7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase
Figures
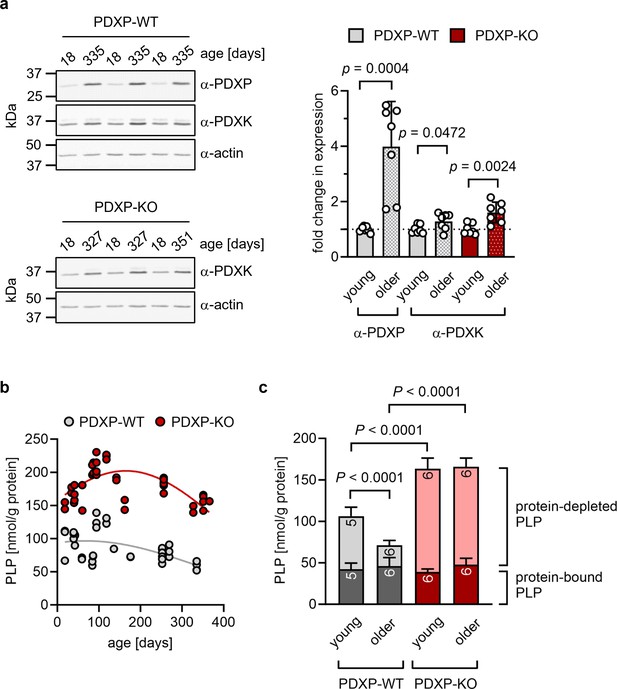
Role of pyridoxal 5’-phosphate phosphatase (PDXP) in hippocampal pyridoxal 5’-phosphate (PLP) homeostasis.
(a) Age-dependent expression of pyridoxal kinase (PDXK) and PDXP in murine hippocampi. Left panels, representative western blots of three hippocampi for each genotype. The same blots were reprobed with α-actin antibodies as a loading control. The age of the investigated mice is indicated above the blots. Right panel, densitometric quantification of hippocampal PDXP and PDXK western blot signals, corrected by the corresponding actin signals. Young mice were 18–42 days of age, older mice were 252–351 days of age; n=7 individual hippocampi were analyzed per group. Data are mean values ± SD. Statistical analysis was performed with unpaired, two-sided t-tests; p-values are indicated. (b) Age-dependent, total PLP concentrations in isolated hippocampi of PDXP-WT and knockout of PDXP (PDXP-KO) mice. PLP was derivatized with semicarbazide and analyzed by HPLC. Each symbol represents the result of the PLP determination in an individual hippocampus. Data were fitted by Gaussian least-squares analyses. (c) Determination of protein-bound and protein-depleted PLP in PDXP-WT and PDXP-KO hippocampal lysates of young (18–42 days of age) and older mice (252–352 days of age). The number of analyzed hippocampi is indicated in the bars. Data are mean values ± SD. Statistical analysis was performed with two-way ANOVA and Tukey’s multiple comparisons test. Significant differences (adjusted p-values) in protein-depleted PLP levels are indicated. The exact age of analyzed mice is listed in Figure 1—source data 3. Source data are available for this figure.
-
Figure 1—source data 1
Western blot quantification (to Figure 1a).
Densitometric quantification of pyridoxal 5’-phosphate phosphatase (PDXP), pyridoxal kinase (PDXK), and actin levels in hippocampi of PDXP-WT mice, and of PDXK and actin levels in hippocampi of knockout of PDXP (PDXP-KO) mice.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Quantification of pyridoxal 5’-phosphate (PLP) levels in hippocampi of pyridoxal 5’-phosphate phosphatase (PDXP)-WT and knockout of PDXP (PDXP-KO) mice (to Figure 1b and c).
High-performance liquid chromatography (HPLC)-based measurements of total PLP concentrations in isolated hippocampi of PDXP-WT and PDXP-KO mice, and of protein-bound and protein-depleted PLP levels in PDXP-WT and PDXP-KO hippocampal lysates.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Analysis of total hippocampal pyridoxal 5’-phosphate (PLP) levels in pyridoxal 5’-phosphate phosphatase (PDXP)-WT and knockout of PDXP (PDXP-KO) mice.
Each value represents the result of the PLP determination in an individual hippocampus. Analysis for statistically significant differences between PLP levels in PDXP-WT and PDXP-KO hippocampi (all ages combined; two-tailed, unpaired t-test) p<0.0001. Bold table entries indicate those hippocampal extracts that were further separated for an analysis of protein-depleted and protein-bound PLP (see Figure 1c). Source data are available for this table.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig1-data3-v1.zip
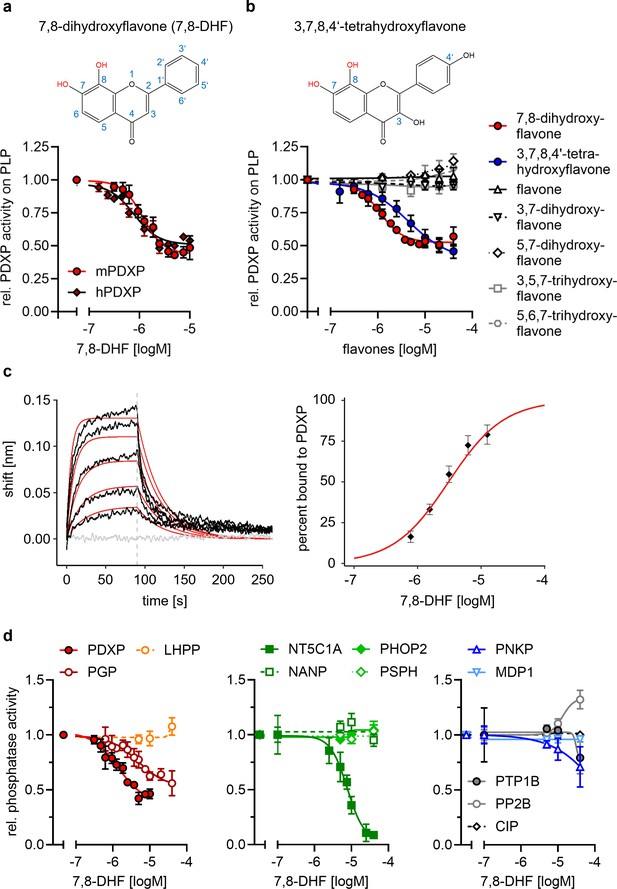
Characterization of the 7,8-dihydroxyflavone (7,8-DHF)/pyridoxal 5’-phosphate phosphatase (PDXP) interaction.
