Protein Tracking: Everything, everywhere, almost at once
Proteins play a role in almost all cellular processes and are essential for maintaining life across species and organisms. This means that their aberrant function is a major cause of disease. If one could look directly inside cells, they would see a seemingly chaotic scene of proteins continuously moving around. The motion of each protein is heterogeneous in time and space, and linked to its role within the cell. It is also heavily influenced by the local cellular environment and interactions with other molecules (Lippincott-Schwartz et al., 2001; Kusumi et al., 2014).
Conventional research techniques average the behavior of a large number of unsynchronized molecules, and thus fail to account for these variable factors, which are essential for understanding the biology of proteins. This is where single-molecule tracking methods come into play (Chenouard et al., 2014 ). Traditional ways for tracking individual molecules rely on advanced fluorescence microscopy, single-particle tracking and super-resolution imaging to directly observe the movement and interactions of proteins (Kusumi et al., 2014; Sahl et al., 2017; Shen et al., 2017).
These approaches provide the required spatiotemporal resolution, but typically can only analyze a few cells under limited conditions, offering a narrow glimpse of the vast and dynamic world of proteins. Although high-throughput microscopy has become much refined, scaling single-particle tracking remains a challenge (Park et al., 2023; Malle et al., 2022). A method that could record the performance of every single protein inside millions of individual cells, as well as thousands of molecular compounds, and analyze how they move, interact and respond to therapeutics, would be a major scientific breakthrough – one that may soon be a reality.
Now, in eLife, Hilary Beck and colleagues at Eikon Therapeutics and University of California Berkeley – including David McSwiggen as first author – report a high-throughput tracking (htSMT) platform that makes it possible to observe and analyze the behavior and movement of single proteins and molecules on an unprecedented scale (McSwiggen et al., 2023). The platform involves a robotic system capable of autonomously handling reagents and collecting sequential microscopy movies that are then computationally processed to obtain the trajectories of individual proteins within cells and even cellular compartments. This system allows users to image over a million cells, track thousands of individual proteins per cell, and screen thousands of compounds in a single day.
McSwiggen et al. then tested the platform on estrogen receptors and investigated how over 5,000 compounds affected their motion, analyzing hundreds of thousands of cells twice in a single day (Figure 1). This revealed a new correlation between the dynamics of estrogen receptors and the ability of their antagonists to suppress the growth of cancer cells, which conventual methods have failed to detect previously. Moreover, the htSMT platform also revealed whether the tested molecules affect estrogen receptors directly or indirectly through other biological targets that are known to modify the receptor. This provides an unprecedented and unbiased analysis of a complex biological pathway.
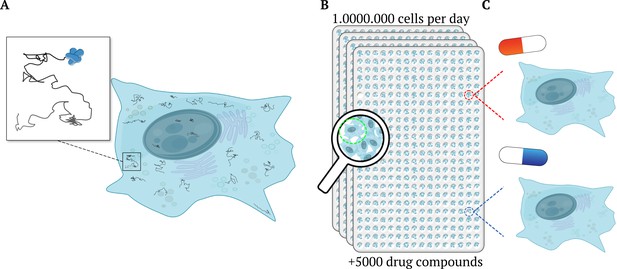
High-throughput single-molecule tracking of proteins across millions of cells.
(A) Schematic illustration of a cell (blue shape) as taken from a high-throughput single-molecule tracking (htSMT) assay (inset), which tracks the motion of multiple proteins within individual cells (squiggly lines). (B) The platform can record the movement of thousands of individual, heterogeneous proteins per cell in over a million cells per day. It does this by automatically collecting a series of images from 384-well plates mounted on a microscope. Each well contains multiples cells and constitutes an independent experiment, enabling researchers to investigate different cell types, and test the effects of various drugs and other molecules and/ or proteins. (C) The htSMT results from cells treated with different drugs (indicated as a red or blue pill) can then be used to assess which treatment is likely to work best.
Image credit: Jacob Kæstel-Hansen and Nikos S. Hatzakis. The figure was – with large modifications – generated with elements from Servier Medical Art and Scidraw.io (10.5281/zenodo.3926549; CC BY 4.0).
Overall, the htSMT platform paves the way for a new era in cellular biology and pharmacology, enabling large-scale, automated observations of how proteins move and interact across millions of cells within 24 hours – a feat that until recently remained in the realm of fantasy. This profound increase in scale, together with advanced analytic tools (Muñoz-Gil et al., 2021; Pinholt et al., 2021), promises to unlock even more unresolved information about complex biological pathways, such as those associated with the estrogen receptor. Adapting htSMT to other proteins and cell systems could help construct unique libraries that ultimately link movement to function. Exploiting the full potential of htSMT will further our understanding of the intricate processes occurring within cells and how protein motion contributes to – and depends on – cellular function. Ultimately this could help researchers design new pharmaceutical treatments for controlling certain diseases.
