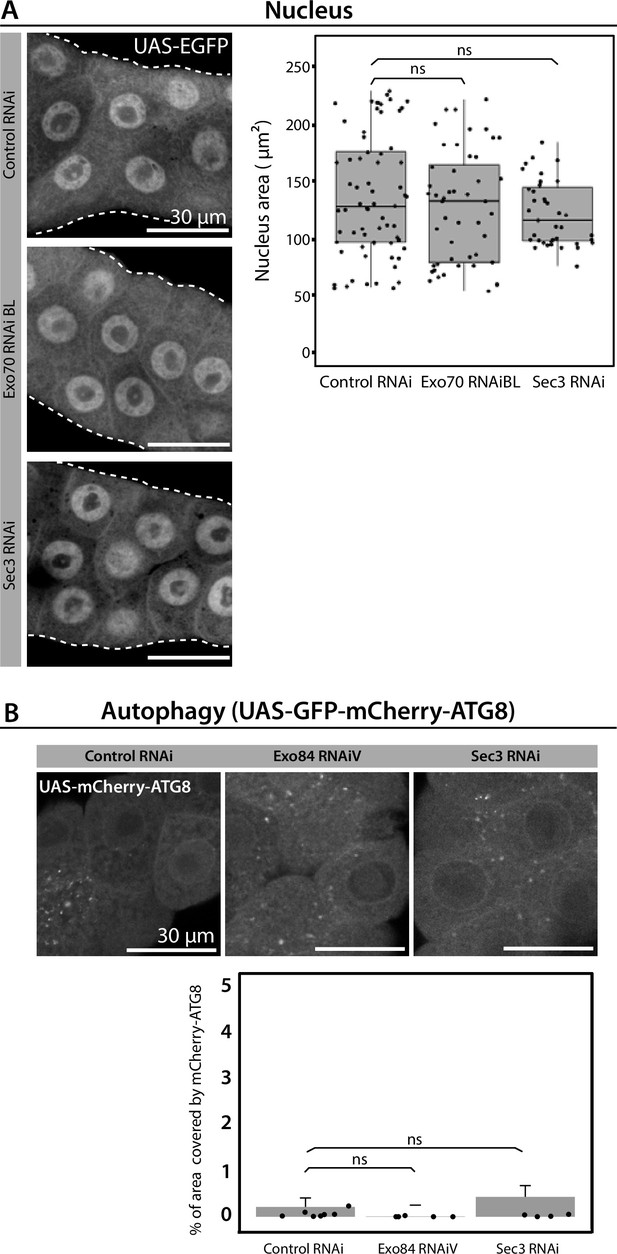
Exocyst down-regulation does not affect nuclei size or autophagy.
(A) Quantification of nuclear area in the indicated genotypes was performed using ImageJ. (B) Quantification of the area coved by mCh-Atg8 foci over the total area of salivary gland analyzed (%) was performed using ImageJ. Statistical analysis was carried-out using one-way analysis of variance (ANOVA) ‘n’ represents the total number of salivary glands analyzed. (A) controlRNAi (whiteRNAi) n = 8; exo70RNAiBL n = 5; sec3RNAi n = 4. (B) controlRNAi (whiteRNAi) n = 8; exo84RNAiV n = 5; sec3RNAi n = 5.
-
Figure 3—figure supplement 6—source data 1
Raw data used to generate Figure 3—figure supplement 6.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig3-figsupp6-data1-v1.xlsx
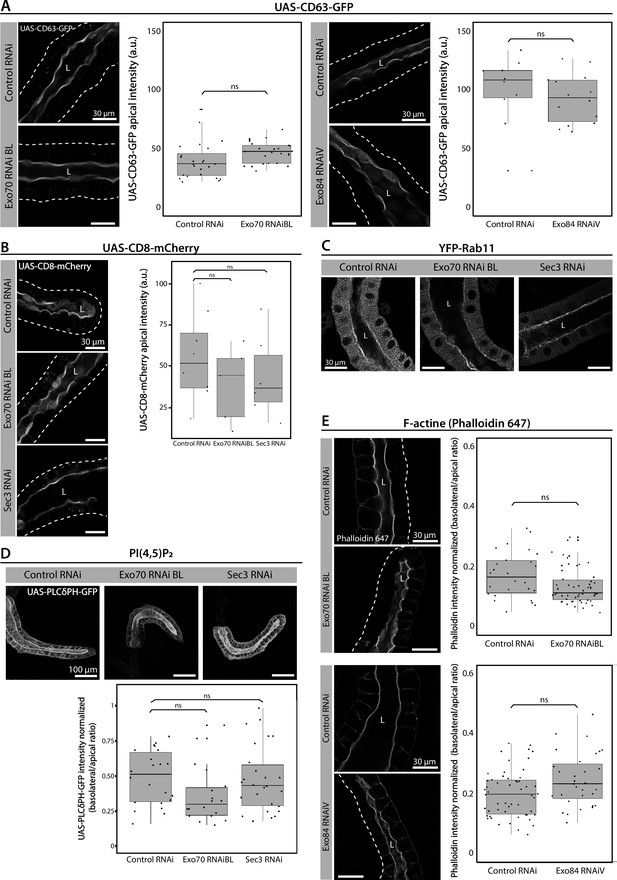
Exocyst down-regulation does not affect apical polarity markers.
(A) CD63 (UAS-CD63-GFP), (B) CD8 (UAS-CD8-mCherry), (C) Rab11 (YFP-Rab11), (D) PI(4,5)P2 (UAS-PLCγ-PH-GFP), and (E) filamentous actin (phalloidine-647) distribution were analyzed in salivary glands of control (UAS-whiteRNAi) or exocyst knock-down salivary glands (UAS-exo70RNAiBL, UAS-sec3RNAi, or UAS-exo84RNAiV). Larvae were grown at 25°C (UAS-exo70RNAiBL or UAS-sec3RNAi) or at 29°C (UAS-exo84RNAiV) to achieve a condition at which exocyst downregulation generate mostly cells with immature secretory granules (SGs). Note that in (A) and (E), for accurate comparisons, the control genotype (UAS-whiteRNAi) was also assayed at different temperatures. Transgenes were expressed with fkh-Gal4. Statistical analysis was performed using (A, B) one-way analysis of variance (ANOVA), (D) Likelihood ratio test followed by Tukey’s test, or (E) Wald test (p-value <0.05, ns = not significant). ‘n’ represents the total number of salivary glands analyzed. (A) controlRNAi (whiteRNAi) n = 6; exo70RNAiBL n = 5; controlRNAi (whiteRNAi) n = 3; exo84RNAiV n = 3. (B) controlRNAi (whiteRNAi) n = 8; exo70RNAiBL n = 5; sec3RNAi n = 6. (D) n = 4. (E) controlRNAi (whiteRNAi) n = 4; exo70RNAiBL n = 10; controlRNAi (whiteRNAi) n = 6; exo84RNAiV n = 4.
-
Figure 3—figure supplement 7—source data 1
Raw data used to generate Figure 3—figure supplement 7.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig3-figsupp7-data1-v1.xlsx
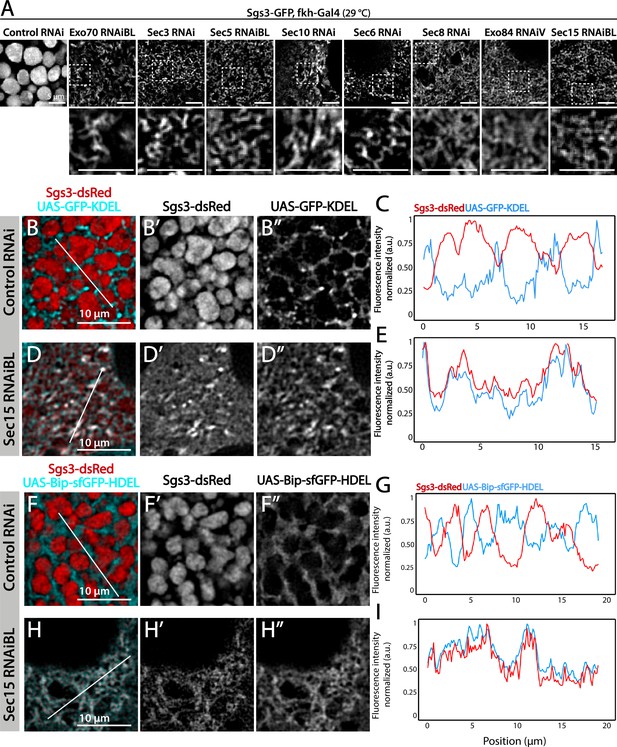
The exocyst is required for Sgs3-GFP exit from endoplasmic reticulum (ER) and secretory granule (SG) biogenesis.
