Structure of the IL-27 quaternary receptor signaling complex
Abstract
Interleukin 27 (IL-27) is a heterodimeric cytokine that functions to constrain T cell-mediated inflammation and plays an important role in immune homeostasis. Binding of IL-27 to cell surface receptors, IL-27Rα and gp130, results in activation of receptor-associated Janus Kinases and nuclear translocation of Signal Transducer and Activator of Transcription 1 (STAT1) and STAT3 transcription factors. Despite the emerging therapeutic importance of this cytokine axis in cancer and autoimmunity, a molecular blueprint of the IL-27 receptor signaling complex, and its relation to other gp130/IL-12 family cytokines, is currently unclear. We used cryogenic-electron microscopy to determine the quaternary structure of IL-27, composed of p28 and Epstein-Barr Virus-Induced 3 (Ebi3) subunits, bound to receptors, IL-27Rα and gp130. The resulting 3.47 Å resolution structure revealed a three-site assembly mechanism nucleated by the central p28 subunit of the cytokine. The overall topology and molecular details of this binding are reminiscent of IL-6 but distinct from related heterodimeric cytokines IL-12 and IL-23. These results indicate distinct receptor assembly mechanisms used by heterodimeric cytokines with important consequences for targeted agonism and antagonism of IL-27 signaling.
Editor's evaluation
Cytokines are small protein signaling molecules with a diverse range of activities in inflammation and immune system function. This manuscript reports the cryo-EM structure of the cytokine interleukin-27 (IL-27) bound to soluble domains of two receptor subunits, IL-27Rα and gp130. IL-27 is a composite cytokine consisting of the protein p28 bound to EBI3, which resembles soluble cytokine receptors such as the receptors for IL-6, IL-11 or CNTF. IL-27 signals predominantly via STAT1 and plays an important role in immune homeostasis. The data provide a detailed molecular view of how IL-27 binds to its receptor.
https://doi.org/10.7554/eLife.78463.sa0Introduction
Cytokines are secreted factors that mediate cell-cell communication in the immune system (Spangler et al., 2015). Binding of cytokines to cell surface receptors, in most cases, leads to activation of receptor-associated Janus Kinase (JAK) proteins, which phosphorylate each other as well as downstream Signal Transducer and Activator of Transcription (STAT) proteins, triggering nuclear translocation and regulation of gene expression. Cytokines can be classified by their use of shared receptors which transduce signals for multiple cytokines within a family. Interleukin-6 signal transducer (IL-6ST), also known as glycoprotein 130 (gp130), is a shared receptor that mediates signaling of multiple cytokines including IL-6, IL-11, and IL-27. Unlike other gp130 family cytokines, IL-27 is a heterodimeric cytokine consisting of a four-helix bundle, IL-27p28 (p28), with similarity to IL-6, complexed with a secreted binding protein, Epstein-Barr Virus-Induced 3 (Ebi3), with homology to type I cytokine receptors (Pflanz et al., 2002). IL-27 signals through a receptor complex consisting of IL-27Rα (TCCR/WSX-1) and gp130 expressed on T cells, NK cells, monocytes, dendritic cells, B cells as well as subsets of endothelial and epithelial cells (Pflanz et al., 2002; Pflanz et al., 2004; Heng et al., 2008; Figure 1A). Binding of IL-27 to its receptor subunits triggers the activation of receptor-associated JAK1 and JAK2 leading to phosphorylation of STAT1 and STAT3 (Lucas et al., 2003; Owaki et al., 2008; Wilmes et al., 2021). Functionally, IL-27 signaling serves to constrain inflammation by antagonizing differentiation of pro-inflammatory Th17 cells (Stumhofer et al., 2006), stimulating T-bet expression in regulatory T cells (Tregs) (Hall et al., 2012), and inducing production of the anti-inflammatory cytokine IL-10 (Stumhofer et al., 2007). The important role of IL-27 in constraining inflammation has prompted the development of antagonist antibodies, one of which is currently being developed as a cancer immunotherapy (Patnaik et al., 2021).