(a) Determination of half-maximal inhibitory constants (IC50) of 7,8-DHF (2D structure shown on top) for purified murine or human PDXP, using pyridoxal 5’-phosphate (PLP) as a substrate. Phosphatase activities in the presence of 7,8-DHF were normalized to the respective enzyme activities measured in the presence of the DMSO solvent control. Data are mean values ± SD of n=3 (human PDXP) and n=4 (murine PDXP) biologically independent experiments. (b) IC50 values of different flavones for purified murine PDXP with PLP as a substrate. Phosphatase activities in the presence of flavones were normalized to the respective enzyme activities in the presence of the DMSO solvent control. All data are mean values ± SD. The inhibition of PDXP by 3,7,8-trihydroxyflavone-4’-hydroxyphenyl (2D structure shown on top) was assessed in n=6 biologically independent experiments. All other data are from n=3 biologically independent experiments. Apparently missing error bars are hidden by the symbols. (c) Biolayer interferometry (BLI) measurements of the interaction of 7,8-DHF with purified murine PDXP. Left panel, example sensorgram overlayed with the global 1:1 binding model (red) and the negative control (gray). The dashed line indicates the start of the dissociation phase. Right panel, steady-state dose-response analysis for 7,8-DHF based on n=4 technically independent measurements. (d) Sensitivity of the indicated phosphatases to 7,8-DHF. Phosphatase activities in the presence of 7,8-DHF were normalized to the respective enzyme activities measured in the presence of the DMSO solvent control. Data are mean values ± SD of n=4 (PGP) or n=3 biologically independent experiments (all other phosphatases). Phosphatase substrates and haloacid dehalogenase (HAD) phosphatase cap types are indicated in parentheses. PDXP, pyridoxal 5’-phosphate phosphatase (pyridoxal 5’-phosphate, C2); PGP, phosphoglycolate phosphatase (2-phosphoglycolate; C2); LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase (imidodiphosphate; C2); NT5C1A, soluble cytosolic 5'-nucleotidase 1A (AMP; C1); NANP, N-acetylneuraminate 9-phosphate phosphatase (6-phosphogluconate; C1); PHOP2, phosphatase orphan 2 (pyridoxal 5’-phosphate; C1); PSPH, phosphoserine phosphatase (O-phospho-L-serine; C1); PNKP, polynucleotide kinase phosphatase (3-phospho-oligonucleotide; C0); MDP1, magnesium-dependent phosphatase-1 (D-ribose-5-phosphate; C0); PTP1B (protein tyrosine phosphatase 1B; EGFR phospho-peptide); PP2B, protein phosphatase 2B/calcineurin (PKA regulatory subunit type II phospho-peptide); CIP, calf intestinal phosphatase (pNPP). Source data are available for this figure.
-
Figure 2—source data 1
Phosphatase activity assays (to Figure 2a).
Effect of 7,8-dihydroxyflavone (7,8-DHF) on murine and human pyridoxal 5’-phosphate phosphatase (PDXP) activity. Data are OD values and normalized data of malachite green assays, using pyridoxal 5’-phosphate (PLP) as a PDXP substrate.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Phosphatase activity assays (to Figure 2b).
Effects of the indicated flavones on the phosphatase activity of murine pyridoxal 5’-phosphate phosphatase (PDXP). Data are OD values and normalized data of malachite green assays, using pyridoxal 5’-phosphate (PLP) as a PDXP substrate.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Biolayer interferometry (BLI) measurements with 7,8-dihydroxyflavone (7,8-DHF) and murine pyridoxal 5’-phosphate phosphatase (PDXP) (to Figure 2c).
Data of the association and dissociation curves for fitting.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig2-data3-v1.xlsx
-
Figure 2—source data 4
Effect of 7,8-dihydroxyflavone (7,8-DHF) on the phosphatase activity of different phosphatases (to Figure 2d).
Data are OD values and normalized data of malachite green assays. For calf intestinal phosphatase (CIP), kinetic data are OD values from pNPP-dephosphorylation assays.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig2-data4-v1.xlsx
-
Figure 2—source data 5
Pyridoxal 5’-phosphate phosphatase (PDXP) inhibitor hits.
Determination of half-maximal inhibitory constants (IC50) of 11 PDXP inhibitory compounds (see InChI Key for chemical substance identification) using purified murine PDXP and pyridoxal 5’-phosphate (PLP) as a substrate. Data marked with an asterisk (*) are results of n=3 biologically independent experiments. Because of the limited quantity of most compounds available for these assays, all other data are results of n=1 determinations. 7,8-DHF, 7,8-dihydroxyflavone.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig2-data5-v1.zip
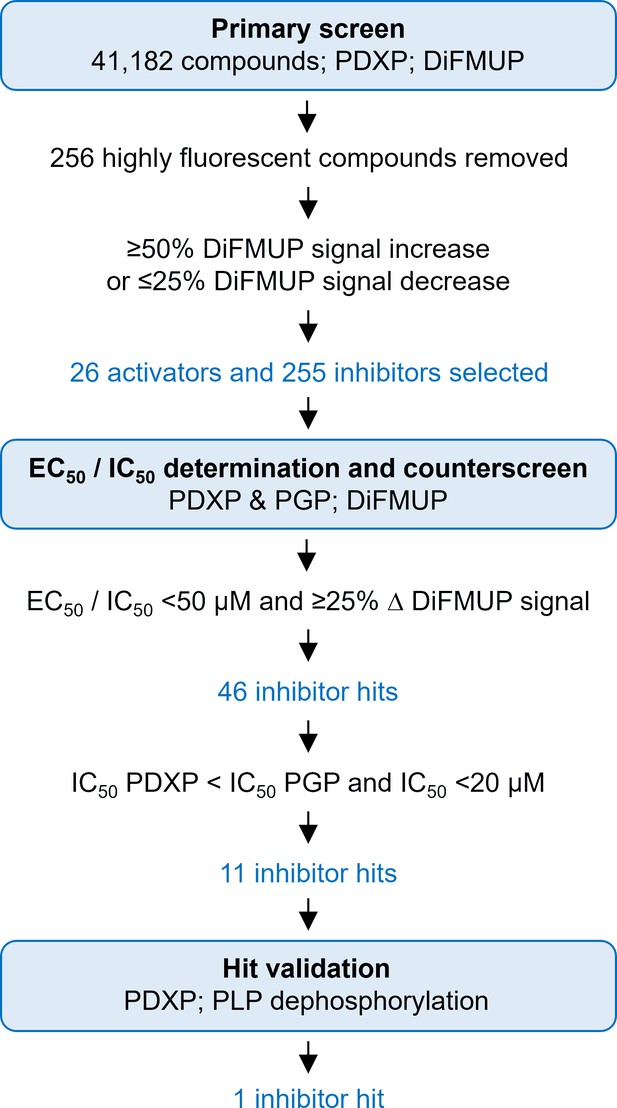
Identification of pyridoxal 5’-phosphate phosphatase (PDXP) inhibitors.