Confocal images of unfixed salivary glands of the indicated genotypes. Larvae were grown at 29°C to achieve maximal activation of the Gal4-UAS system, and maximal downregulation of exocyst subunits. (A) In control salivary glands (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi) 116–120 AEL, Sgs3-GFP was packed in mature SGs. Salivary glands expressing RNAis against each of the exocyst subunits, where Sgs3-GFP exhibited a reticular distribution are shown. SGs were not formed in these cells. Scale bar 5 μm. The ER, labeled with GFP-KDEL (UAS-GFP-KDEL), (B, D) or Bip-sfGFP-HDEL (UAS-Bip-sfGFP-HDEL), (F, H) distributed in between SGs in control salivary glands (B, F); in sec15RNAi salivary glands (fkh-Gal4/UAS-sec15RNAiBL) SGs did not form and the Sgs3-dsRed signal overlapped with the ER markers (D, H). (C, E, G, I) Two-dimensional line scans of fluorescence intensity across the white lines in panels B, D, F, and H of Sgs3-dsRed and the ER markers. In all cases, transgenes were expressed using fkh-Gal4. Scale bar 10 μm.
-
Figure 4—source data 1
Raw data used to generate Figure 4C.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig4-data1-v1.xlsx
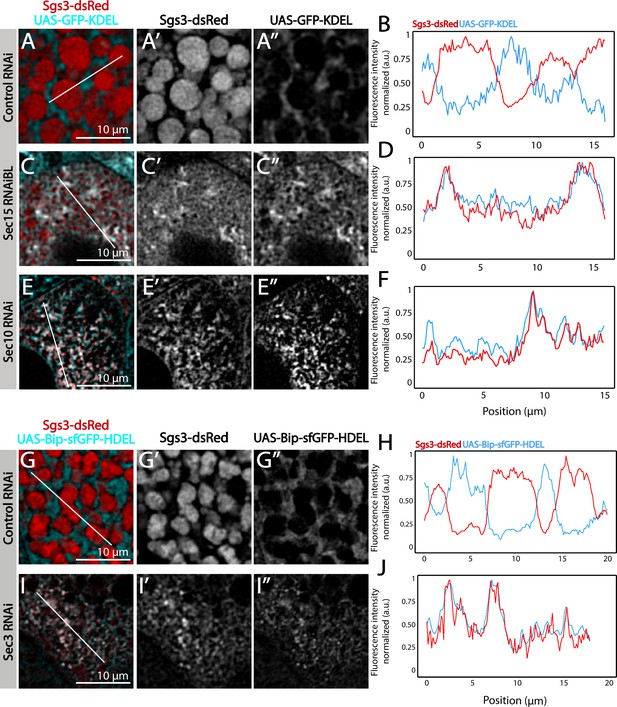
The exocyst is required for Sgs3-GFP exit from endoplasmic reticulum (ER) and secretory granule (SG) biogenesis.
Confocal images of unfixed salivary glands of the indicated genotypes. Larvae were grown at 29°C to achieve maximal activation of the Gal4-UAS system, and maximal downregulation of exocyst complex subunits. The ER was labeled with GFP-KDEL (UAS-GFP-KDEL), (A, C, E) or with Bip-sfGFP-HDEL (UAS-Bip-sfGFP-HDEL) (G, I). In control salivary glands (fkh-Gal4/UAS-whiteRNAi) (A, G), the ER displayed a network-like appearance localized in between mature SGs that contain Sgs3-dsRed, and thus no colocalization between the ER marker and Sgs3-dsRed was detected (B, H). Upon expression of sec15RNAi (fkh-Gal4/UAS-sec15RNAiBL) (C), sec10RNAi (UAS-sec10RNAi; fkh-Gal4) (E), or sec3RNAi (UAS-sec3RNAi; fkh-Gal4) SGs did not form, and Sgs3-dsRed was retained in the ER, as demonstrated by colocalization of both markers in the two-dimensional line scan analysis (D, F, J). RNAis were expressed using fkh-Gal4. Scale bar 10 μm.
-
Figure 4—figure supplement 1—source data 1
Raw data used to generate Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig4-figsupp1-data1-v1.xlsx
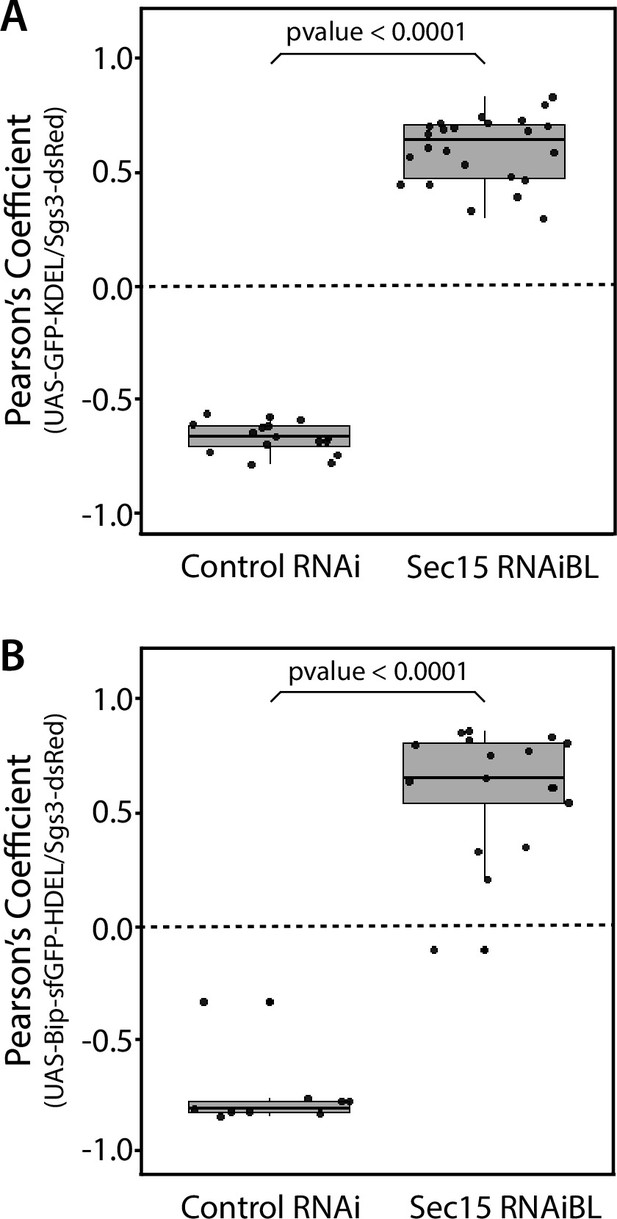
Quantification of colocalization between endoplasmic reticulum (ER) markers and Sgs3.
Pearson’s coefficient to evaluate colocalization between the ER markers UAS-GFP-KDEL (A) or UAS-Bip-sfGFP-HDEL (B) and Sgs3-dsRed in control salivary glands (UAS-whiteRNAi) or after exocyst knock-down (UAS-sec15RNAiBL). Larvae were grown at 29°C. Statistical analysis was performed using one-way analysis of variance (ANOVA). Analysis corresponds to data shown in Figure 4. ‘n’ represents the total number of salivary glands analyzed. (A) n = 3, (B) n = 2.
-
Figure 4—figure supplement 2—source data 1
Raw data used to generate Figure 4—figure supplement 2B.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig4-figsupp2-data1-v1.xlsx
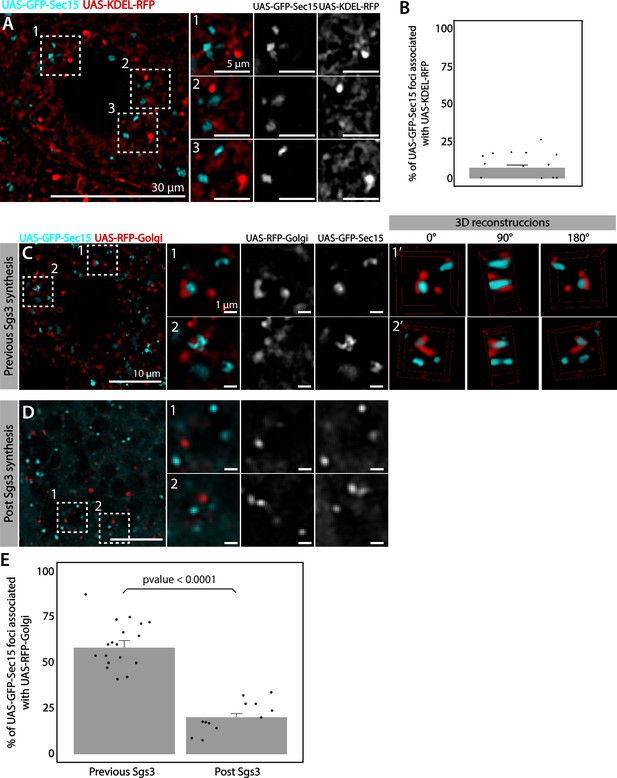
The exocyst localizes at the Golgi complex before Sgs3 synthesis.