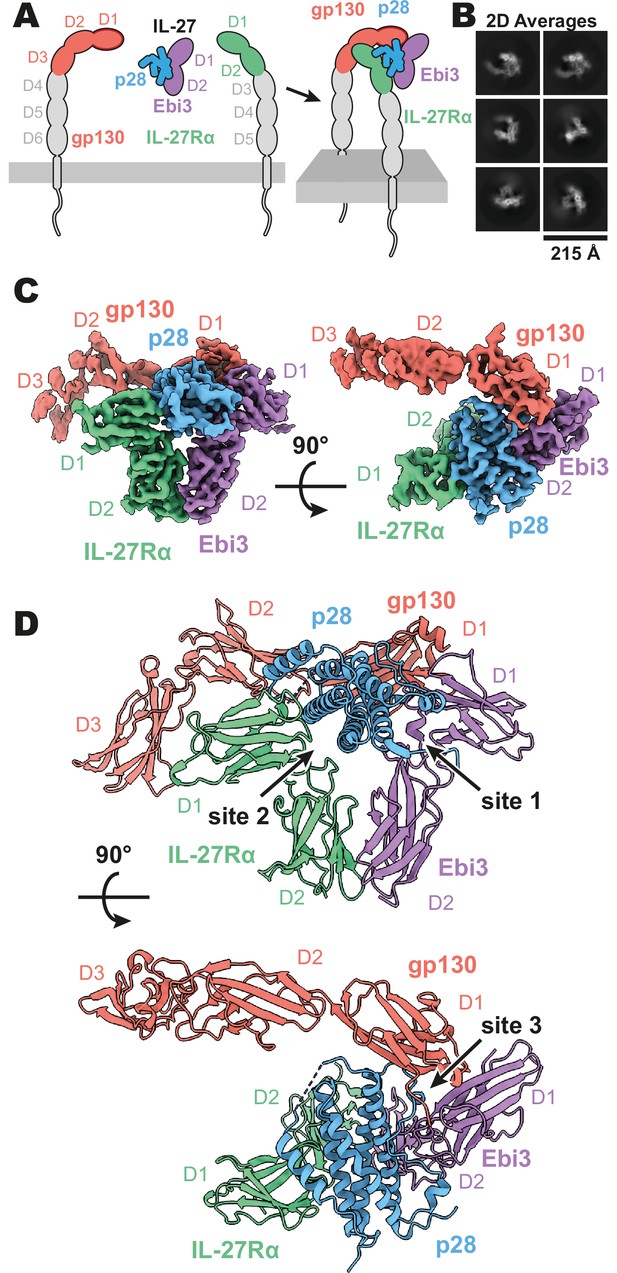
Composition and cryogenic-electron microscopy (cryoEM) structure of human interleukin 27 (IL-27) quaternary complex.
(A) Cartoon representation of the components of the IL-27 quaternary signaling complex. gp130 (red), p28 (blue), Epstein-Barr Virus-Induced 3 (Ebi3; purple), IL-27Rα (green), with domains excluded from the imaged constructs in gray. The D1 Ig domain of gp130 is represented by red, with an additional dark red outline to distinguish it from FNIII domains in gp130, Ebi3, and IL-27Rα. (B) Reference-free 2D averages from cryoEM of the IL-27 quaternary complex. (C) Refined and sharpened cryoEM density maps of IL-27 quaternary complex, colored as in (A). (D) Ribbon representation of the atomistic modeling of IL-27 quaternary complex, colored as in (A).
Although IL-27 is a member of the gp130 family, its heterodimeric composition is similar to IL-12 and IL-23, which are cytokines that share a p40 subunit analogous to Ebi3. In this system, the p40 subunit of IL-12 and IL-23 directly engages the shared receptor IL-12Rβ1 in a manner distinct from IL-6 and related cytokines (Glassman et al., 2021a). Given the diverse receptor assembly mechanisms used by cytokine receptors, we sought to determine the structure of IL-27 in complex with its signaling receptors, gp130 and IL-27Rα, by cryoEM.