A primary screen was conducted using 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) as an artificial substrate. Out of 41,182 screened compounds, 256 compounds were discarded that showed very high autofluorescence (as recognized by elevated fluorescence at the start of the kinetic curve); 26 compounds showed statistically significant PDXP activation, and 255 compounds showed PDXP inhibition (as recognized by an elevated or decreased slope of the kinetic curve, respectively). The average Z’-factor of the screen was 0.75±0.112. These 281 compounds were selected for DiFMUP-based concentration-dependent validation, and the 46 most potent compounds were selected. A counter-screening was conducted in parallel, also in a concentration-dependent fashion, against the PDXP paralog and closest relative phosphoglycolate phosphatase (PGP). The 11 compounds that were inactive against PGP were validated in a secondary assay, using the PDXP substrate pyridoxal 5’-phosphate (PLP). One PDXP inhibitor hit blocked PLP dephosphorylation by ≥50%. Source data are available for this figure.
-
Figure 2—figure supplement 1—source data 1
Screening campaign for pyridoxal 5’-phosphate phosphatase (PDXP) inhibitors: IC50 data of the PDXP_PDXP primary screen.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig2-figsupp1-data1-v1.xlsx
-
Figure 2—figure supplement 1—source data 2
Screening campaign for pyridoxal 5’-phosphate phosphatase (PDXP) inhibitors: IC50 data of the PGP_PDXP counter-screen.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig2-figsupp1-data2-v1.xlsx
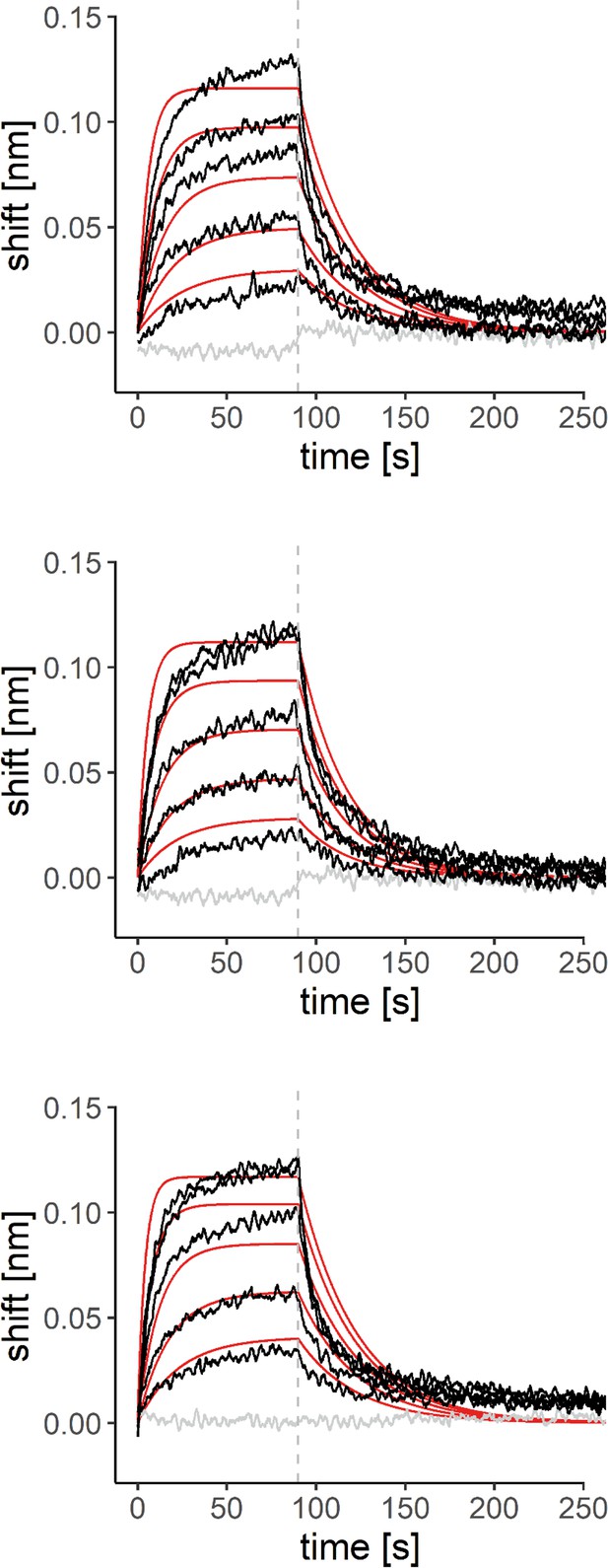
Biolayer interferometry (BLI) measurements of the interaction of 7,8-dihydroxyflavone (7,8-DHF) with purified murine pyridoxal 5’-phosphate phosphatase (PDXP).
Sensorgrams of three additional experiments overlayed with the global 1:1 binding model (red) and the negative control (gray). The dashed line indicates the start of the dissociation phase. Source data are available for this figure.