Confocal images of unfixed salivary glands expressing GFP-Sec15 and (A) the endoplasmic reticulum (ER) marker KDEL-RFP or (C) the trans-Golgi marker RFP-Golgi. Sec15 foci did not localize at or associate with the ER (A, B). (C) Sec15 foci were found in close association with trans-Golgi complex cisternae. Examples of association events are shown in insets 1 and 2, including different angles of three-dimensional reconstruction stacks. (D) Sec15 foci and trans-Golgi complex association was lost after Sgs3 synthesis has begun. (E) Quantification of Sec15 foci-Golgi complex association events before and after the onset of Sgs3 synthesis (Wald test, p-value <0.05). ‘n’ represents the number of salivary glands analyzed, n = 4.
-
Figure 5—source data 1
Raw data used to generate Figure 5B and E.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig5-data1-v1.xlsx
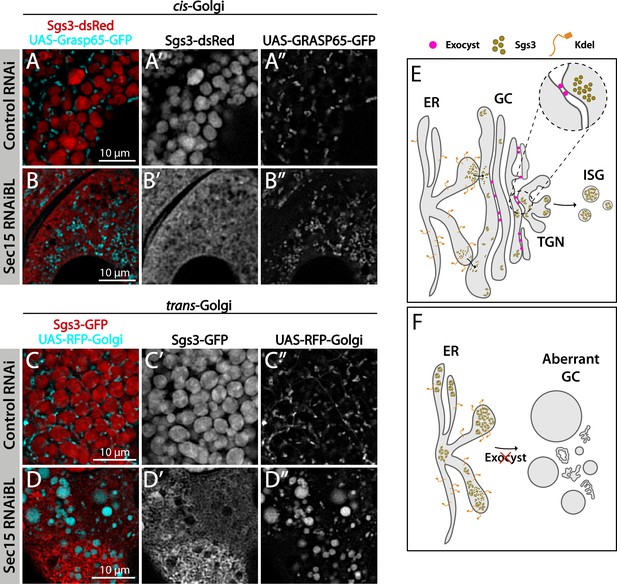
The exocyst is required to maintain normal Golgi complex morphology.
Confocal images of unfixed salivary glands of the indicated genotypes. Larvae were grown at 29°C to achieve maximal activation of the Gal4-UAS system and maximal downregulation of exocyst subunits. In control larvae (fkh-Gal4/UAS-whiteRNAi), Sgs3-dsRed (A) or Sgs3-GFP (C) were in mature secretory granules (SGs). In sec15RNAi salivary glands (fkh-Gal4/UAS-sec15RNAiBL) Sgs3 was retained in a mesh-like structure (B, D), also shown in Figure 4. (A, B) The cis-Golgi complex was labeled with Grasp65-GFP (UAS-Grasp65-GFP), and the trans-Golgi with RFP-Golgi (UAS-RFP-Golgi), (C, D). The morphology of the cis- and trans-Golgi complexes changed dramatically in sec15-knock-down cells (B”, D”), in comparison to controls (A”, C”). Transgenes were expressed with fkh-Gal4. Scale bar 10 μm. (E) Model of Sgs3 transit from the endoplasmic reticulum (ER) through the cis- and trans-Golgi complex to sprouting SGs from the trans-Golgi complex. (F) The exocyst is needed for tethering the Golgi complex cisternae and to support Golgi complex structure. In the absence of the exocyst Golgi cisternae disconnect, cis- and trans-Golgi become dysfunctional, resulting in Sgs3 retention at the ER.
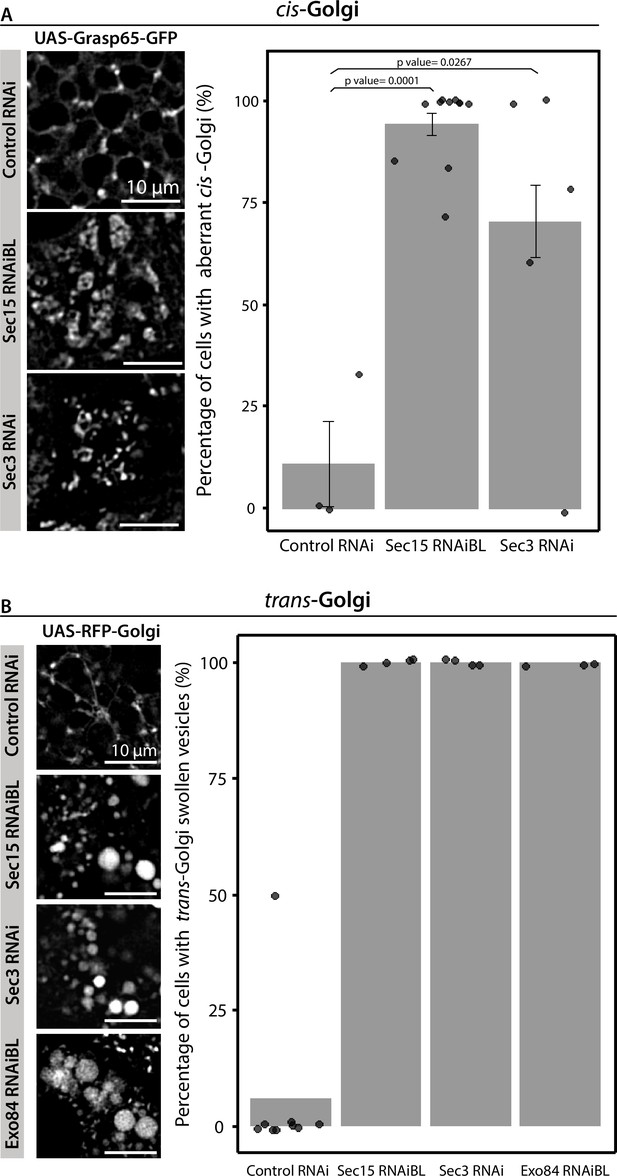
The exocyst is required to maintain the typical morphology of the Golgi complex.
Confocal images of unfixed salivary glands of the indicated genotypes. Larvae were grown at 29°C to achieve maximal activation of the Gal4-UAS system, and maximal downregulation of exocyst subunits. cis-Golgi (UAS-Grasp65-GFP) (A) and trans-Golgi (UAS-RFP-Golgi) (B) markers were analyzed In control salivary glands (fkh-Gal4/UAS-whiteRNAi) or in exocyst-down-regulated salivary glands (UAS-sec15RNAiBL, UAS-sec3RNAi, or UAS-exo84RNAiV). RNAis were expressed using fkh-Gal4. Scale bar 10 μm. Bar graphs show the quantification of the penetrance of the Golgi complex phenotype. Statistical analysis was performed using a Likelihood ratio test followed by Tukey’s test (p-value <0.05, ns = not significant). ‘n’ represents the total number of salivary glands analyzed. (A) controlRNAi (whiteRNAi) n = 3; sec15RNAiBL n = 11; sec3RNAi n = 3. (B) controlRNAi (whiteRNAi) n = 9; sec15RNAiBL n = 4; sec3RNAi n = 4; exo84RNAiV n = 3.