Here we report the 3.47 Å resolution structure of the complete IL-27 receptor complex. In this structure, the central IL-27 cytokine engages IL-27Rα through a composite interface consisting of p28 and the D2 domain of Ebi3. A conserved tryptophan residue at the tip of p28 then engages the D1 domain of gp130 to facilitate quaternary complex assembly. This receptor assembly mechanism bears striking resemblance to that of IL-6 but is distinct from IL-12 and IL-23, indicating divergent receptor binding modes for heterodimeric cytokines.
Results
CryoEM structure of the IL-27 quaternary complex
Interleukin 27 (p28/Ebi3) signals through a receptor complex composed of IL-27Rα and the shared receptor gp130 (Figure 1A). The extracellular domain of IL-27Rα consists of five fibronectin type III (FNIII) domains (D1–5) of which the N-terminal D1–D2 constitute a cytokine-binding homology region (CHR). gp130 has a similar domain architecture but with the addition of an N-terminal immunoglobin (Ig) domain (D1). Initial attempts at reconstituting a soluble receptor complex through mixing of IL-27, IL-27Rɑ D1–D2, and gp130 D1–D3 resulted in dissociation on size-exclusion chromatography due to the low affinity of gp130. To stabilize the complex, we expressed p28 fused to gp130 through a long (20 a.a.) flexible linker which enabled us to purify the complete IL-27 receptor complex by size exclusion chromatography. Importantly, the long linker did not constrain the binding mode, with ~110 Å of unresolved residues spanning a distance of 38 Å between termini (Figure 1—figure supplement 2), but rather raised the effective concentration of gp130. The complex was vitrified and subject to single-particle cryoEM analysis. The quaternary complex was determined to a resolution of 3.47 Å (Figure 1—figure supplement 1 and Table 1). All domains of IL-27 and IL-27Rα, as well as D1 of gp130 had well resolved cryoEM density (Figure 1B-C, Figure 1—figure supplement 1E), with local resolution estimates near and exceeding 3 Å in this region (Figure 1—figure supplement 1B). The local resolution for the D2 density of gp130 is lower, yet able to be built with confidence, while D3 density was interpretable for domain placement (Figure 1C, Figure 1—figure supplement 1B).
CryoEM data collection, refinement, and validation statistics.
| IL-27 Complex (PDB 7U7N/EMD-26382) | |
|---|---|
| Data collection and processing | |
| Magnification | 105,000 |
| Voltage (keV) | 300 |
| Electron exposure (e-/Å2) | 60 |
| Defocus range (µm) | –0.8 to –2.0 |
| Pixel size (Å) | 0.839 |
| Symmetry imposed | C1 |
| Initial particle images | 6,387,370 |
| Final particle images | 548,147 |
| Map resolution FSC threshold (Å) | 0.143 |
| Map resolution (Å) | 3.47 |
| Refinement | |
| Initial model used (PDB) | AlphaFold |
| Model resolution FSC threshold (Å) | 0.143 |
| Model resolution (Å) | 1.9 |
| Map sharpening B-factor (Å2) | 189.6 |
| Model Composition | |
| Non-hydrogen atoms | 7,284 |
| Protein residues | 884 |
| Ligands | 16 |
| B-factors (Å2) | |
| Protein | 102.97 |
| Ligand | 114.62 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.610 |
| Validation | |
| MolProbity score | 1.60 |
| Clashscore | 8.81 |
| EMringer score | 2.33 |
| Rotamer outliers (%) | 0.89 |
| Ramachandran plot | |
| Favored (%) | 97.37 |
| Allowed (%) | 2.63 |
| Outliers (%) | 0.00 |
The IL-27 quaternary complex exhibits a molecular architecture containing ‘sites 1–3’, as seen in other gp130 family cytokines (Boulanger et al., 2003) and as hypothesized previously (Skiniotis et al., 2008; Figure 1D). In this structure, IL-27 bridges IL-27Rα and gp130 to initiate downstream signaling through JAK1 and JAK2 (Ferrao et al., 2016; Pradhan et al., 2010). Within IL-27, the four-helical bundle of p28 packs against the hinge between Ebi3 D1 and D2 in a site 1 interaction. Opposite Ebi3, IL-27Rα D1-D2 engages the helical face of p28 in an interaction stabilized by stem contact between the D2 domains of IL-27Rα and Ebi3 to form a site 2 interaction. On the posterior face of IL-27, the D1 domain of gp130 binds to a conserved tryptophan at the tip of p28 to make a classical site 3 interaction (Boulanger et al., 2003).