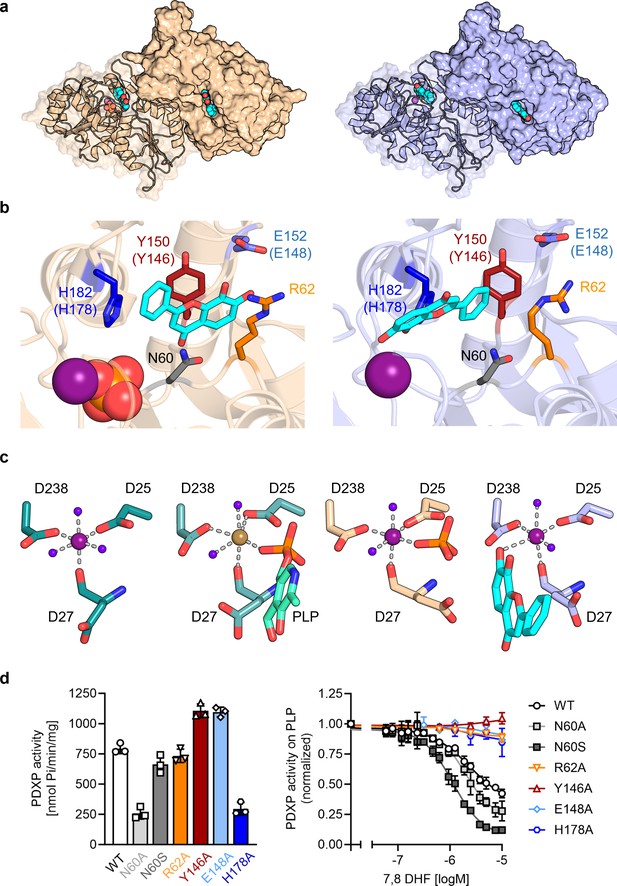
X-ray crystal structures of human pyridoxal 5’-phosphate phosphatase (PDXP) in complex with 7,8-dihydroxyflavone (7,8-DHF).
(a) The models were refined to a resolution of 1.5 Å for full-length human 7,8-DHF-PDXP with phosphate (PDB code 9EM1, colored in wheat yellow, left panel) and 1.5 Å for full-length human 7,8-DHF-PDXP without phosphate (PDB code 8S8A, colored in light blue, right panel). One protomer of each homodimeric PDXP is shown in cartoon representation and the other protomer in surface representation. 7,8-DHF is displayed in sphere representation with its C-atoms in cyan. Mg2+ ions are shown as deep purple spheres and phosphate ions are shown in sphere representation with the phosphorous atom in orange. (b) Orientation of 7,8-DHF in the active sites of human 7,8-DHF-PDXP in the presence or absence of phosphate. Structural details of bound 7,8-DHF and adjacent residues of the active sites are shown. Left, phosphate-containing 7,8-DHF-PDXP (wheat yellow, cartoon representation). Right, phosphate-free 7,8-DHF-PDXP (light blue, cartoon representation). 7,8-DHF is shown in stick representation (cyan C-atoms). The corresponding amino acids in murine PDXP are given in parentheses (see also Figure 3—figure supplement 1e and f). (c) Comparison of the Mg2+ coordination spheres. From left to right: human apo-PDXP (PDB: 2P27), human PDXP in complex with pyridoxal 5’-phosphate (PLP) (PDB: 2CFT), human PDXP in complex with 7,8-DHF in the presence of phosphate (PDB: 9EM1), human PDXP in complex with 7,8-DHF in the absence of phosphate (PDB: 8S8A). The catalytically essential Mg2+ is shown as a deep purple sphere. In 2CFT, Mg2+ was exchanged for Ca2+, which is shown here as a light brown-colored sphere. Water molecules are shown as blue spheres. (d) Verification of 7,8-DHF-PDXP interactions. Left panel, phosphatase activity of purified PDXP or the indicated PDXP variants. Data are mean values ± SD of n=3 biologically independent experiments. Right panel, determination of the IC50 values of 7,8-DHF for purified PDXP or the indicated PDXP variants. Data are mean values ± SD of n=3 biologically independent experiments. Apparently missing error bars are hidden by the symbols.
-
Figure 3—source data 1
Phosphatase activity and 7,8-dihydroxyflavone (7,8-DHF) sensitivity of pyridoxal 5’-phosphate phosphatase (PDXP) and PDXP variants (to Figure 3g).
Data are OD values and normalized data of malachite green assays, using pyridoxal 5’-phosphate (PLP) as a PDXP substrate.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig3-data1-v1.xlsx
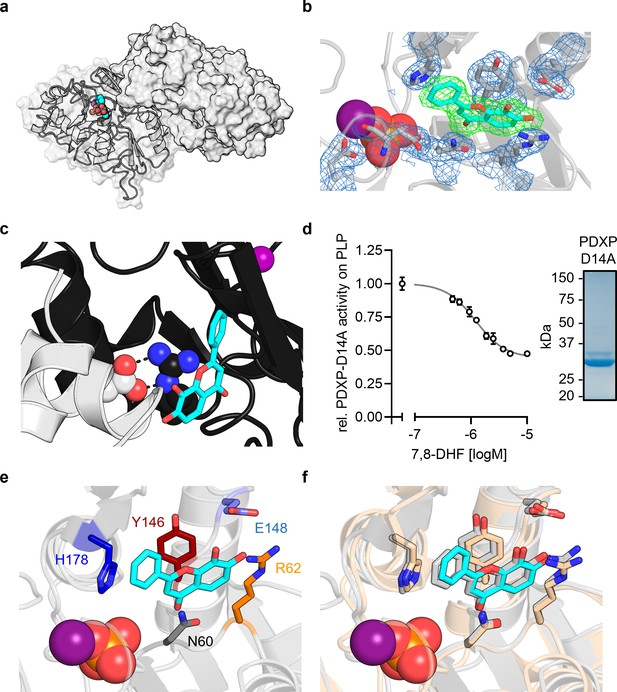
X-ray crystal structures of murine pyridoxal 5’-phosphate phosphatase (PDXP) in complex with 7,8-dihydroxyflavone (7,8-DHF).
