Gene Fusion: Decoding the identity of rare tumors
Gene fusions are common mutations that have been linked to several types of cancer, most notably chronic myelogenous leukemia, which is a result of the BCR gene on chromosome 22 fusing with the ABL gene on chromosome 9 (Nowell and Hungerford, 1960). Identifying fusion genes, and investigating the molecular behavior of the chimeric fusion proteins that arise from these mutations, may help with the diagnosis, prognosis and treatment of certain cancers.
The fusion of two genes on chromosome 12 – NAB2 and STAT6 – has been found in a rare form of cancer, known as a solitary fibrous tumor, which can form in almost any part of the body (Thway et al., 2016). While the initial tumor can often be removed, around 40% of them can recur or metastasize, at which point they become untreatable.
In most healthy tissues, NAB2 and STAT6 function independently. NAB2 is a transcriptional co-regulator that – being physiologically sequestered in the cytoplasm – can restrict the nuclear activity of two transcription factors involved in cell proliferation, EGR1 and EGR2 (Svaren et al., 1996), whereas STAT6 is a transcription factor that travels to the nucleus to activate gene expression (Figure 1; Hu et al., 2021). However, the molecular function of the NAB2-STAT6 fusion protein remains obscure. Now, in eLife, Alessandro Gardini and colleagues at the Wistar Institute and the University of Pennsylvania – including Connor Hill as first author – report the results of studies that help clarify the role of this fusion protein in solitary fibrous tumors (Hill et al., 2024).
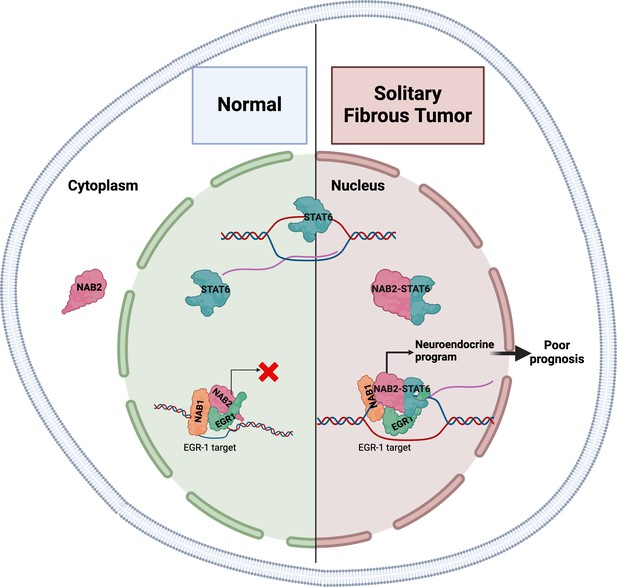
The gene expression patterns in solitary fibrous tumors are characteristic of neuronal development.
In normal healthy tissues (left), the NAB2 protein is mostly located in the cytoplasm, and thus cannot co-activate EGR1 in the nucleus (red X), a transcription factor involved in cell proliferation. NAB1 is also involved in this process. STAT6 is a transcription factor that travels to the nucleus to activate gene expression. In solitary fibrous tumors (right), the NAB2-STAT6 fusion protein readily travels to the nucleus, where it binds to EGR1, and the entire complex binds to promoters and enhancers normally targeted by EGR1 (black arrow). This increases the expression of these gene targets and results in the tumors having gene expression patterns characteristic of neuronal development.
Image credit: This image was created using Biorender.
The researchers used a combination of cell-based studies, analyses of genome-wide binding patterns of specific proteins within tumor samples, and a comparative analysis of gene expression signatures. They found that compared to adjacent healthy tissue, primary solitary fibrous tumors had gene expression patterns characteristic of neuronal development, in particular, the targets of ERG1 were enriched in the tumors. Immune and cell signaling pathways were also significantly downregulated. To validate these findings, Hill et al. used cells derived from an osteosarcoma (a type of bone cancer) to create an in vitro model that expressed the NAB2-STAT6 fusion protein. This model also showed gene expression patterns characteristic of neuronal development and increased levels of NAB1, NAB2 and EGR1 bolstered by the fusion protein were also observed (Figure 1).
In both cell lines and samples from patients with cancer, the fusion protein and EGR1 bound to EGR1-targeted promoters and enhancers, increasing their accessibility and expression of the corresponding genes. The binding appeared to be mediated by the NAB2 portion, while the STAT6 portion was responsible for translocating the fusion protein to the cell nucleus.