-
Figure 6—figure supplement 1—source data 1
Raw data used to generate Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig6-figsupp1-data1-v1.xlsx
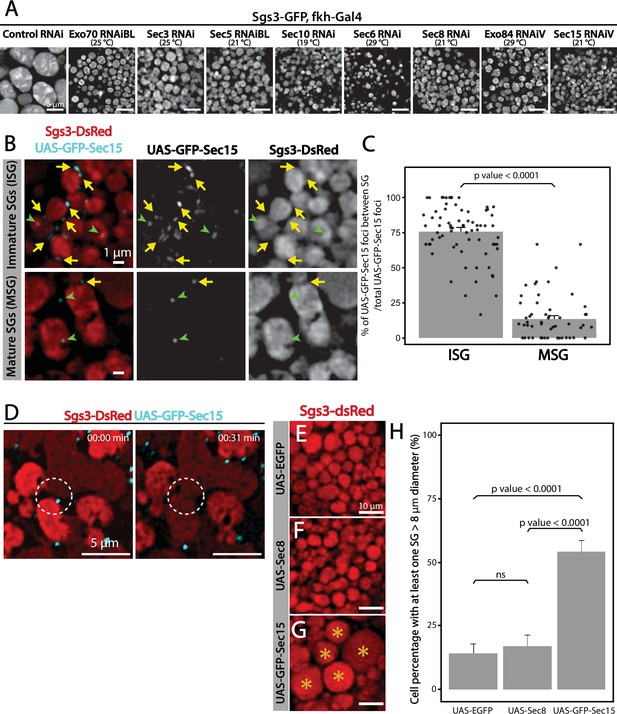
The exocyst is required for secretory granule (SG) homotypic fusion.
(A) Confocal images of unfixed salivary glands of the indicated genotypes. In control salivary glands (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi), Sgs3-GFP was packed in mature SGs. RNAis targeting any of the subunits of the exocyst were expressed at the indicated temperatures, giving rise to salivary cells with immature granules. Scale bar 5 μm. (B) GFP-Sec15 (cyan) and Sgs3-dsRed (red) were expressed in salivary glands (sgs3-dsRed/UAS-GFP-sec15; fkh-Gal4) at 18°C. GFP-Sec15 foci were mostly found in between SGs (yellow arrows) in cells bearing immature SGs (upper panel) and not in cells with mature SGs (lower panel), in which most foci did not localize in between SGs (green arrowheads). Scale bar 1 μm. (C) Quantification of Sec15 foci in between SGs relative to the total number of Sec15 foci (%) (Wald test, p-value <0.05). Eight salivary glands with ISGs and five salivary glands with MSGs were analyzed. (D) Still panels of two different frames of Video 6 showing that during a homotypic fusion event, GFP-Sec15 accumulated precisely at the contact site between two neighboring SGs (dotted circle); scale bar 5 μm. (E–G) Expression of EGFP (sgs3-dsRed; UAS-EGFP, fkh-Gal4) (E) or Sec8 (sgs3-dsRed/UAS-Sec8; fkh-Gal4) (F) did not affect SG size (red). Expression of GFP-Sec15 (G) (sgs3-dsRed/UAS -GFP-sec15; fkh-Gal4) generated giant SGs (asterisks). Scale bar 10 μm. (H) Quantification of the percentage of salivary gland cells with at least one SG larger than 8 μm diameter; (Likelihood ratio test followed by Tukey’s test, p-value <0.05). ‘n’ represents the number of salivary glands analyzed. UAS-EGFP n = 7; UAS-Sec8 n = 5; UAS-GFP-Sec15 n = 5. Transgenes were expressed using fkh-Gal4. ns = not significant.
-
Figure 7—source data 1
Raw data used to generate Figure 7.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig7-data1-v1.xlsx
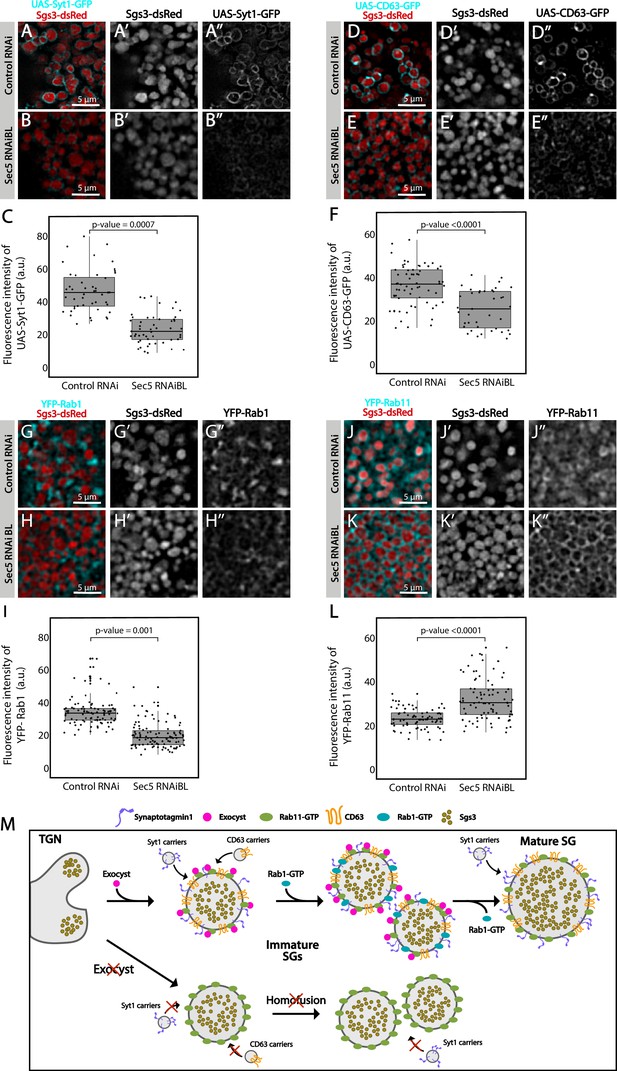
The exocyst mediates the acquisition of secretory granule (SG) maturation factors.
Confocal images of unfixed salivary glands of the indicated genotypes. Larvae were grown at 21°C to reduce the activity of the Gal4-UAS system, and to generate a maximal proportion of cells with immature SGs. Recruitment of (A, B) Syt1-GFP (UAS-Syt1-GFP); (D, E) CD63-GFP (UAS-CD63-GFP); (G, H) YFP-Rab1; (J, K) YFP-Rab11 was analyzed in control salivary glands expressing whiteRNAi (fkh-Gal4/UAS-whiteRNAi) and in salivary glands expressing sec5RNAiBL (fkh-Gal4/UAS-sec5RNAiBL). Fluorescent intensity around SGs of each of the analyzed maturation factors was quantified using the ImageJ software and plotted (C, F, I, L). Comparison of fluorescent intensity among genotypes and statistical analysis was performed using one-way analysis of variance (ANOVA). ‘n’ represents the number of salivary glands: (C) control RNAi n = 4, sec5RNAiBL n = 5; (F) control RNAi n = 5, sec5RNAiBL n = 4; (I) control RNAi n = 6, sec5RNAiBL n = 9; (L) control RNAi n = 11, sec5RNAiBL n = 8. Transgenes were expressed using fkh-Gal4. Scale bar 5 μm. (M) Proposed model of exocyst-dependent SG homotypic fusion and maturation. The exocyst complex is required for homotypic fusion between immature SGs. Immature SGs incorporate Syt-1, CD63, and Rab1 in an exocyst-dependent manner. Syt-1 continues to be recruited to SGs after homotypic fusion. The exocyst complex also inhibits the incorporation of an excess of Rab11 around SGs.
-
Figure 8—source data 1
Raw data used to generate Figure 8.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig8-data1-v1.xlsx
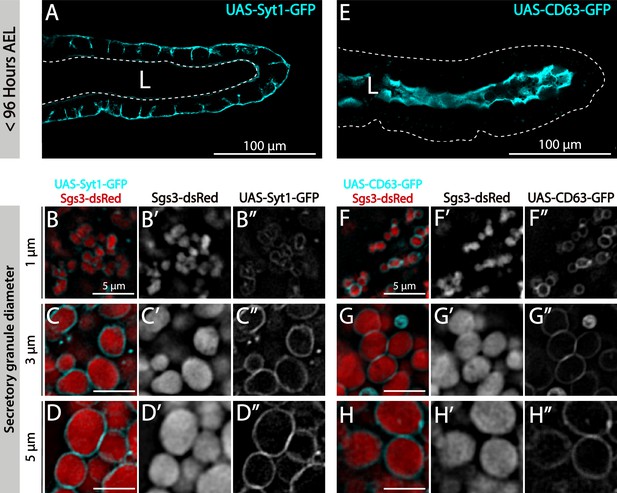
Incorporation of maturation markers to secretory granules (SGs) in wild-type salivary glands.
Incorporation of the SG maturation markers Syt1 (sgs3-dsRed/UAS-syt1-GFP; fkh-Gal4) (A–D) and CD63 (sgs3-dsRed/UAS-CD63-GFP; fkh-Gal4) (E–H) to developing granules. Prior to SG biogenesis (<96 h AEL) Syt1 localized at the basolateral plasma membrane of salivary cells (A), while CD63 localized at the apical plasma membrane (E). Scale bar 100 μm. Both markers localized at the membrane of immature SGs and mature SGs (B, D, F–H). Scale bar 5 μm. Note that Syt1 signal intensity increases around SGs as they mature (compare B”, C”, D”), whereas CD63 signal remains fairly stable during SG maturation (compare F”, G”, H”).