(a) X-ray crystal structure of the assembled homodimeric, full-length murine PDXP in gray cartoon (protomer A) and surface (protomer B) representation. The model was refined to a resolution of 2.0 Å (PDB code 8QFW). 7,8-DHF is shown in sphere representation with its C-atoms in cyan. The Mg2+ ion is shown as deep purple sphere. The phosphate ion is shown as spheres with the phosphorous atom in orange. (b) The 2Fo − Fc electron density map of the depicted amino acids is contoured at a root mean square deviation (RMSD) of 1.0 in blue mesh and superimposed with the refined model. The Fo − Fc polder electron density map of 7,8-DHF is contoured at an RMSD of 3.0 in green mesh. (c) A salt bridge between Arg62 in the B-protomer (in black) and Asp14 of a symmetry-related A-protomer (in gray) blocks the 7,8-DHF binding site in the crystal lattice of murine PDXP. 7,8-DHF (in stick representation with cyan C-atoms) is modeled based on the A-protomer. (d) In vitro phosphatase activity of the purified PDXP-D14A variant in the presence of 7,8-DHF. Data are mean values ± SD of n=3 technically independent experiments. Apparently missing error bars are hidden by the symbols. The purity of PDXP-D14A is shown in the Coomassie blue-stained gel on the right. (e) Structural details of bound 7,8-DHF and adjacent residues of the active site of mPDXP (in gray cartoon representation). (f) Superimposition of the 7,8-DHF binding sites in the phosphate-containing murine PDXP (shown in gray) and human PDXP (in wheat yellow) structures. The corresponding amino acids are shown as gray or wheat yellow-colored sticks. 7,8-DHF bound to murine or human PDXP is shown as sticks colored in gray or cyan, respectively. The position of the Mg2+ and phosphate ions is based on human PDXP.
-
Figure 3—figure supplement 1—source data 1
Effect of 7,8-dihydroxyflavone (7,8-DHF) on the phosphatase activity of pyridoxal 5’-phosphate phosphatase (PDXP)-D14A.
Data are OD values and normalized data of malachite green assays, using pyridoxal 5’-phosphate (PLP) as a PDXP substrate.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig3-figsupp1-data1-v1.xlsx
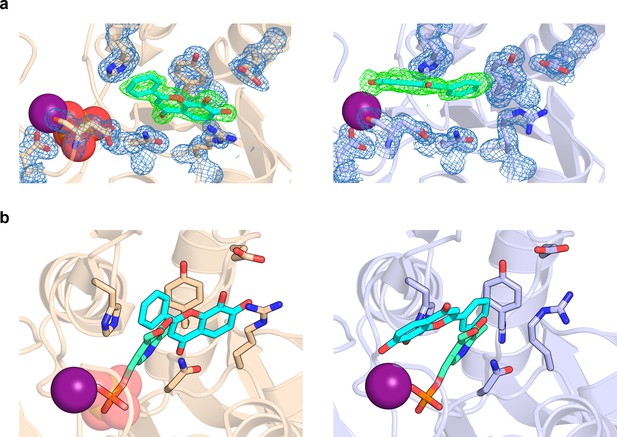
7,8-Dihydroxyflavone (7,8-DHF) coordination in pyridoxal 5’-phosphate phosphatase (PDXP).
(a) The 2Fo − Fc electron density map of the depicted amino acids is contoured at a root mean square deviation (RMSD) of 1.0 in blue mesh and superimposed with the refined model. The Fo − Fc polder electron density map of 7,8-DHF is contoured at an RMSD of 3.0 in green mesh. The Mg2+ ion is shown as a deep purple sphere. The phosphate ion is shown as a sphere with the phosphorous atom in orange. Left panel, human PDXP with phosphate (cartoon representation in wheat yellow); right panel, human PDXP without phosphate (cartoon representation in light blue). (b) Comparison of the 7,8-DHF and pyridoxal 5’-phosphate (PLP) binding sites in human PDXP with phosphate (wheat yellow, left panel) or human PDXP without phosphate (light blue, right panel). 7,8-DHF is shown in stick representation with cyan C-atoms. PLP (in stick representation with green C-atoms) was modeled based on a superposition of the human PDXP-PLP complex (PDB code 2CFT).

Alignment of human and murine pyridoxal 5’-phosphate phosphatase (PDXP).
Protein sequences of human PDXP (UniProtKB Q96GD0) and murine PDXP (UniProtKB P60487) were aligned with the EMBL-EBI multiple sequence alignment tool Clustal Omega version 1.2.4. PDXP residues found to engage in 7,8-dihydroxyflavone (7,8-DHF) interactions (highlighted in red color) are identical in human and murine PDXP.
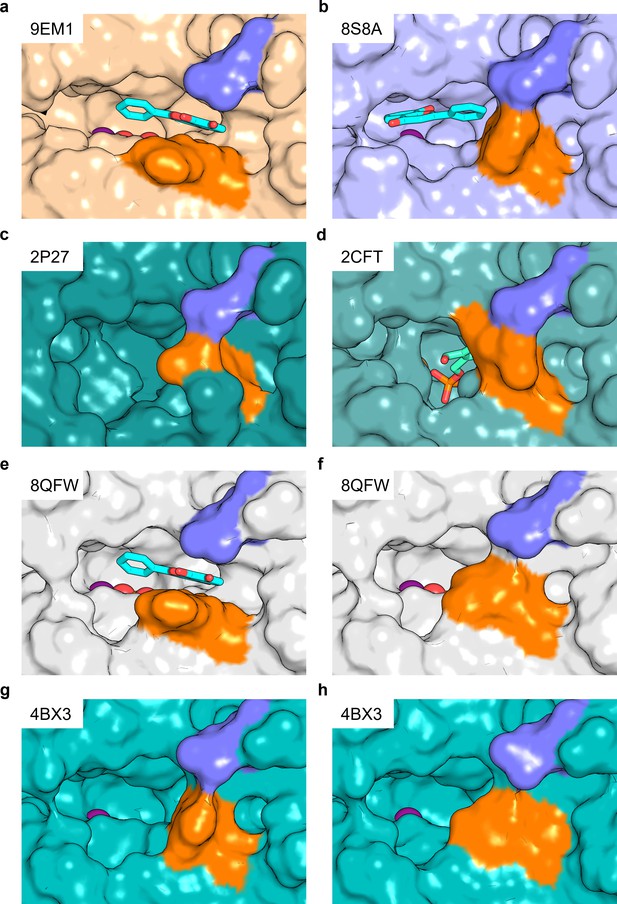
Salt bridge formation between Glu152 (Glu148) and Arg62 gates the active site entrance in pyridoxal 5’-phosphate phosphatase (PDXP).
