Sumoylation of the human histone H4 tail inhibits p300-mediated transcription by RNA polymerase II in cellular extracts
Figures
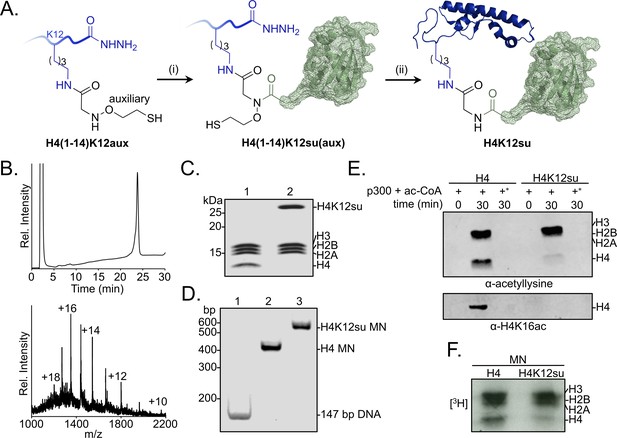
Sumoylation inhibits p300-mediated H4 acetylation in octamer and mononucleosome substrates.
(A) Synthetic scheme for H4K12su. (i) An H4(1–14)K12aux peptide was ligated with a SUMO-3 (2–91) C47S α-thioester. (ii) The sumoylated H4(1–14) peptidyl hydrazide containing the auxiliary was converted to a C-terminal α-thioester and ligated with H4(15–102) A15C. The auxiliary was then reductively cleaved from the ligation product. Cys15 in the final ligation product was desulfurized to the native Ala15 to yield site-specifically sumoylated H4K12su. (B) C4 analytical RP-HPLC trace of purified H4K12su (top). ESI-MS of purified H4K12su (bottom). Calculated mass 21,596.7 Da. Observed, 21,594.2 ± 3.4 Da. (C) Coomassie-stained 15% SDS-PAGE of reconstituted octamers containing wild-type (wt) H4 or H4K12su. (D) Ethidium bromide stained 5% TBE gel of mononucleosomes containing wt H4 or H4K12su. (E) Western blots of p300 assay products with octamer substrates containing wt H4 or H4K12su, probed with a site-independent pan-acetyllysine antibody (top) and an H4K16ac-specific antibody (bottom). An asterisk indicates assays with heat-inactivated p300 to exclude non-enzymatic acetylation. (F) Fluorogram of p300 assay products with [3H]-acetyl-CoA as the co-factor and mononucleosome substrates containing wt H4 or H4K12su.
-
Figure 1—source data 1
Unedited intact SDS-PAGE gels for all gel images shown in Figure 1.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig1-data1-v2.pdf
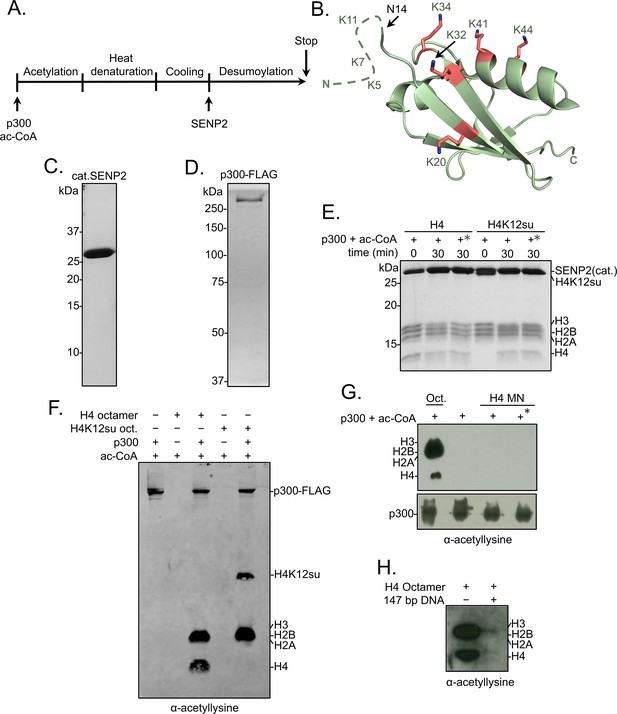
Histone octamer and mononucleosome acetylation by p300.
(A) Scheme outlining the p300 histone acetyltransferase assay with octamer and mononucleosome substrates. (B) Cartoon representation of SUMO-3 showing all surface-exposed lysine residues in stick representation. The dashed line represents N-terminal residues not observed in the structure. PDB code 1U4A. (C) Coomassie-stained SDS-PAGE of purified catalytic domain of SENP2, cat.SENP2, consisting of residues 365–590. (D) Coomassie-stained SDS-PAGE of purified p300-FLAG from HEK293T cells. (E) Coomassie-stained SDS-PAGE corresponding to the HAT assay shown in Figure 1E. Asterisk indicates heat-inactivated p300 was used. (F) Histone acetylation assay with octamers containing wild-type (wt) H4 or H4K12su. Autoacetylation of p300 was observed with a pan-acetyllysine antibody. (G) Histone acetylation assay with p300 and wt H4 containing octamers and mononucleosomes. No cat.SENP2 was used in this assay. The asterisk indicates pre-incubation of p300-FLAG with acetyl-CoA for 1 hr to allow for the build-up of activating p300-autoacetylation prior to the addition of mononucleosome substrate (Thompson et al., 2004) (H) Histone acetylation assay with wt H4 octamer with or without equimolar amounts of 147 bp Widom 601 double-stranded DNA (dsDNA).