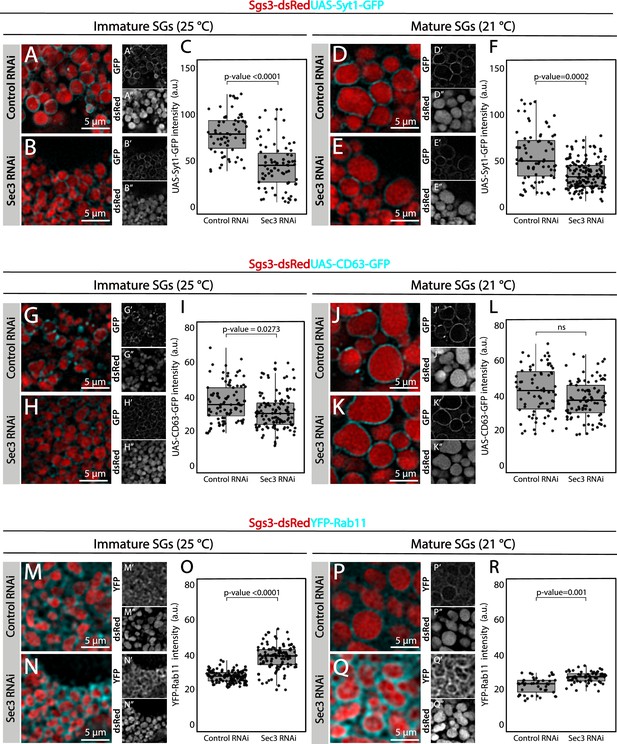
The exocyst mediates the acquisition of secretory granule (SG) maturation factors.
Maturation proteins Syt-1 and CD63 were gradually incorporated to the membrane of SG. Confocal images of unfixed salivary glands of the indicated genotypes. Larvae were grown at 25 or 21°C to modulate the activity of the Gal4-UAS system and generate a maximal proportion of immature (A, B, G, H, M, N) or mature SGs (D, E, J, K, P, Q), respectively. (A, B, D, E) Recruitment of Syt1-GFP (UAS-Syt1-GFP); of (G, H, J, K) CD63-GFP (UAS-CD63-GFP); or of (M, N, P, Q) YFP-Rab11 was analyzed in control salivary glands expressing whiteRNAi (fkh-Gal4/UAS-whiteRNAi) and in salivary glands expressing sec3RNAi (UAS-sec3RNAi; fkh-Gal4). SGs were labeled with Sgs3-dsRed. Fluorescent intensity around SGs of each of the analyzed maturation factors was quantified using the ImageJ software and plotted (C, F, I, L, O, R). Comparison of fluorescence intensity among genotypes, and statistical analysis were performed using one-way analysis of variance (ANOVA). ‘n’ represents the number of salivary glands: (C) controlRNAi n = 7, sec3RNAi n = 6; (F) controlRNAi n = 7, sec3RNAi n = 13; (I) controlRNAi n = 12, sec3RNAi n = 9; (L) controlRNAi n = 8, sec3RNAi n = 8; (O) controlRNAi n = 7, sec3RNAi n = 10; (R) controlRNAi n = 7, sec3RNAi n = 4. Transgenes were expressed using fkh-Gal4. ns = not significant. Scale bar 5 μm.
-
Figure 8—figure supplement 2—source data 1
Raw data used to generate Figure 8—figure supplement 2O and R.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig8-figsupp2-data1-v1.xlsx
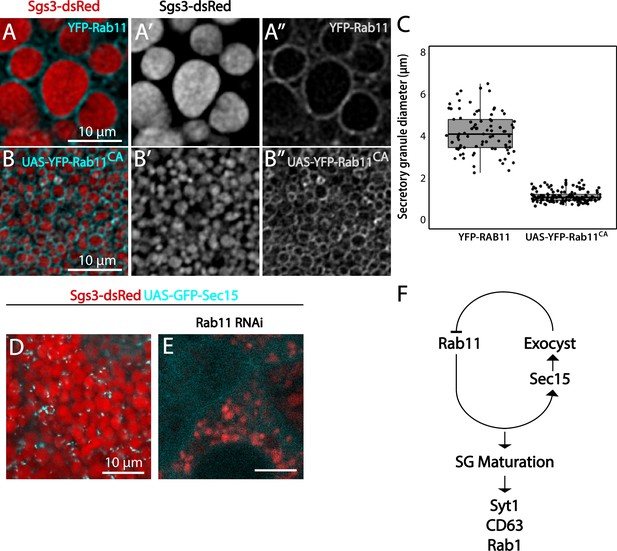
The exocyst negatively regulates incorporation of Rab11 to secretory granules (SGs).
Salivary gland cells expressing Sgs3-dsRed and endogenously tagged YFP-Rab11 (A), or overexpressing a constitutively active form of Rab11 at 29°C (fkh-Gal4/UAS-YFP-Rab11CA) (B). YFP-Rab11 localized around mature SGs (A). Overexpression of Rab11CA arrested SG maturation (B), as shown in the quantification of SG diameter (C). YFP-Rab11, n = 6; UAS-YFP-RAB11CA, n = 5. n = number of salivary glands. (D) In control salivary glands, GFP-Sec15 localized as foci on the periphery of mature SGs (sgs3-dsRed/UAS-GFP-sec15; fkh-Gal4). (E) Knock-down of Rab11 (sgs3-dsRed; UAS-rab11RNAi/fkh-Gal4) generated immature SGs and prevented GFP-Sec15 from localizing on SGs. (F) Proposed mechanism of genetic interactions between Rab11 and Sec15. A single-negative feedback loop regulates the levels of these proteins on the SGs, and recruitment of the maturation factors Syt-1, CD63, and Rab1. Scale bar 10 μm.
-
Figure 8—figure supplement 3—source data 1
Raw data used to generate Figure 8—figure supplement 3C.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig8-figsupp3-data1-v1.xlsx
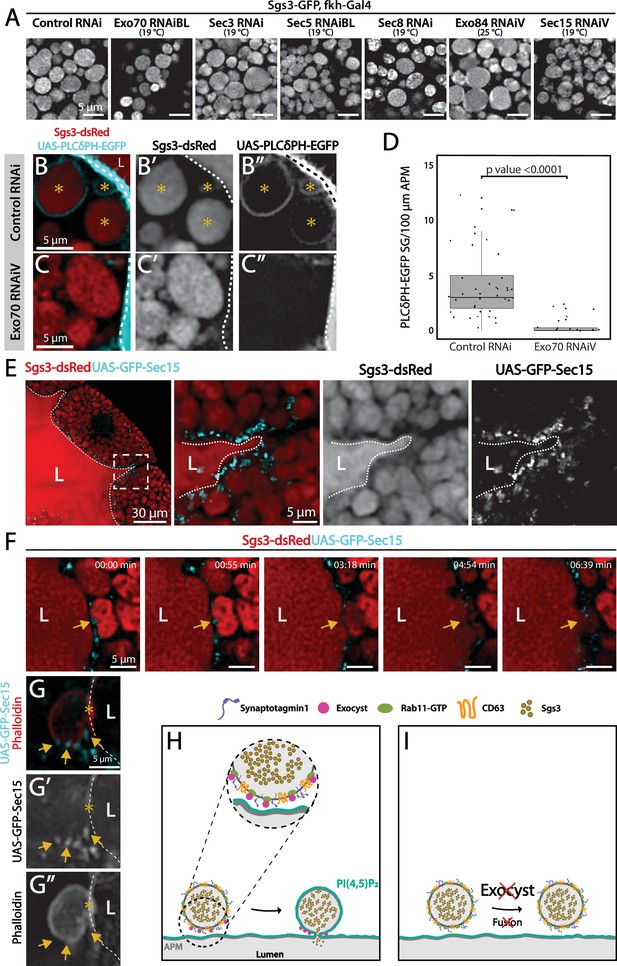
The exocyst is required for secretory granule (SG) fusion with the plasma membrane during regulated exocytosis.