Shown are views of the active site entrance in (a) hPDXP + 7,8-DHF with PO43-, (b) hPDXP + 7,8-DHF without PO43-, (c) apo-hPDXP, (d) hPDXP + pyridoxal 5’-phosphate (PLP), (e) mPDXP + 7,8-DHF with PO43, chain A, (f) mPDXP + 7,8-DHF with PO43-, chain B (inhibitor-free), (g) apo-mPDXP, chain A, (h) apo-mPDXP, chain B. The cap domain residue Glu152 in hPDXP (corresponding to Glu148 in mPDXP) is shown in blue, and the core domain residue Arg62 in hPDXP and mPDXP is shown in orange. 7,8-Dihydroxyflavone (7,8-DHF) is shown in stick representation with C-atoms in cyan. PLP is shown in stick representation with C-atoms in light green. The phosphate is shown in sphere representation with the phosphorous atom in orange. Mg2+ is shown as a deep purple sphere. hPDXP, human PDXP; mPDXP, murine PDXP. The respective PDB entries are indicated.
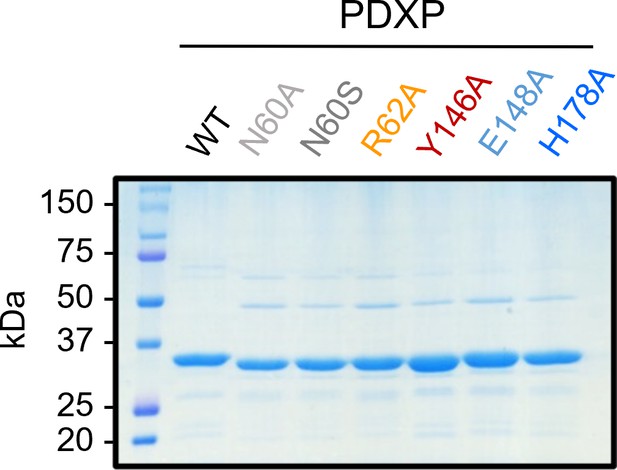
Purity of the employed pyridoxal 5’-phosphate phosphatase (PDXP) and PDXP variants.
A Coomassie blue-stained gel is shown. Source data are available for this figure.
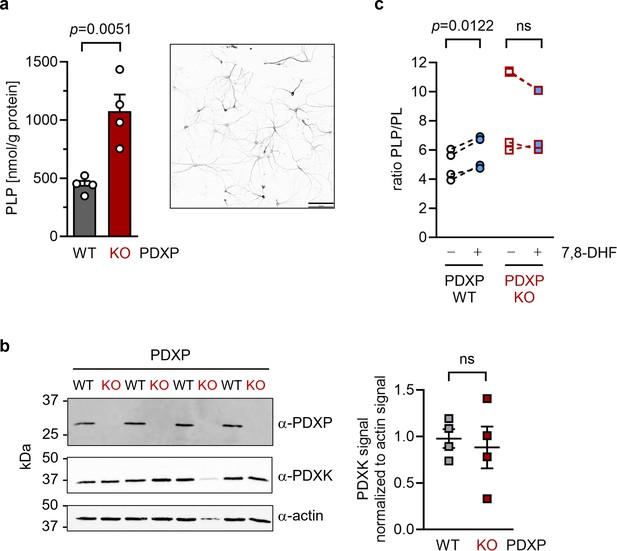
Effect of 7,8-dihydroxyflavone (7,8-DHF) on the pyridoxal 5’-phosphate (PLP)/PL ratio in cultured hippocampal neurons from WT or knockout of pyridoxal 5’-phosphate phosphatase (PDXP-KO) mice.
(a) Effect of long-term PDXP deficiency on total PLP levels in hippocampal neurons. Data are mean values ± SE of n=4 biologically independent experiments. Statistical significance was assessed with a two-tailed, unpaired t-test. A representative image of primary hippocampal neurons stained for the neuronal marker protein MAP2 is shown in the insert (pixel intensities were color-inverted for better visualization). Scale bar, 100 µm. (b) Western blot analysis of PDXP and pyridoxal kinase (PDXK) expression in hippocampal neuron samples shown in (a). The same blots were reprobed with α-actin antibodies as a loading control. The densitometric quantification of PDXK signals is shown on the right; data are mean values ± SE of n=4 biologically independent experiments. (c) Effect of 7,8-DHF (20 µM, 45 min) or the DMSO solvent control (0.02% vol/vol, 45 min) on the PLP/PL ratio in hippocampal neurons of PDXP-WT or PDXP-KO mice. Source data are available for this figure.
-
Figure 4—source data 1
Quantification of pyridoxal 5’-phosphate (PLP) and PLP/PL levels in hippocampal neurons (to Figure 4a and c).
Data are from high-performance liquid chromatography (HPLC)-based measurements.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Quantification of western blots (to Figure 4b).
Densitometric quantification of pyridoxal kinase (PDXK) and actin levels in hippocampal neurons derived from pyridoxal 5’-phosphate phosphatase (PDXP)-WT and knockout of PDXP (PDXP-KO) mice.
- https://cdn.elifesciences.org/articles/93094/elife-93094-fig4-data2-v1.xlsx
Tables
Kinetic constants of pyridoxal 5’-phosphate phosphatase (PDXP)-catalyzed pyridoxal 5’-phosphate (PLP) hydrolysis in the presence of 7,8-dihydroxyflavone (7,8-DHF).
| 7,8-DHF [µM] | 0 | 1.0 | 1.5 | 2.0 | 3.0 | 5.0 | 10.0 |
|---|---|---|---|---|---|---|---|
| KM [µM] | 14.98 ±1.28 | 18.54 ±6.24 | 20.20 ±6.19 | 18.97 ±5.15 | 24.83 ±2.61 | 32.96 ±2.13 | 30.61 ±2.57 |
| vmax [µmol/min/mg] | 1.08 ±0.04 | 0.95 ±0.01 | 0.89 ±0.04 | 0.85 ±0.02 | 0.80 ±0.04 | 0.81 ±0.05 | 0.74 ±0.06 |
| kcat [s–1] | 0.57 ±0.02 | 0.5 ±0.01 | 0.47 ±0.02 | 0.45 ±0.01 | 0.42 ±0.02 | 0.43 ±0.03 | 0.39 ±0.03 |
| kcat/KM [s–1∙M–1] (×10–4) | 3.93 ±0.29 | 3.27 ±0.84 | 2.75 ±0.67 | 2.72 ±0.66 | 1.72 ±0.10 | 1.31 ±0.06 | 1.29 ±0.01 |
-
The data are mean values ± SEM of n=3 technically independent experiments, except for the solvent control samples (n=6). Curves were fitted and parameters KM (Michaelis-Menten constant); vmax (maximum enzyme velocity); kcat (turnover number) were derived using the Michaelis-Menten model in GraphPad Prism 9.5.1. The kcat values were calculated from the maximum enzyme velocities using a molecular mass of 31,828 Da for PDXP. DMSO concentrations were kept constant (0.1% DMSO under all conditions, including the solvent control samples). Source data are available for this table.