-
Figure 1—figure supplement 1—source data 1
Unedited intact SDS-PAGE gels and western blot membranes for all gel and western blot images shown in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig1-figsupp1-data1-v2.pdf
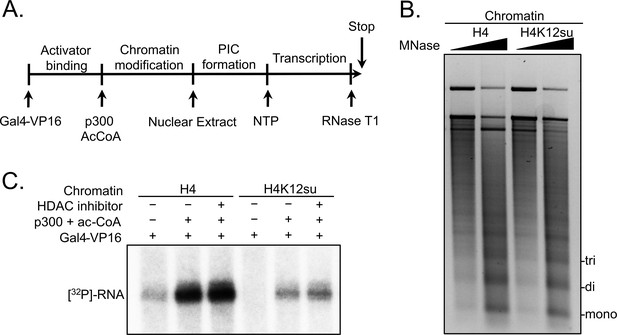
Histone H4 sumoylation inhibits in vitro transcription from chromatinized plasmid templates.
(A) Scheme outlining steps during the in vitro transcription assay with chromatinized plasmids, nuclear extracts, activator Gal4-VP16 and p300. (B) Micrococcal nuclease digestion analysis of plasmids chromatinized with wild-type (wt) H4 or H4K12su indicating the similar occupancy and spacing of mononucleosomes. (C) Autoradiogram of 32P-labeled 365 base RNA transcript generated from p300-mediated transcription from chromatinized templates containing wt H4 or H4K12su in the presence or absence of the histone deacetylase (HDAC) inhibitor, trichostatin A.
-
Figure 2—source data 1
Unedited intact TBE gel for chromatin digestion gel shown in Figure 2.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig2-data1-v2.pdf
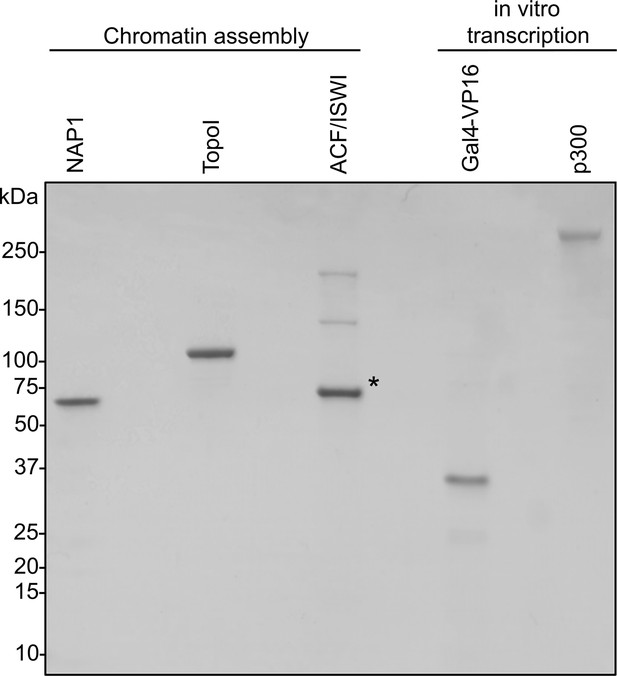
Coomassie-stained SDS-PAGE of chromatin assembly proteins and in vitro transcription components.
Asterisk indicates BSA used as a stabilizer.
-
Figure 2—figure supplement 1—source data 1
Unedited intact SDS-PAGE gel showing protein components of the in vitro transcription assay.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig2-figsupp1-data1-v2.pdf
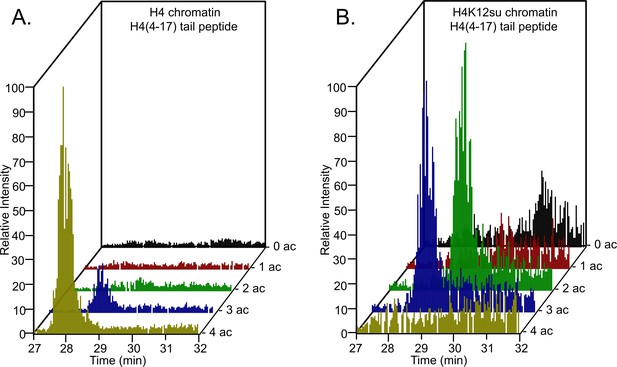
Comparison of H4 tail acetylation by p300 in chromatinized plasmid templates with activator Gal4-VP16.
(A) Extracted ion chromatograms of all H4(4–17) tryptic peptides obtained after SDS-PAGE resolution and in-gel trypsination of acetylated chromatin containing wild-type (wt) H4. (B) Extracted ion chromatograms of all H4(4–17) tryptic peptides obtained after SDS-PAGE resolution, in-gel desumoylation and trypsination of acetylated chromatin containing H4K12su. The extracted m/z of each spectrum is centered on the [M + 2 H]2+ precursor ion.
-
Figure 3—source data 1
Excel file listing all mass spectral data plotted in Figure 3, Tables 1–4; and Figure 3—figure supplements 2–7.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig3-data1-v2.xlsx
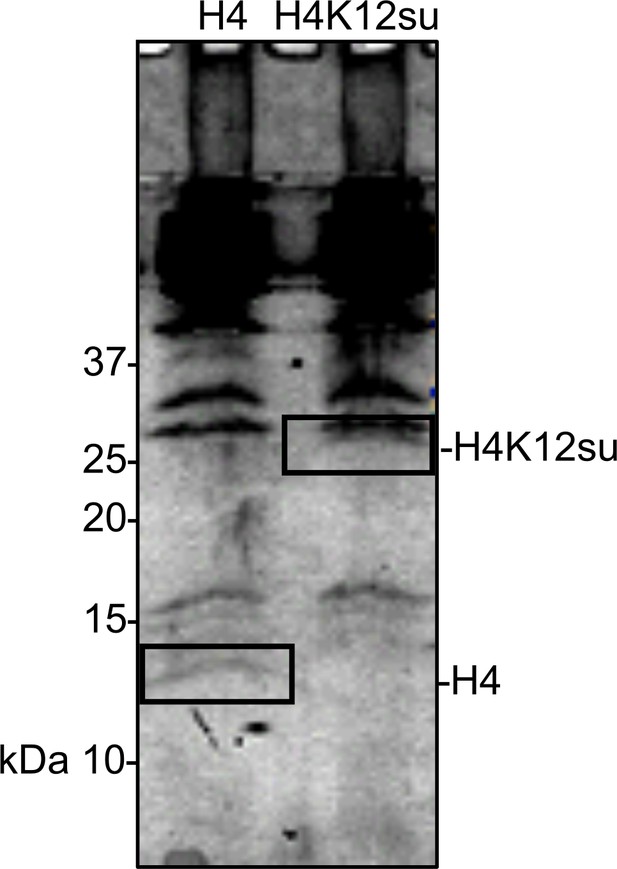
Coomassie-stained SDS-PAGE of histone acetylation assay on chromatinized plasmids containing wild-type (wt) H4 or H4K12su with p300 and activator Gal4-VP16.