Confocal images of unfixed salivary glands of the indicated genotypes. Larvae were grown at the indicated temperatures to attain levels of RNAi-mediated silencing that bring about maximal proportion of cells with mature, exocytosis incompetent SGs. (A) Sgs3-GFP localized within SGs. Control SGs (sgs3-GFP, fkh-Gal4/UAS-cherryRNAi) were indistinguishable from SGs of salivary cells in which a subunit of the exocyst has been knocked-down. Scale bar 5 μm. (B) In control salivary glands (fkh-Gal4/UAS-whiteRNAi), the PI(4,5)P2 reporter UAS-PLCγ-PH-GFP labels the plasma membrane (dotted line) and also the SGs that have already fused with the plasma membrane (asterisks). (C) SGs of cells expressing exo70RNAiV (UAS-exo70RNAiV; fkh-Gal4) were not labeled with the reporter, indicating that these SGs failed to fuse with the plasma membrane. Scale bar 5 μm. (D) The number of mature SGs positive for PLCγ-PH-EGFP per 100 μm of linear plasma membrane was quantified in the indicated genotypes; Exo70 knock-down reduced SG–plasma membrane fusion Wald test (p-value <0.05); 7 salivary glands per genotype were analyzed. (E) During SG exocytosis the exocyst complex, labeled with GFP-Sec15 (cyan) localized as dots in contact sites between SGs and the apical plasma membrane (dotted line) (sgs3-dsRed/UAS-GFP-sec15; fkh-Gal4). (F) Still panels of Video 7 showing a fusion event between a mature SG and the plasma membrane (arrow); a dot of GFP-Sec15 indicating the position of the exocyst (arrow) was positioned just at the site where fusion was taking place. Scale bar 5 μm. (G) Confocal image of a fixed salivary gland. A mature SG that has fused with the plasma membrane (dotted line), and thus became labeled with phalloidin, displayed dots of GFP-Sec15 on its side (arrow), just next to the fusion point (asterisk). Transgenes were expressed with fkh-Gal4. The salivary gland lumen is indicated with ‘L’. Scale bar 5 μm. (H) Model of the role of the exocyst complex during SG–plasma membrane fusion during regulated exocytosis. The exocyst sits on the membrane of the SG, and tethers the granule to the plasma membrane, favoring the action of fusion molecules. (I) Upon loss of the exocyst complex, mature SGs cannot contact the plasma membrane and fusion does not occur.
-
Figure 9—source data 1
Raw data used to generate Figure 9.
- https://cdn.elifesciences.org/articles/92404/elife-92404-fig9-data1-v1.xlsx
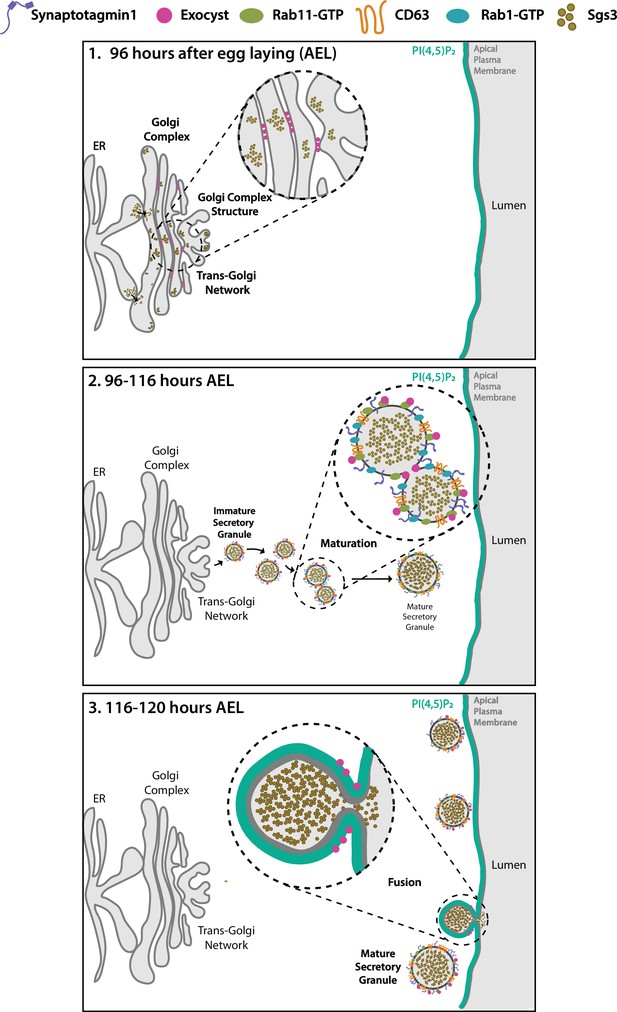
Proposed model of the action of the exocyst in maintenance of normal Golgi complex structure, maturation, and exocytosis of secretory granules (SGs) in Drosophila larval salivary gland cells.
(1) Before SG biogenesis (<96 h AEL), the exocyst (pink dots) localizes at the Golgi complex, where it is required to maintain the normal Golgi structure. The mucine Sgs3 (brown dots) moves through the secretory pathway from the endoplasmic reticulum to the Golgi complex, from where immature SGs containing the mucine sprout out. (2) After sprouting, SGs undergo maturation (96–116 h AEL). During maturation, the exocyst localizes in between immature SGs, where it is required for homotypic fusion. The exocyst is also required for incorporation of maturation factors to the membrane of SGs. These maturation factors include Syt-1 (purple line), DC63 (orange line), Rab11 (green oval), and Rab1 (light blue oval). At this stage, the exocyst no longer localizes at the Golgi complex. (3) When maturation has been completed SGs fuse with the apical plasma membrane and exocytosis takes place. During exocytosis (116–120 h AEL), the exocyst localizes at mature SGs, in contact with the apical plasma membrane (APM), where it is required for tethering and subsequent fusion, prior to release of the SG content to the salivary gland lumen.

Criteria utilized to define Sec15 focithat were“associated” or“not associated” withthe trans-Golgi network in the experiments of Figure 5C-E of the manuscript.
When the distance between maximal intensities of GFP-Sec15 and Golgi-RFP signals was equal or less than 0.6 µm, the signals were considered “associated” (upper panels). When the distance was more than 0.6 µm, the signals were considered “not associated” (lower panels).

Criteria utilized to define Sec15 focithat were“associated” or“not associated” withthe ERin the experiments of Figure 5A-Bof the manuscript.
When the distance between maximal intensities of GFP-Sec15 and KDEL-RFP signals was equal or less than 0.6 µm, the signals were considered “associated”. When the distance was more than 0.6 µm, the signals were considered “not associated”.

The exocyst complex associates with immature SGs but not with mature SGs.
(A) GFP-Sec15 foci (cyan) and SGs (red) are shown in cells bearing Immature SGs or (B) with mature SGs. Yellow arrows indicate GFP-Sec15 foci localized in between SGs; green arrowheads indicate GFP-Sec15 foci that arenot in between SGs. (C) Quantification of the percentage (%) of Sec15 foci localized in between SGs respect to the total number of Sec15 foci in cells filled with immature SGs (ISG)vs cells with mature SGs (MSG).

Conceptual model of RNAi expression at different temperatures , remaining levels of mRNA/protein levels and phenotypes obtained at each temperature.
Videos
Real-time imaging showing association between the exocyst and the trans-Golgi network.
A Drosophila early third instar larva salivary gland (~168 hr at 18°C equivalent to ~72–96 hr at 25°C) expressing the trans-Golgi marker RFP-Golgi (red) and the exocyst maker GFP-Sec15 (cyan (UAS-GFP-sec15/UAS-RFP-Golgi; fkh-Gal4)) was imaged. Three-dimensional reconstruction of six slides with 0.44 μm spacing was generated using ImageJ. Frame acquisition time was 2.25 s. A total of 78 frames per channel were acquired. Total video time is 77 s. Crop size is 8 μm.
Three-dimensional reconstruction showing association between the exocyst and the trans-Golgi network.
A salivary gland from an early third instar larva (~168 hr at 18°C equivalent to ~72–96 hr at 25°C) was imaged. The trans-Golgi marker RFP-Golgi (red) and the exocyst maker GFP-Sec15 (cyan) were in close association (UAS-GFP-sec15/UAS-RFP-Golgi; fkh-Gal4). Three-dimensional reconstruction of 33 slides with 0.1 μm spacing was generated using ImageJ. Crop size is 5.57 μm. Corresponds to images shown in Figure 5C-1’.
3D recontruction of GFP-Sec15 foci (cyan) associated to trans-Golgi marker (red).
Three-dimensional reconstruction showing association between the exocyst and immature secretory granules (SGs).