-
Table 1—source data 1
Kinetic constants of pyridoxal 5’-phosphate phosphatase (PDXP)-catalyzed pyridoxal 5’-phosphate (PLP) hydrolysis in the presence of 7,8-dihydroxyflavone (7,8-DHF).
Data are OD values of malachite green assays, using PLP as a PDXP substrate, and derived kinetic constants.
- https://cdn.elifesciences.org/articles/93094/elife-93094-table1-data1-v1.xlsx
Data collection and refinement statistics.
| mPDXP-7,8-DHF with phosphate(8QFW) | hPDXP-7,8-DHF with phosphate(9EM1) | hPDXP-7,8-DHF without phosphate(8S8A) | |
|---|---|---|---|
| Data collection | |||
| Space group | I23 | P43212 | P43212 |
| a, b, c (Å) | 167.01, 167.01, 167.01 | 53.96, 53.96, 211.75 | 54.04, 54.04, 212.49 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 47.21–2.00 (2.07–2.00) | 48.08–1.50 (1.53–1.50) | 48.17–1.50 (1.53–1.50) |
| Rsym* | 0.190 (4.101) | 0.126 (4.893) | 0.081 (3.670) |
| Rpim† | 0.030 (0.652) | 0.018 (0.689) | 0.017 (0.731) |
| CC1/2 | 1.00 (0.459) | 0.991 (0.526) | 0.999 (0.579) |
| <I/σI>‡ | 20.7 (1.1) | 24.6 (1.4) | 17.5 (1.1) |
| Completeness | 0.998 (0.973) | 1.00 (1.00) | 1.00 (1.00) |
| Redundancy | 41.0 (38.5) | 50.7 (51.0) | 25.6 (26.0) |
| Refinement | |||
| Resolution (Å) | 20.00–2.00 (2.07–2.00) | 38.16–1.50 (1.55–1.50) | 48.165–1.50 (1.55–1.50) |
| R-work§ | 0.1838 (0.300) | 0.1702 (0.2740) | 0.1817 (0.2938) |
| R-free¶ | 0.21 (0.322) | 0.1939 (0.3217) | 0.2050 (0.2955) |
| RMS deviations in | |||
| Bond lengths (Å) | 0.002 | 0.012 | 0.005 |
| Bond angles (°) | 0.50 | 1.08 | 0.80 |
| Chiral centers (Å3) | 0.038 | 0.065 | 0.046 |
| Planar groups (Å) | 0.005 | 0.014 | 0.010 |
| Estimated coordinate error (Å) | 0.26 | 0.16 | 0.19 |
| Ramachandran statistics (%) | 98.95/1.05/0 | 98.63/1.37/0 | 98.63/1.37/0 |
-
Numbers in parentheses refer to the highest resolution data shell.
-
Ramachandran statistics reflect the percentage of residues in favored/allowed/outlier regions. Source data (raw diffraction images) have been deposited in the Xtal Raw Data Archive and can be accessed under the XRDA entries 8QFW (https://xrda.pdbj.org/entry/8qfw), 9EM1 (https://xrda.pdbj.org/entry/9em1), and 8S8A (https://xrda.pdbj.org/entry/8s8a).
-
*
Rsym = ΣhklΣi | Ii – <I> |/ΣhklΣiIi where Ii is the ith measurement and <I> is the weighted mean of all measurements of I.
-
†
Rpim = Σhkl1/(N – 1)½ Σi|Ii(hkl) – I(hkl)|/ΣhklΣiI(hkl), where N is the redundancy of the data and I(hkl) the average intensity.
-
‡
<I/σI> indicates the average of the intensity divided by its standard deviation.
-
§
Rwork = Σhkl ||Fo| – |Fc||/Σhkl|Fo| where Fo and Fc are the observed and calculated structure factor amplitudes.
-
¶
Rfree same as R for 5% of the data randomly omitted from the refinement. The number of reflections includes the Rfree subset.
Alignment of murine and human pyridoxal 5’-phosphate phosphatase (PDXP) structures.