Gel bands excised for tandem mass spectrometry (MS-MS) analysis are indicated.
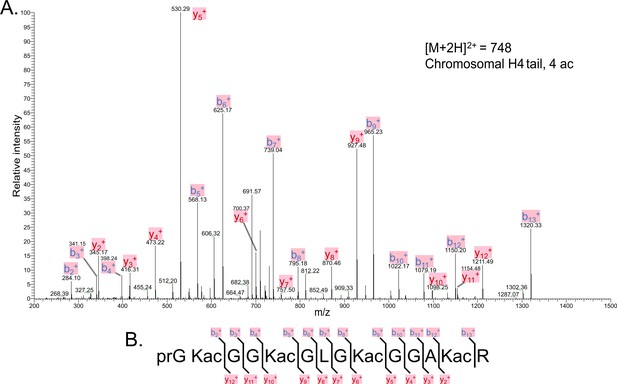
Tandem MS of tetra-acetylated tryptic peptide H4(4–17).
(A) Representative tandem mass spectrometry (MS-MS) spectrum of tetra-acetylated tryptic peptide H4(4–17) generated after in vitro acetylation of chromatinized plasmids containing wild-type (wt) H4 with p300 and activator Gal4-VP16. (B) Peptide fragment-ion map of the tetra-acetylated H4(4–17) peptide indicating all ions identified over three spectra.
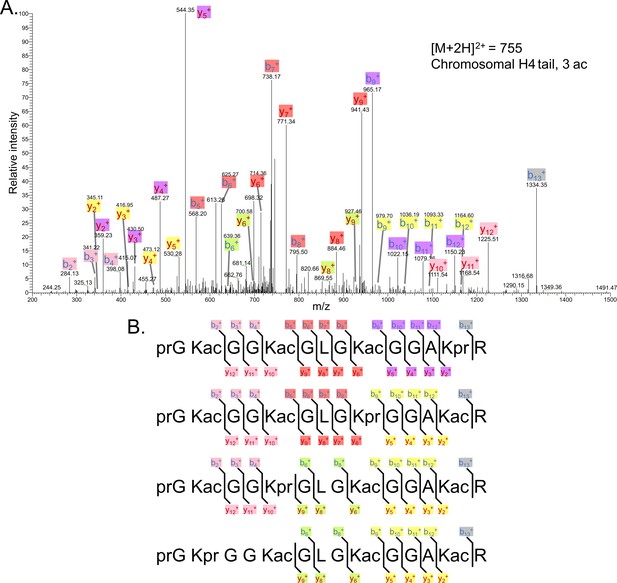
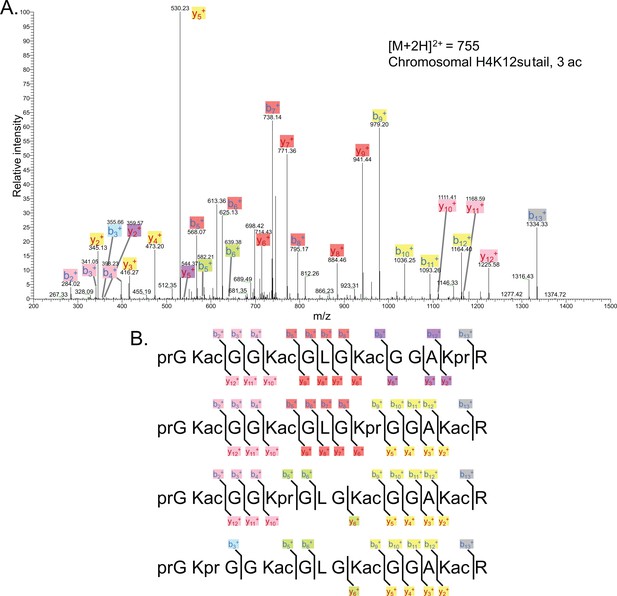
Tandem MS of tri-acetylated tryptic peptide H4(4–17).
(A) Representative tandem mass spectrometry (MS-MS) spectrum of tri-acetylated tryptic peptide H4(4–17) generated after in vitro acetylation of chromatinized plasmids containing wild-type (wt) H4 with p300 and activator Gal4-VP16. (B) Peptide fragment-ion maps of the four possible tri-acetylated H4(4–17) peptide patterns, indicating all ions identified over three spectra.