A salivary gland from a third instar larva (~186 hr at 18°C equivalent to ~100 hr at 25°C) was imaged. SGs with Sgs3-dsRed (red) and the exocyst marker GFP-Sec15 (cyan) were in close association (sgs3-dsRed/UAS-GFP-sec15; fkh-Gal4). Three-dimensional reconstruction of 6 slides with 0.4 μm spacing was generated using ImageJ. Crop size is 10 μm.
Three-dimensional reconstruction showing association between the exocyst and immature secretory granules (SGs).
A salivary gland from a third instar larva (~186 hr at 18°C equivalent to ~104 hr at 25°C) was imaged. SGs with Sgs3-dsRed (red) and the exocyst marker GFP-Sec15 (cyan) were in close association (sgs3-dsRed/UAS-GFP-sec15; fkh-Gal4). Three-dimensional reconstruction of 12 slides with 0.45 μm spacing was generated using ImageJ. Crop size is 10 μm.
Real-time imaging of a fusion event between secretory granules (SGs).
A Drosophila salivary gland expressing Sgs3-dsRed and GFP-Sec15 (sgs3-dsRed/UAS-GFP-sec15; fkh-Gal4) in which a fusion event between SGs was captured (circle). Note that the exocyst (GFP-Sec15) localized at the fusion point between two SGs. The movie was not deconvolved, and represents a maximum intensity projection of three optical slices (total range: 1,6 μm). Scale bar 5 μm.
Real-time imaging of a fusion event between a secretory granule (SG) and the plasma membrane.
A Drosophila salivary gland expressing Sgs3-dsRed and GFP-Sec15 (sgs3-dsRed/UAS-GFP-sec15; fkh-Gal4) in which a fusion event between SGs and the plasma membrane was captured (circle). The granule content was released to the gland lumen (L). The movie shows a single slice of 32 frames comprising a total time of 0.32 s. The movie was not deconvolved. Scale bar 5 μm.
Tables
Quantification of secretory granule (SG) diameter at the indicated time intervals after egg laying.
SGs from salivary gland distal-most cells were analyzed. Columns display: Hours AEL: hours after egg laying; n: number of salivary glands analyzed; number of cells analyzed; number of SGs measured; mean diameter; median diameter; minimum diameter and maximum diameter.
| Hours AEL | n | Cells quantified | SGquantified | Meandiameter (μm) | Mediandiameter(μm) | Minimum diameter (μm) | Maximum diameter (μm) |
|---|---|---|---|---|---|---|---|
| 96–100 | 3 | 7 | 57 | 0.92 | 0.91 | 0.62 | 1.49 |
| 100–104 | 3 | 18 | 173 | 2.22 | 2.11 | 0.89 | 4.76 |
| 104–108 | 3 | 12 | 112 | 2.53 | 2.53 | 1.50 | 5.78 |
| 108–112 | 4 | 13 | 96 | 3.18 | 3.18 | 1.78 | 4.81 |
| 112–116 | 3 | 10 | 86 | 4.22 | 4.22 | 1.16 | 6.59 |
| 116–120 | 3 | 15 | 107 | 4.48 | 4.45 | 2.47 | 7.13 |
List of the Drosophila lines utilized in this work.
Stock number and repository center from where each line was obtained are indicated.
| Line | Number | Stock center |
|---|---|---|
| Sgs3-GFP | 5885 | Bloomington |
| Sgs3-dsRed | - | A.J. Andres' Lab |
| YFP-Rab11 | 62549 | Bloomington |
| YFP-Rab1 | 62539 | Bloomington |
| UAS-dicer2 | 24650 | Bloomington |
| UAS-White-RNAi | 33613 | Bloomington |
| UAS-Cherry-RNAi | 35785 | Bloomington |
| UAS-Rab11-RNAi | 27730 | Bloomington |
| UAS-Sec3-RNAi | 35806 | Vienna |
| UAS-Sec5-RNAi | 27526 | Bloomington |
| UAS-Sec5-RNAi | 28873 | Vienna |
| UAS-Sec6-RNAi | 27314 | Bloomington |
| UAS-Sec8-RNAi | 45032 | Vienna |
| UAS-Sec10-RNAi | 27483 | Bloomington |
| UAS-Sec15-RNAi | 27499 | Bloomington |
| UAS-Sec15-RNAi | 35161 | Vienna |
| UAS-Exo84-RNAi | 108650 | Vienna |
| UAS-Exo84-RNAi | 28712 | Bloomington |
| UAS-Exo70-RNAi | 27867 | Vienna |
| UAS-Exo70-RNAi | 28041 | Bloomington |
| UAS-PLCγ-PH-EGFP | 58362 | Bloomington |
| UAS-Synaptotagmin1-GFP | 6925 | Bloomington |
| UAS-Sec8 | 9556 | Bloomington |
| UAS-CD63-GFP | 91390 | Bloomington |
| UAS-Myr-Tomato | 32221 | Bloomington |
| UAS-GFP-KDEL | 9898 | Bloomington |
| UAS-Bip-SfGFP-HDEL | 64749 | Bloomington |
| UAS-RFP-Golgi | 30908 | Bloomington |
| UAS-GRASP65-GFP | 8508 | Bloomington |
| UAS-EGFP | 5430 | Bloomington |
| UAS-GFP-Sec15 | 39685 | Bloomington |
| UAS-YFP-Rab11CA | 9791 | Bloomington |
| tubP-GAL80[ts] | 7017 | Bloomington |
| UAS-mCD8-mCherry | 27391 | Bloomington |
| UAS-mito-GFP | 8443 | Bloomington |
| UASp-GFP-mCherry-Atg8a | 37749 | Bloomington |
Raw data for experiments of Figure 3C and Figure 3—figure supplement 1A.
| Figure 3C | ||||||
|---|---|---|---|---|---|---|
| Genotype | Temperature (°C) | Number of glands analyzed | Number of distal cells analyzed | Phenotype | % of phenotype | Standard deviation |
| Control RNAi | 29 | 4 | 12 | Mesh-like structure | 0 | 0 |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 25 | 5 | 13 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 21 | 5 | 17 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 19 | 5 | 15 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| Exo70 RNAi BL | 29 | 11 | 43 | Mesh-like structure | 12.73 | 28.67 |
| SG immature | 87.27 | 28.67 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 5 | 23 | Mesh-like structure | 0 | 0 | |
| SG immature | 95.24 | 12.6 | ||||
| SG mature | 4.76 | 12.6 | ||||
| 21 | 7 | 23 | Mesh-like structure | 0 | 0 | |
| SG immature | 90.48 | 25.2 | ||||
| SG mature | 9.52 | 25.2 | ||||
| 19 | 4 | 15 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| Sec3 RNAi | 29 | 7 | 31 | Mesh-like structure | 69.52 | 42.84 |
| SG immature | 24.05 | 32.37 | ||||
| SG mature | 6.43 | 11.07 | ||||
| 25 | 7 | 20 | Mesh-like structure | 16.67 | 28.87 | |
| SG immature | 78.57 | 28.41 | ||||
| SG mature | 4.76 | 12.6 | ||||
| 21 | 4 | 11 | Mesh-like structure | 0 | 0 | |
| SG immature | 27.08 | 35.6 | ||||
| SG mature | 72.92 | 35.6 | ||||
| 19 | 9 | 32 | Mesh-like structure | 0 | 0 | |
| SG immature | 33.52 | 33.75 | ||||
| SG mature | 66.48 | 33.75 | ||||
| Sec5 RNAi BL | 29 | 4 | 14 | Mesh-like structure | 96.43 | 7.14 |
| SG immature | 3.57 | 7.14 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 12 | 45 | Mesh-like structure | 62.5 | 38.35 | |
| SG immature | 37.5 | 38.35 | ||||
| SG mature | 0 | 0 | ||||
| 21 | 9 | 20 | Mesh-like structure | 27.78 | 44.1 | |
| SG immature | 72.22 | 44.1 | ||||
| SG mature | 0 | 0 | ||||
| 19 | 6 | 21 | Mesh-like structure | 12.22 | 19.05 | |
| SG immature | 84.44 | 18.22 | ||||
| SG mature | 3.33 | 8.16 | ||||
| Sec10 RNAi | 29 | 9 | 26 | Mesh-like structure | 77.78 | 44.1 |
| SG immature | 22.22 | 44.1 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 6 | 34 | Mesh-like structure | 59.17 | 46.95 | |
| SG immature | 40.83 | 46.95 | ||||
| SG mature | 0 | 0 | ||||
| 21 | 7 | 23 | Mesh-like structure | 20.24 | 34.65 | |
| SG immature | 79.76 | 34.65 | ||||
| SG mature | 0 | 0 | ||||
| 19 | 6 | 20 | Mesh-like structure | 0 | 0 | |
| SG immature | 100 | 0 | ||||
| SG mature | 0 | 0 | ||||
| Sec6 RNAi | 29 | 9 | 34 | Mesh-like structure | 66.67 | 50 |
| SG immature | 33.33 | 50 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 4 | 7 | Mesh-like structure | 75 | 50 | |
| SG immature | 25 | 50 | ||||
| SG mature | 0 | 0 | ||||
| Sec8 RNAi | 29 | 8 | 37 | Mesh-like structure | 41.16 | 35.88 |
| SG immature | 58.84 | 35.88 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 6 | 20 | Mesh-like structure | 8.33 | 20.41 | |
| SG immature | 80.56 | 30.58 | ||||
| SG mature | 11.11 | 27.22 | ||||
| 21 | 7 | 26 | Mesh-like structure | 3.57 | 9.45 | |
| SG immature | 96.43 | 9.45 | ||||
| SG mature | 0 | 0 | ||||
| 19 | 6 | 22 | Mesh-like structure | 0 | 0 | |
| SG immature | 96.67 | 8.16 | ||||
| SG mature | 3.33 | 8.16 | ||||
| Exo84 RNAi V | 29 | 8 | 33 | Mesh-like structure | 2.08 | 5.89 |
| SG immature | 60.42 | 32.49 | ||||
| SG mature | 37.5 | 33.46 | ||||
| 25 | 4 | 14 | Mesh-like structure | 0 | 0 | |
| SG immature | 27.92 | 28 | ||||
| SG mature | 72.08 | 28 | ||||
| 21 | 5 | 15 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 19 | 4 | 8 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| Sec15 RNAi V | 29 | 4 | 17 | Mesh-like structure | 100 | 0 |
| SG immature | 0 | 0 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 4 | 16 | Mesh-like structure | 87.5 | 25 | |
| SG immature | 12.5 | 25 | ||||
| SG mature | 0 | 0 | ||||
| 21 | 6 | 25 | Mesh-like structure | 43.33 | 46.33 | |
| SG immature | 56.67 | 46.33 | ||||
| SG mature | 0 | 0 | ||||
| 19 | 5 | 14 | Mesh-like structure | 16.67 | 23.57 | |
| SG immature | 53.33 | 36.13 | ||||
| SG mature | 30 | 44.72 | ||||
Salivary gland developmental staging.