| 7,8-DHF-mPDXP (+P), protomer A | 7,8-DHF-mPDXP (– P), protomer B | |
|---|---|---|
| 7,8-DHF-mPDXP (+P), protomer B | 0.43 Å | |
| Apo-mPDXP, protomer A | 0.50 Å | 0.71 Å |
| Apo-mPDXP, protomer B | 0.60 Å | 0.69 Å |
| 7,8-DHF-hPDXP (+P) | 7,8-DHF-hPDXP (–P) | |
| 7,8-DHF-hPDXP (–P) | 0.37 Å | |
| Apo-hPDXP | 0.54 Å | 0.45 Å |
| PLP-hPDXP | 0.39 Å | 0.29 Å |
-
Cα atom-based alignment of the structures representing murine apo-PDXP (PDB: 4BX3), 7,8-dihydroxyflavone (7,8-DHF)-bound murine PDXP (with inhibitor-bound protomer A and inhibitor-free protomer B; PDB: 8QFW), human apo-PDXP (PDB: 2P27), 7,8-DHF-bound human PDXP with phosphate (+P) (PDB: 9EM1), 7,8-DHF-bound human PDXP without phosphate (–P) (PDB: 8S8A) and pyridoxal 5’-phosphate (PLP)-bound human PDXP (PDB: 2CFT); mPDXP, murine PDXP; hPDXP, human PDXP. Root mean square deviations are indicated.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Pdxp | UniProtKB | P60487 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) pLysS | Stratagene Europe/VWR | AGLS200132 | |
| Genetic reagent (M. musculus; male) | Pdxptm1Goh; C57Bl/6J | Ozgene Ltd.; Jeanclos et al., 2019 | Floxed Pdxp mice | |
| Genetic reagent (M. musculus; female) | B6.FVB-Tg(EIIa-cre)C5379Lmgd/J | Jackson Labs | RRID:MGI:2174520 | Ubiquitous Cre deleter |
| Genetic reagent (M. musculus; male and female) | Pdxptm1Goh × EIIa-cre | Jeanclos et al., 2019 | Pdxp-deficient mice | |
| Biological sample (M. musculus) | Primary hippocampal neurons | This paper | From embryos of Pdxp-deficient or floxed Pdxp control mice | |
| Biological sample (M. musculus) | Hippocampi | This paper | Freshly isolated tissues from Pdxp-deficient or floxed Pdxp control mice | |
| Antibody | Anti-MAP2 (mouse monoclonal) | Millipore | Cat# MAB3418, RRID:AB_94856 | IF (1:500) |
| Antibody | Anti-actin (mouse monoclonal) | Sigma-Aldrich | Cat# MAB1501, RRID:AB_2223041 | WB (1:5000) |
| Antibody | Anti-PDXP (rabbit monoclonal) | Cell Signaling Technology | Cat# 4686, RRID:AB_2162520 | WB (1:1000) |
| Antibody | Anti-PDXK (rabbit polyclonal) | Sigma-Aldrich | Cat# AV53615, RRID:AB_1855158 | WB (1:1000) |
| Antibody | Anti-PNPO (rabbit polyclonal) | Thermo Fisher Scientific | Cat# PA5-26400, RRID:AB_2543900 | WB (1:1000) |
| Recombinant DNA reagent | pGEX-4T-1 (plasmid) | This paper | N-terminally GST-tagged, human PDXP | |
| Recombinant DNA reagent | pET-SUMO (plasmid) | This paper | N-terminally His6-SUMO-tagged human PDXP | |
| Recombinant DNA reagent | pET-M11 (plasmid) | EMBL Heidelberg | N-terminally His6-tagged, human SenP2 | |
| Recombinant DNA reagent | pET-M11 (plasmid) | Jeanclos et al., 2022 | Murine HAD phosphatases (PDXP, PGP, LHPP, NT5C1A, NANP, PHOP2, PSPH, PNKP, MDP1) | |
| Sequence-based reagent | Pdxp_F | This paper | PCR primers | TCGACCATGGCGCGCTGCGAGCGG |
| Sequence-based reagent | Pdxp_R | This paper | PCR primers | AAAAGTGAATTCTCAGTCCTCCAGCCCCTC |
| Sequence-based reagent | Pdxp-D14A_F | This paper | PCR primers | GCCCTGCGCGCCGTGCTGGGCCAGGCGCAG |
| Sequence-based reagent | Pdxp-D14A_R | This paper | PCR primers | GCCCAGCACGGCGCGCAGGGCCGCGCCGCG |
| Sequence-based reagent | Pdxp-N60A_F | This paper | PCR primers | TTCGTGAGCAACGCCAGCCGGCGCGCG |
| Sequence-based reagent | Pdxp-N60A_R | This paper | PCR primers | CGCGCGCCGGCTGGCGTTGCTCACGAA |
| Sequence-based reagent | Pdxp-N60S_F | This paper | PCR primers | TTCGTGAGCAACAGCAGCCGGCGCGCG |
| Sequence-based reagent | Pdxp-N60S_R | This paper | PCR primers | CGCGCGCCGGCTGCTGTTGCTCACGAA |
| Sequence-based reagent | Pdxp-R62A_F | This paper | PCR primers | AGCAACAACAGCGCGCGCGCGCGGCCC |
| Sequence-based reagent | Pdxp-R62A_R | This paper | PCR primers | GGGCCGCGCGCGCGCGCTGTTGTTGCT |
| Sequence-based reagent | Pdxp-Y146A_F | This paper | PCR primers | GTGCTCGTAGGCGCCGACGAGCAGTTT |
| Sequence-based reagent | Pdxp-Y146A_R | This paper | PCR primers | AAACTGCTCGTCGGCGCCTACGAGCAC |
| Sequence-based Reagent | Pdxp-E148A_F | This paper | PCR primers | GTAGGCTACGACGCGCAGTTTTCCTTC |
| Sequence-based reagent | Pdxp-E148A_R | This paper | PCR primers | GAAGGAAAACTGCGCGTCGTAGCCTAC |
| Sequence-based reagent | Pdxp-H178A_F | This paper | PCR primers | CGCGACCCTTGGGCCCCGCTCAGCGAC |
| Sequence-based reagent | Pdxp-H178A_R | This paper | PCR primers | GTCGCTGAGCGGGGCCCAAGGGTCGCG |
| Peptide, recombinant protein | Bovine brain calcineurin | Sigma-Aldrich | Cat# C1907 | PP2B |
| Peptide, recombinant protein | Phosphopeptide from PKA regulatory subunit type II | Sigma-Aldrich | Cat# 207008 | DLDVPIPGRFDRRVpSVAAE; PP2B substrate |
| Peptide, recombinant protein | Recombinant human PTP1B | Cayman Chemical | Cat# 10010896 | Amino acids 1–321 |
| Peptide, recombinant protein | EGFR phosphopeptide with Tyr992 autophosphorylation site | Santa Cruz Biotechnology | Cat# sc-3126 | DADEpYLIPQQG; PTP1B substrate |
| Peptide, recombinant protein | Calf intestinal alkaline phosphatase | NEB | Cat# M0525S | |
| Commercial assay or kit | EZ-Link NHS-PEG4-Biotin | Thermo Fisher | Cat# 21455 | |
| Chemical compound, drug | Flavone; 3,7-dihydroxyflavone; 5,7-dihydroxyflavone; 3,5,7-trihydroxyflavone; 5,6,7-trihydroxyflavone; 7,8-dihydroxyflavone | Sigma-Aldrich | Cat# F2003; Cat# 419826; Cat# 95082; Cat# 282200; Cat# 465119; Cat# D5446 | |
| Chemical compound, drug | 3,7,8,4’-Tetrahydroxyflavone | Ambinter | Cat# AMB30621919 | |
| Software, algorithm | Prism version 9.5.1 | GraphPad Prism | RRID:SCR_002798 | |
| Software, algorithm | OriginPro 2021b | OriginLab | RRID:SCR_014212 | |
| Other | Super Streptavidin Biosensors | Sartorius | Cat# 18-5057 | For biolayer interferometry experiments |