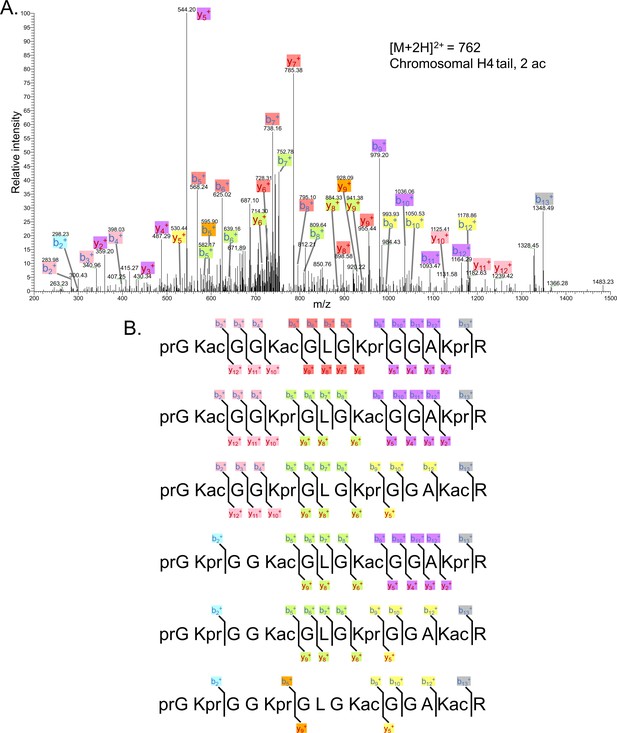
Tandem MS of di-acetylated tryptic peptide H4(4–17).
(A) Representative tandem mass spectrometry (MS-MS) spectrum of di-acetylated tryptic peptide H4(4–17) generated after in vitro acetylation of chromatinized plasmids containing wild-type (wt) H4 with p300 and activator Gal4-VP16. (B) Peptide fragment-ion maps of the six possible di-acetylated H4(4–17) peptide patterns, indicating all ions identified over two spectra.
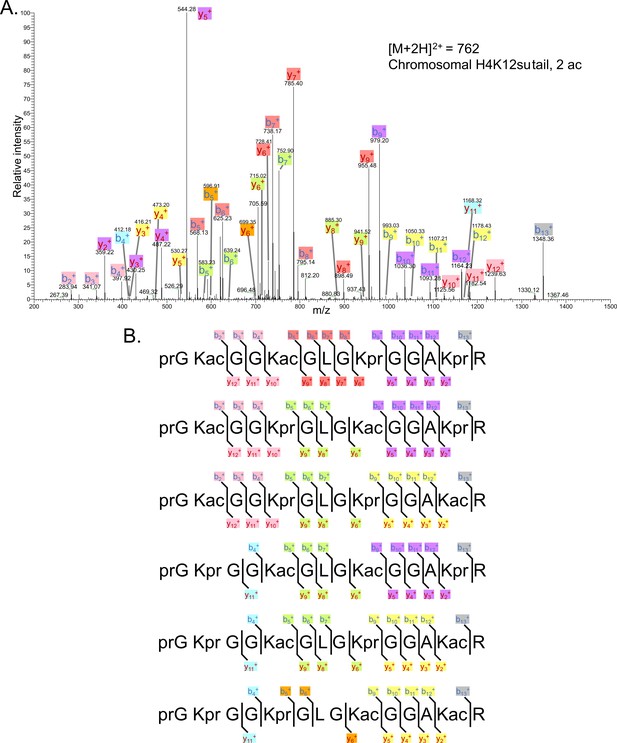
Representative tandem mass spectrometry (MS-MS) spectrum of tri-acetylated tryptic peptide H4(4–17) generated after in vitro acetylation of chromatinized plasmids containing H4K12su with p300 and activator Gal4-VP16 followed by in-gel desumoylation.
(B) Peptide fragment-ion maps of the four possible tri-acetylated H4(4–17) peptide patterns, indicating all ions identified over three spectra. Acetylation on K12 in H4K12su is not possible due to presence of SUMO-3 at K12.
Representative tandem mass spectrometry (MS-MS) spectrum of di-acetylated tryptic peptide H4(4–17) generated after in vitro acetylation of chromatinized plasmids containing H4K12su with p300 and activator Gal4-VP16 followed by desumoylation.
(B) Peptide fragment-ion maps of the six possible di-acetylated H4(4–17) peptide species, indicating all ions identified over three spectra.
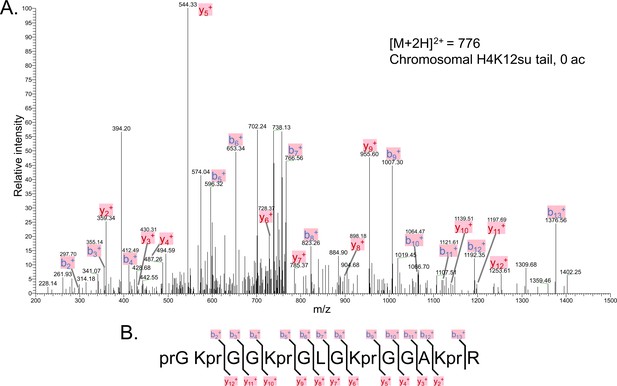
The tandem mass spectrometry (MS-MS) spectrum of unacetylated tryptic peptide H4(4–17) after in vitro acetylation of chromatinized plasmids containing H4K12su with p300 and activator Gal4-VP16 followed by desumoylation.
(B) Peptide fragment-ion map of the unmodified H4(4–17) peptide, indicating identified ions.
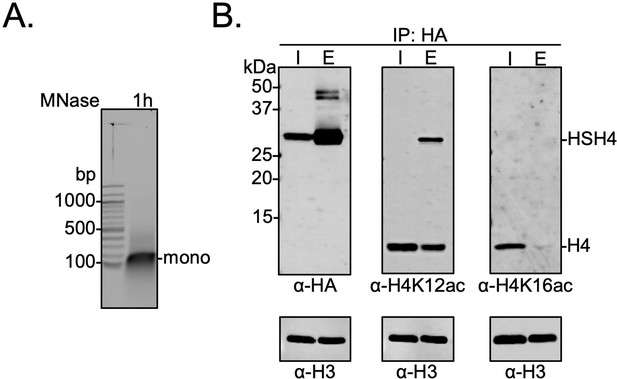
Biochemical crosstalk between H4 sumoylation and acetylation in HEK293T cells.