Salivary glands analyzed in experiments of the indicated figures. Columns display: Experimental temperature, larval hours of development after egg laying (AEL), equivalent hours of development at 25°C (based on SG phenotype and salivary gland general appearance), presence or absence of Sgs3 in salivary glands, at the developmental time studied, and expected stage of SGs (mature or immature).
| Experimental temperature (°C) | Hours of development at the experimental temperature (h AEL) | Equivalent hours of development at 25°C (h AEL) | Presence of Sgs3 | Expected developmental stage of SGs in wild type | ||
|---|---|---|---|---|---|---|
| Figure 2 | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 3 | 19, 21, 25, 29 | ~216, ~144, ~120, ~96 | 116–120 | Yes | Mature | |
| Figure 4 | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 5 | 18 | ~120–168 | 72–96 | No | - | |
| Figure 6 | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 7A, E–H | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 8A, D, G, J | 21 | ~108 | 96–104 | Yes | Immature | |
| Figure 8B, E, H, K | 21 | ~144 | 116–120 | Yes | Mature | |
| Figure 9A–D | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 9E–G | 18 | ~240 | 116–120 | Yes | Mature | |
| Figure 3—figure supplement 1 | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 3—figure supplement 2 | 19, 25, or 29 | ~216, ~120, ~96 | 112–120 | Yes | Mature | |
| Figure 3—figure supplement 3A, D, G | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 3—figure supplement 3B, E, H | 120 hr at 18°C and 36 hr at 29°C | ~156 | 116–120 | Yes | Mature | |
| Figure 3—figure supplement 4 | 18 | ~240 | 116–120 | Yes | Mature | |
| Figure 3—figure supplement 5 | 25 | ~72 | ~72 | No | - | |
| Figure 3—figure supplement 6 | 25 | ~72 | ~72 | No | - | |
| Figure 3—figure supplement 7A, E | 25 or 29 | ~72 or ~50 | ~72 | No | - | |
| Figure 3—figure supplement 7B–D | 25 | ~72 | ~72 | No | - | |
| Figure 4—figure supplement 1 | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 4—figure supplement 2 | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 6—figure supplement 1 | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 8—figure supplement 1A, E | 25 | 72–96 | 72–96 | No | - | |
| Figure 8—figure supplement 1B, F | 25 | 100–104 | 100–104 | Yes | Immature | |
| Figure 8—figure supplement 1C, G | 25 | 108–112 | 108–112 | Yes | Mature | |
| Figure 8—figure supplement 1D, H | 25 | 116–120 | 116–120 | Yes | Mature | |
| Figure 8—figure supplement 2A, G, M | 25 or 21 | 96–104 or ~108 | 96–104 | Yes | Immature | |
| Figure 8—figure supplement 2B, H, N | 25 or 21 | 116–120 or ~144 | 116–120 | Yes | Mature | |
| Figure 8—figure supplement 2D, E, J, K, P, Q | 25 or 21 | 116–120 or ~144 | 116–120 | Yes | Mature | |
| Figure 8—figure supplement 3A, B, E | 29 | ~96 | 116–120 | Yes | Mature | |
| Figure 8—figure supplement 3D | 29 | ~72 | 96–104 | Yes | Immature | |
| Figure 3—figure supplement 1A | ||||||
| Genotype | Temperature | Number of glands analyzed | Number of distal cells analyzed | Phenotype | % of phenotype | Standard deviation |
| Control RNAi | 29 | 4 | 12 | Mesh-like structure | 0 | 0 |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 25 | 5 | 13 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 21 | 5 | 17 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 19 | 5 | 15 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| Exo70 RNAi V | 29 | 5 | 17 | Mesh-like structure | 0 | 0 |
| SG immature | 39.43 | 42.11 | ||||
| SG mature | 60.57 | 42.11 | ||||
| 25 | 6 | 15 | Mesh-like structure | 0 | 0 | |
| SG immature | 8.33 | 20.41 | ||||
| SG mature | 91.67 | 20.41 | ||||
| 21 | 6 | 17 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| 19 | 4 | 15 | Mesh-like structure | 0 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 100 | 0 | ||||
| Sec5 RNAi V | 29 | 5 | 25 | Mesh-like structure | 53.33 | 50.55 |
| SG immature | 46.67 | 50.55 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 5 | 12 | Mesh-like structure | 20 | 27.39 | |
| SG immature | 70 | 27.39 | ||||
| SG mature | 10 | 22.36 | ||||
| 21 | 6 | 21 | Mesh-like structure | 8.33 | 20.41 | |
| SG immature | 87.5 | 20.92 | ||||
| SG mature | 4.17 | 10.21 | ||||
| 19 | 6 | 27 | Mesh-like structure | 5.56 | 13.61 | |
| SG immature | 91.11 | 14.4 | ||||
| SG mature | 3.33 | 8.16 | ||||
| Exo84 RNAi BL | 29 | 7 | 38 | Mesh-like structure | 93.2 | 12.85 |
| SG immature | 6.8 | 12.85 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 5 | 22 | Mesh-like structure | 90 | 22.36 | |
| SG immature | 10 | 22.36 | ||||
| SG mature | 0 | 0 | ||||
| 21 | 6 | 27 | Mesh-like structure | 95.83 | 10.21 | |
| SG immature | 4.17 | 10.21 | ||||
| SG mature | 0 | 0 | ||||
| 19 | 5 | 21 | Mesh-like structure | 66.43 | 41.31 | |
| SG immature | 33.57 | 41.31 | ||||
| SG mature | 0 | 0 | ||||
| Sec15 RNAi BL | 29 | 5 | 25 | Mesh-like structure | 100 | 0 |
| SG immature | 0 | 0 | ||||
| SG mature | 0 | 0 | ||||
| 25 | 6 | 26 | Mesh-like structure | 94.44 | 13.61 | |
| SG immature | 5.56 | 13.61 | ||||
| SG mature | 0 | 0 | ||||
| 21 | 5 | 14 | Mesh-like structure | 100 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 0 | 0 | ||||
| 19 | 5 | 20 | Mesh-like structure | 100 | 0 | |
| SG immature | 0 | 0 | ||||
| SG mature | 0 | 0 | ||||