References
-
Objective comparison of particle tracking methodsNature Methods 11:281–289.https://doi.org/10.1038/nmeth.2808
-
Tracking single molecules at work in living cellsNature Chemical Biology 10:524–532.https://doi.org/10.1038/nchembio.1558
-
Studying protein dynamics in living cellsNature Reviews. Molecular Cell Biology 2:444–456.https://doi.org/10.1038/35073068
-
Objective comparison of methods to decode anomalous diffusionNature Communications 12:6253.https://doi.org/10.1038/s41467-021-26320-w
-
Fluorescence nanoscopy in cell biologyNature Reviews Molecular Cell Biology 18:685–701.https://doi.org/10.1038/nrm.2017.71
-
Single particle tracking: from theory to biophysical applicationsChemical Reviews 117:7331–7376.https://doi.org/10.1021/acs.chemrev.6b00815
Article and author information
Author details
Publication history
Copyright
© 2024, Kæstel-Hansen and Hatzakis
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,565
- views
-
- 167
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cancer Biology
- Cell Biology
Testicular microcalcifications consist of hydroxyapatite and have been associated with an increased risk of testicular germ cell tumors (TGCTs) but are also found in benign cases such as loss-of-function variants in the phosphate transporter SLC34A2. Here, we show that fibroblast growth factor 23 (FGF23), a regulator of phosphate homeostasis, is expressed in testicular germ cell neoplasia in situ (GCNIS), embryonal carcinoma (EC), and human embryonic stem cells. FGF23 is not glycosylated in TGCTs and therefore cleaved into a C-terminal fragment which competitively antagonizes full-length FGF23. Here, Fgf23 knockout mice presented with marked calcifications in the epididymis, spermatogenic arrest, and focally germ cells expressing the osteoblast marker Osteocalcin (gene name: Bglap, protein name). Moreover, the frequent testicular microcalcifications in mice with no functional androgen receptor and lack of circulating gonadotropins are associated with lower Slc34a2 and higher Bglap/Slc34a1 (protein name: NPT2a) expression compared with wild-type mice. In accordance, human testicular specimens with microcalcifications also have lower SLC34A2 and a subpopulation of germ cells express phosphate transporter NPT2a, Osteocalcin, and RUNX2 highlighting aberrant local phosphate handling and expression of bone-specific proteins. Mineral disturbance in vitro using calcium or phosphate treatment induced deposition of calcium phosphate in a spermatogonial cell line and this effect was fully rescued by the mineralization inhibitor pyrophosphate. In conclusion, testicular microcalcifications arise secondary to local alterations in mineral homeostasis, which in combination with impaired Sertoli cell function and reduced levels of mineralization inhibitors due to high alkaline phosphatase activity in GCNIS and TGCTs facilitate osteogenic-like differentiation of testicular cells and deposition of hydroxyapatite.
-
- Cell Biology
- Genetics and Genomics
A glaucoma polygenic risk score (PRS) can effectively identify disease risk, but some individuals with high PRS do not develop glaucoma. Factors contributing to this resilience remain unclear. Using 4,658 glaucoma cases and 113,040 controls in a cross-sectional study of the UK Biobank, we investigated whether plasma metabolites enhanced glaucoma prediction and if a metabolomic signature of resilience in high-genetic-risk individuals existed. Logistic regression models incorporating 168 NMR-based metabolites into PRS-based glaucoma assessments were developed, with multiple comparison corrections applied. While metabolites weakly predicted glaucoma (Area Under the Curve = 0.579), they offered marginal prediction improvement in PRS-only-based models (p=0.004). We identified a metabolomic signature associated with resilience in the top glaucoma PRS decile, with elevated glycolysis-related metabolites—lactate (p=8.8E-12), pyruvate (p=1.9E-10), and citrate (p=0.02)—linked to reduced glaucoma prevalence. These metabolites combined significantly modified the PRS-glaucoma relationship (Pinteraction = 0.011). Higher total resilience metabolite levels within the highest PRS quartile corresponded to lower glaucoma prevalence (Odds Ratiohighest vs. lowest total resilience metabolite quartile=0.71, 95% Confidence Interval = 0.64–0.80). As pyruvate is a foundational metabolite linking glycolysis to tricarboxylic acid cycle metabolism and ATP generation, we pursued experimental validation for this putative resilience biomarker in a human-relevant Mus musculus glaucoma model. Dietary pyruvate mitigated elevated intraocular pressure (p=0.002) and optic nerve damage (p<0.0003) in Lmx1bV265D mice. These findings highlight the protective role of pyruvate-related metabolism against glaucoma and suggest potential avenues for therapeutic intervention.