(A) Extended micrococcal nuclease digestion of chromatin to generate mononucleosomes that were detected by the presence of ~150 bp DNA in 1.5% agarose gels. (B) Immunoprecipitation (IP) from HEK293 cells transfected with HA-Su3(ΔGG)-H4 (HSH4). Input (I) and eluate (E) lanes correspond to undigested bulk chromatin and eluted HA-tagged mononucleosomes containing HSH4. Antibodies targeting H4K12ac and H4K16ac were employed to detect the degree of wild-type (wt) H4 and HSH4 acetylation in HA-tagged mononucleosomes. Total histone H3 in each sample was employed as an equal loading control.
-
Figure 4—source data 1
Unedited intact SDS-PAGE gels and western blot membranes for all gels and western blot images shown in Figure 4.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig4-data1-v2.pdf
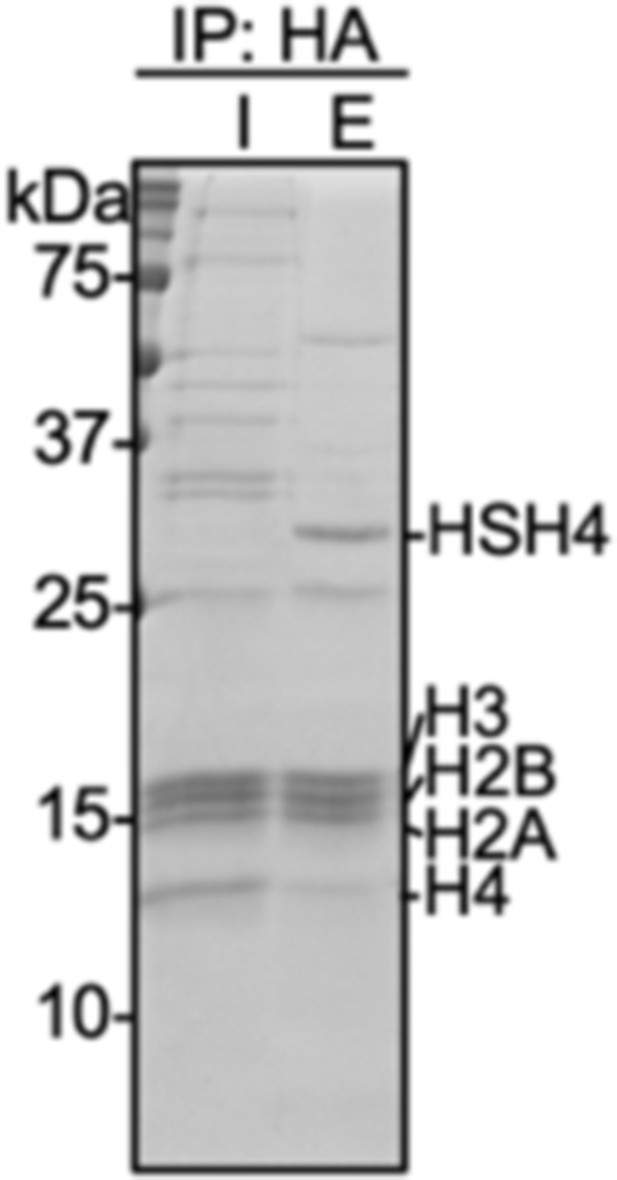
Coomassie-stained SDS-PAGE gel of input (I) and elution (E) samples from immunoprecipitation with anti-HA magnetic beads of micrococcal nuclease digested nuclear extracts prepared from HEK293T cells transfected with HA-Su3(ΔGG)-H4.
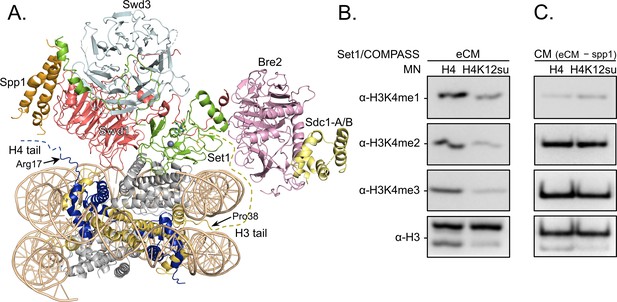
H3K4 methylation by the extended catalytic module (eCM) of the complex of proteins associated with Set1 (COMPASS) methyltransferase complex is inhibited by H4 sumoylation.
(A) Structure of the COMPASS eCM bound to a mononucleosome (PDB code 6UGM). The disordered H3 and H4 tails are shown in gold and blue, respectively, with the last observable amino acid indicated. Dotted lines indicate missing N-terminal amino acids. Spp1 (orange) suppressor of PRP protein 1. Swd3 (light blue), Set1 complex WD40 repeat protein 3. Swd1 (red), Set1 complex WD40 repeat protein 1. Set1 (green), SET domain protein 1. Bre2 (pink), brefeldin-A sensitivity protein 2. Sdc1-A/B (yellow), suppressor of CDC25 protein 1. (B) Western blots of the products from methylation assays with mononucleosome substrates containing wild-type (wt) H4 or H4K12su and the COMPASS eCM complex. Mono-, di-, and trimethylated states of H3K4 were detected by the indicated modification-specific antibodies. (C) Western blots of the products from methylation assays with mononucleosome substrates containing wt H4 or H4K12su and the COMPASS catalytic module. Mono-, di-, and trimethylated states of H3K4 were detected by the indicated modification-specific antibodies.