Hill et al. then compared the gene expression signature of the solitary fibrous tumors with existing datasets from The Cancer Genome Atlas, which revealed a striking similarity between solitary fibrous tumors and neuroendocrine tumors (arising from cells that can release hormones in response to signals from the nervous system), including glioblastoma. Moreover, an analysis of registered survival rates indicated a significantly worse outcome for cancer patients with a gene signature indicative of solitary fibrous tumors.
The study of Hill et al. provides valuable insights into the dynamics of solitary fibrous tumors and their unique neural-like gene expression signature that might be relevant for other, more common cancers. Their neuroendocrine identity driven by NAB2-STAT6 fusion highlights their similarity to other neuroendocrine malignancies, such as pheochromocytoma (affecting adrenal glands) and oligodendroglioma (affecting specific glial cells in the brain), raising the question of their cell-of-origin (Hill et al., 2024; Davanzo et al., 2018; Demicco et al., 2012).
Future research on the molecular consequences of the fusion of NAB2 and STAT6 may help refine cancer diagnosis and inform drug development. Targeting NAB2-STAT6 or its downstream pathways could help prevent the recurrence of this cancer, or serve as a strategy for patients who are not candidates for surgery. However, more research is needed to confirm these hypotheses. Being the first of its kind, the study of Hill et al. significantly advances our molecular understanding of solitary fibrous tumors, a critical first step toward targeted precision medicine approaches.
References
-
Solitary fibrous tumorTranslational Gastroenterology and Hepatology 3:94.https://doi.org/10.21037/tgh.2018.11.02
-
The JAK/STAT signaling pathway: from bench to clinicSignal Transduction and Targeted Therapy 6:402.https://doi.org/10.1038/s41392-021-00791-1
-
Chromosome studies on normal and leukemic human leukocytesJournal of the National Cancer Institute 25:85–109.
-
NAB2, A corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuliMolecular and Cellular Biology 16:3545–3553.https://doi.org/10.1128/MCB.16.7.3545
-
The current status of solitary fibrous tumor: diagnostic features, variants, and geneticsInternational Journal of Surgical Pathology 24:281–292.https://doi.org/10.1177/1066896915627485
Article and author information
Author details
Publication history
Copyright
© 2024, Yuan et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 468
- views
-
- 41
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cancer Biology
- Cell Biology
The endoplasmic reticulum (ER) is an essential sensing organelle responsible for the folding and secretion of almost one-third of eukaryotic cells' total proteins. However, environmental, chemical, and genetic insults often lead to protein misfolding in the ER, accumulating misfolded proteins, and causing ER stress. To solve this, several mechanisms were reported to relieve ER stress by decreasing the ER protein load. Recently, we reported a novel ER surveillance mechanism by which proteins from the secretory pathway are refluxed to the cytosol to relieve the ER of its content. The refluxed proteins gain new prosurvival functions in cancer cells, thereby increasing cancer cell fitness. We termed this phenomenon ER to CYtosol Signaling (or ‘ERCYS’). Here, we found that in mammalian cells, ERCYS is regulated by DNAJB12, DNAJB14, and the HSC70 cochaperone SGTA. Mechanistically, DNAJB12 and DNAJB14 bind HSC70 and SGTA - through their cytosolically localized J-domains to facilitate ER-protein reflux. DNAJB12 is necessary and sufficient to drive this phenomenon to increase AGR2 reflux and inhibit wt-p53 during ER stress. Mutations in DNAJB12/14 J-domain prevent the inhibitory interaction between AGR2-wt-p53. Thus, targeting the DNAJB12/14-HSC70/SGTA axis is a promising strategy to inhibit ERCYS and impair cancer cell fitness.
-
- Cancer Biology
- Cell Biology
DNA base lesions, such as incorporation of uracil into DNA or base mismatches, can be mutagenic and toxic to replicating cells. To discover factors in repair of genomic uracil, we performed a CRISPR knockout screen in the presence of floxuridine, a chemotherapeutic agent that incorporates uracil and fluorouracil into DNA. We identified known factors, such as uracil DNA N-glycosylase (UNG), and unknown factors, such as the N6-adenosine methyltransferase, METTL3, as required to overcome floxuridine-driven cytotoxicity. Visualized with immunofluorescence, the product of METTL3 activity, N6-methyladenosine, formed nuclear foci in cells treated with floxuridine. The observed N6-methyladenosine was embedded in DNA, called 6mA, and these results were confirmed using an orthogonal approach, liquid chromatography coupled to tandem mass spectrometry. METTL3 and 6mA were required for repair of lesions driven by additional base-damaging agents, including raltitrexed, gemcitabine, and hydroxyurea. Our results establish a role for METTL3 and 6mA in promoting genome stability in mammalian cells, especially in response to base damage.