-
Figure 5—source data 1
Unedited western blot membranes for all western blot images shown in Figure 5.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig5-data1-v2.pdf
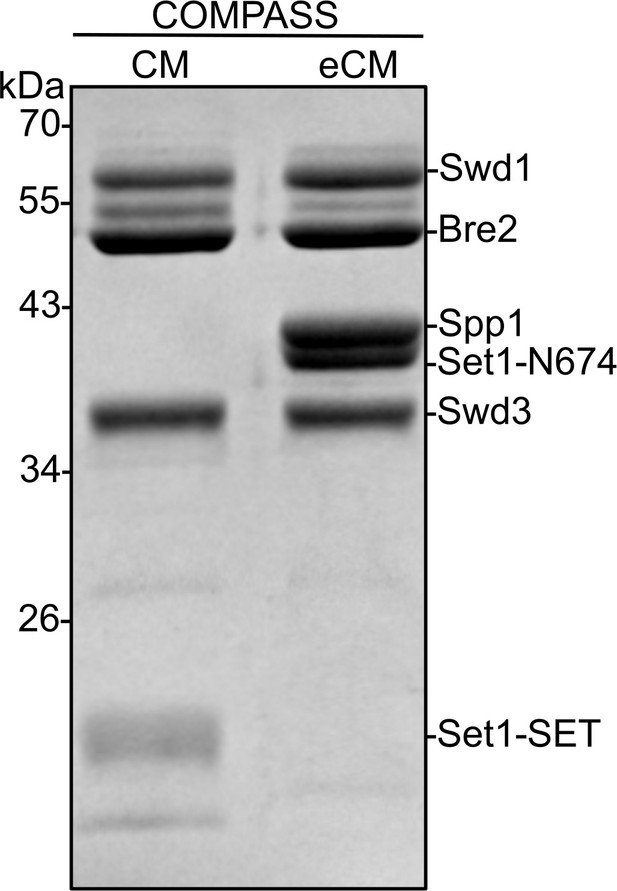
Coomassie-stained SDS-PAGE gel of recombinant Kluyveromyces lactis complex of proteins associated with Set1 (COMPASS) catalytic module (CM) and COMPASS extended catalytic module (eCM) sub-complexes used in H3K4 methylation assays.
The Sdc1 subunit (10 kDa) is not observed on this gel due to its size. Set1-SET is the catalytic domain. Set1-N674 includes the nSET domain, beginning at residue 674.
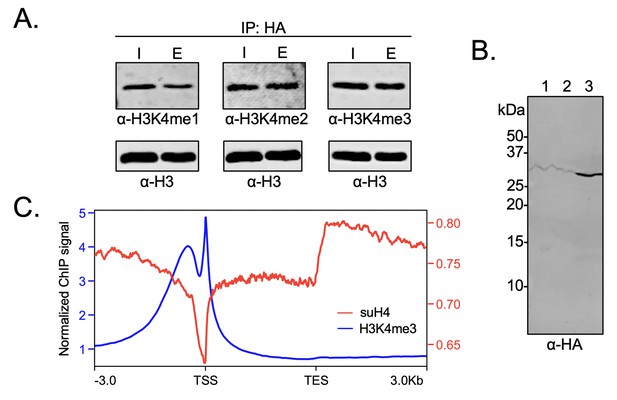
Biochemical crosstalk between H4 sumoylation and H3 methylation in HEK293 cells.
(A) Immunoprecipitation (IP) from HEK293 cells transfected with HA-Su3(ΔGG)-H4 (HSH4). Input (I) and eluate (E) lanes correspond to undigested bulk chromatin and eluted HA-tagged mononucleosomes containing HSH4. Antibodies targeting H3K4me1/2/3 were employed to detect the degree of H3K4 methylation in HA-tagged mononucleosomes. Total histone H3 in each sample was employed as an equal loading control. (B) FLAG-HA-Su3(ΔGG)-H4(Δ1–11) (suH4) protein expression in HEK293 cells and its localization to chromatin within 4 hr. Lanes: 1, Cytoplasmic fraction. 2, Nucleoplasmic fraction. 3, Chromatin fraction. (C) RPMK normalized chromatin immunoprecipitation (ChIP)-seq signals for SUMO-H4 (red, right y-axis) and H3K4me3 (blue, left y-axis) are plotted across length normalized gene bodies for 19,531 UniProt annotated protein coding genes plus 3 kb upstream of the transcription start site (TSS) and 3 kb downstream of the transcription end site (TES).
-
Figure 6—source data 1
Unedited intact gels and western blot membranes for all gels and western blot images shown in Figure 6.
- https://cdn.elifesciences.org/articles/67952/elife-67952-fig6-data1-v2.pdf
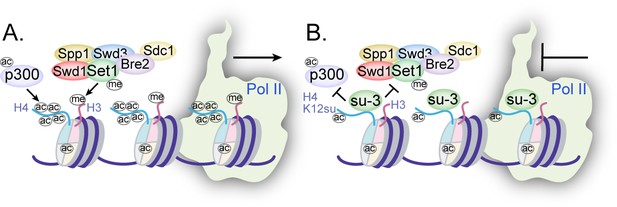
Mechanisms of chromatin regulation by H4K12su.
(A) Transcription from chromatinized templates containing wild-type (wt) H4 is accompanied by acetylation of all four histones by p300, and with the methylation of the H3 tail by the complex of proteins associated with Set1 (COMPASS)/SET1 complexes. (B) The inhibition of PolII-mediated transcription from chromatinized templates containing H4K12su is accompanied by reduced p300-mediated H4 tail acetylation. H4K12su also inhibits H3 tail methylation by the extended catalytic module of the COMPASS complex. For clarity, only one of two histone tails, each, is shown for H3 and H4.
Tables
H4(4–17) tail peptides acetylated by p300 in chromatinized plasmid templates with activator Gal4-VP16*,†.
| H4(4–17) peptide | [M] | [M + 2 H]2+ | PSMH4 | PSMH4K12su |
|---|---|---|---|---|
| prGKprGGKprGLGKprGGAKprR | 1549.89 | 775.95 | n.d. | 1 |
| prGKprGGKacGLGKprGGAKprR | 1535.88 | 768.95 | n.d. | n.d. |
| prGKacGGKacGLGKprGGAKprR | 1521.86 | 761.94 | 2 | 3 |
| prGKacGGKacGLGKacGGAKprR | 1507.85 | 754.93 | 5 | n.d.‡ |
| prGKacGGKacGLGKprGGAKacR | 1507.85 | 754.93 | n.d. | 3§ |
| prGKacGGKacGLGKacGGAKacR | 1493.83 | 747.92 | 7 | n.d.‡ |
-
*
Peptides were chemically propionylated before and after trypsinization to cap unmodified lysine side-chains and newly generated N-termini.
-
†
Tandem mass spectrometry (MS-MS) spectra observed contained major fragments for the shown modification pattern over other potential patterns, however, no singly acetylated peptides were observed for wild-type (wt) H4.
-
‡
Acetylation at K12 is not possible for H4K12su.
-
§
The triply acetylated peptide from H4K12su is blocked from acetylation at K12, but is propionylated after in-gel desumoylation. PSM = peptide spectral match. n.d. = not detected.
Comparisons of relative ion intensities of characteristic fragment ions from an enzymatically di-acetylated and chemically propionylated H4(4–17) peptide, [M + 2 H]2+ = 762 Da, after activator and p300-mediated acetylation of chromatinized plasmids containing wild-type (wt) H4*.
| Species | Ion | % of Total ion intensity | Avg. ratio | |
|---|---|---|---|---|
| K5ac, K8ac | b5+ | 1.316 | 1.654 | 12.4 |
| K5pr, K8pr | 0.072 | 0.253 | ||
| K12ac, K16ac | y9+ | 0.042 | 0.306 | 0.2 |
| K12pr, K16pr | 3.176 | 0.971 | ||
-
*
Only two unique spectra were observed and analyzed for the doubly acetylated and propionylated H4(4–17) tail peptide from wild-type (wt) H4 chromatin.
Comparisons of relative ion intensities of characteristic fragment ions from an enzymatically tri-acetylated and chemically propionylated H4(4–17) peptide, [M + 2 H]2+ = 755 Da, after activator and p300-mediated acetylation of chromatinized plasmids containing wild-type (wt) H4*.
| Species | Ion | % of Total ion intensity | Avg. ratio | ||
|---|---|---|---|---|---|
| K5ac, K8ac, K12ac | b9+ | 3.032 | 4.938 | 3.981 | 24.1 ± 13.3 |
| K5ac, K8ac, K12pr | 0.339 | 0.096 | 0.147 | ||
| K5ac, K8ac, K12ac | b10+ | 0.875 | 0.862 | 1.109 | 10.3 ± 5.1 |
| K5ac, K8ac, K12pr | 0.112 | 0.152 | 0.064 | ||
| K12pr, K16ac | y8+ | 0.271 | n.d. | 0.012 | 0.2 |
| K12ac, K16pr | 0.885 | 1.005 | 1.263 | ||
| K12pr, K16ac | y9+ | n.d. | 0.123 | 0.054 | 0.02 |
| K12ac, K16pr | 2.587 | 4.442 | 6.296 | ||
-
*
Three unique spectra corresponding to the tri-acetylated and propionylated H4(4–17) tail peptide from wild-type (wt) H4 chromatin were analyzed. Error reported is standard deviation of the mean. n.d. = not detected.
Comparisons of relative ion intensities of characteristic fragment ions from an enzymatically di-acetylated, desumoylated, and chemically propionylated H4(4–17) peptide, [M + 2 H]2+ = 762 Da, after activator and p300-mediated acetylation of chromatinized plasmids containing H4K12su*.
| Species | Ion | % of Total ion intensity | Avg. ratio | ||
|---|---|---|---|---|---|
| K5ac, K8ac | b5+ | 2.096 | 2.308 | 1.770 | 71.6 |
| K5pr, K8pr | n.d. | 0.071 | 0.016 | ||
| K5ac, K8ac | b6+ | 2.603 | 2.835 | 2.900 | 33.1 |
| K5pr, K8pr | 0.053 | n.d. | 0.170 | ||
| K12pr, K16ac | y6+ | 0.038 | 0.080 | n.d. | 0.04 |
| K12pr, K16pr | 1.227 | 1.378 | 1.375 | ||
| K12pr, K16ac | y8+ | 0.027 | 0.013 | 0.138 | 0.1 ± 0.09 |
| K12pr, K16pr | 0.815 | 0.732 | 0.677 | ||
-
*
Three unique spectra corresponding to the di-acetylated and propionylated H4(4–17) tail peptide from H4K12su chromatin were analyzed. Error reported is standard deviation of the mean. n.d. = not detected.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Acetyl-CoA | Roche | 10101893001 | Co-factor for p300 enzyme |
| Commercial assay or kit | Calcium Phosphate Transfection Kit | Thermo | K278001 | Mammalian cell transfection |
| Commercial assay or kit | Lipofectamine 3000 | Invitrogen | L3000001 | Mammalian cell transfection |
| Other | Anti-DYKDDDDK G1 (mouse monoclonal) Affinity Resin | GenScript | L00432 | Antibody-conjugated resin for IP |
| Other | HisPur Ni-NTA resin | Thermo | 88221 | Affinity purification resin |
| Other | Anti-HA (mouse monoclonal) magnetic beads | Pierce | 88836; RRID:AB_2861399 | Antibody-conjugated resin for IP |
| Peptide, recombinant protein | Micrococcal nuclease solution | Thermo | 88216 | Digestion of dsDNA |
| Other | cOmplete, Mini, EDTA-free protease inhibitor cocktail | Roche | 11836170001 | Protease inhibitor cocktail |
| Chemical compound, drug | [3H]-acetyl-CoA | American Radiolabeled Chemicals | ART0213B | Radioactive co-factor for p300 |
| Peptide, recombinant protein | Pierce Trypsin Protease, MS Grade | Thermo | 90057 | Protein cleavage C-terminal to Arg/Lys |
| Chemical compound, drug | Propionic anhydride | Sigma-Aldrich | P51478 | Propionylation of peptide lysines and N-terminus |
| Other | DMEM | Gibco | 11956118 | Cell culture medium |
| Other | DPBS | Gibco | 14190250 | Cell culture PBS buffer |
| Other | Fetal bovine serum | Gibco | 16000044 | Cell culture medium additive |
| Other | Amplify fluorographic reagent | GE Amersham | NAMP100 | Tritium decay signal amplifier |
| Other | Kodak GBX developer and fixer | Carestream Health | 1900943 | Immunoblot imaging reagents |
| Chemical compound, drug | Trifluoroacetic acid | Alfa Aesar | AA31771-36 | Peptide synthesis reagent |
| Chemical compound, drug | Formic acid | Acros Organics | AC147932500 | Ion-pairing agent for HPLC |
| Chemical compound, drug | Acetonitrile (ACN) | Fisher | A996 | Solvent for HPLC |
| Other | C18 Zip tip | Millipore | ZTC18S096 | Peptide purification |
| Chemical compound, drug | Glacial acetic acid | Fisher | A38C-212 | Additive for HPLC solvent |
| Strain, strain background (Escherichia coli) | E. coli BL21(DE3) competent cells | Thermo | FEREC0114 | Chemically competent cells |
| Strain, strain background (Escherichia coli) | E. coli DH5α competent cells | NEB | C2987HVIAL | Chemically competent cells |
| Cell line (Homo sapiens) | HEK 293T | ATCC | CRL-3216; RRID:CVCL_0063 | For transient transfection |
| Cell line (Homo sapiens) | Flp-In T-Rex 293 cell line | Invitrogen | R78007 | Stable cell line generation |
| Recombinant DNA reagent | pST100-20xNCP601a | Gift from Dr Robert K McGinty | – | Plasmid containing 20 repeats of Widom 601 sequence |
| Recombinant DNA reagent | pcDNA3.1-p300-His6 | Addgene | 23252; RRID:Addgene_23252 | Plasmid for full-length p300 |
| Recombinant DNA reagent | pET28a-His6-SENP2(365–590) | Addgene | 16357; RRID:Addgene_16357 | Plasmid for SUMO protease catalytic domain |
| Recombinant DNA reagent | pcDNA3.1-HA-SUMO-3(ΔGG)-H4 | GenScript | – | This study; generated plasmid containing indicated CDS |
| Recombinant DNA reagent | pcDNA5-FLAG-HA-SUMO-3(ΔGG)H4(Δ1–11) | This study | – | This study; plasmid generated containing indicated CDS |
| Sequence-based reagent | p300_Ctrm_FLAG_R | IDT | – | 5’-ATC CTT GTA ATC GTG TAT GTC TAG TGT ACT C-3’ |
| Sequence-based reagent | p300_Ctrm_FLAG_F | IDT | – | 5’-GAT GAC GAT AAA TAG TGA TAC TAA GCT TAA GTT TAA AC-3’ |
| Antibody | Rabbit polyclonal anti-acetyllysine antibody | Millipore | AB3879; RRID:AB_11214410 | WB (1:2000) dilution |
| Antibody | Rabbit polyclonal anti-H4K16ac antibody | Active Motif | 39167; RRID:AB_2636968 | WB (1:2000) dilution |
| Antibody | Rabbit polyclonal anti-H4K12ac antibody | Active Motif | 39066 | WB (1:2000) dilution |
| Antibody | Rabbit monoclonal anti-H3K4me1 | Cell Signaling Technology | 5326; RRID:AB_10695148 | WB (1:2000) dilution |
| Antibody | Rabbit polyclonal anti-H3K4me2 | Abcam | ab7766; RRID:AB_2560996 | WB (1:2000) dilution |
| Antibody | Rabbit polyclonal anti-H3K4me3 | Abcam | ab8580; RRID:AB_306649 | WB 1:2000 dilution |
| Antibody | Rabbit polyclonal anti-Histone H3 | Abcam | ab1791; RRID:AB_302613 | WB (1:2000) dilution |
| Antibody | Mouse monoclonal anti-Histone H3 | Abcam | ab24834; RRID:AB_470335 | WB (1:2000) dilution |
| Antibody | Rabbit monoclonal anti-HA | Cell Signaling Technology | 3724; RRID:AB_1549585 | WB (1:2000) dilution |
| Antibody | Mouse monoclonal anti-FLAG | Sigma-Aldrich | F1804; RRID:AB_262044 | WB (1:2000) dilution |
| Antibody | Anti-rabbit monoclonal, HRP conjugated | GE Healthcare | NA934; RRID:AB_2722659 | WB (1:40000) dilution |
| Antibody | IRDye 680RD Goat polyclonal anti-Rabbit IgG | Li-COR Biosciences | 926–68071; RRID:AB_10956166 | WB (1:15000) dilution |
| Antibody | IRDye 800CW Goat polyclonal anti-Rabbit IgG | Li-COR Biosciences | 926–32211; RRID:AB_621843 | WB (1:15000) dilution |
| Antibody | IRDye 800CW Goat polyclonal anti-Mouse IgG | Li-COR Biosciences | 926–32210; RRID:AB_621842 | WB (1:15000) dilution |