Site 1–3 interface architecture of the IL-27 quaternary complex
A site 1 interface between p28 and Ebi3 is used to form the heterodimeric cytokine IL-27. In this interaction, the alpha helices A and D, the AB loop, and the C-terminus of p28 pack tightly against the hinge region between D1 and D2 of Ebi3 (Figure 2A, B). This interaction is mediated by a variety of hydrogen bonds between p28 and the hinge region of Ebi3, particularly along alpha helices A and D of p28. At the center of this interface is p28 R219 which is coordinated by D207 and T209 in Ebi3. Extending from this region, a hydrophobic patch of residues on the C-terminus and AB loop of p28 (F94, W97, L223, W232) packs against residues in the Ebi3 hinge (F97, F157, I160). The extensive and hydrophobic nature of this interface is consistent with the observation that co-expression of p28 and Ebi3 is required for efficient cytokine secretion (Pflanz et al., 2002).
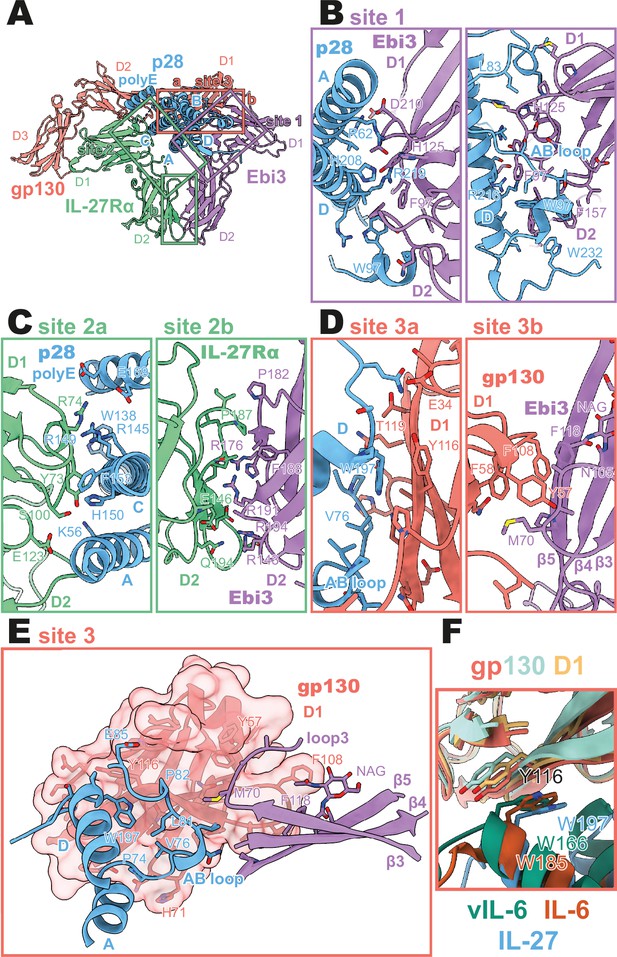
Binding interfaces of the human interleukin 27 (IL-27) quaternary complex.
(A) Ribbon representation of the IL-27 quaternary signaling complex, containing gp130 (red), p28 (blue), Epstein-Barr Virus-Induced 3 (Ebi3; purple), and IL-27Rα (green). Regions containing sites 1, 2, and 3 are boxed in purple, green, and red, respectively. (B) Two views of the site 1 interface, colored as in (A). (C) The site 2 interface, composed of a site 2a, p28 to IL-27Rα, interaction, and a site 2b, Ebi3 to IL-27Rα, interaction. All proteins are colored as in (A). (D,E) The site 3 interface, composed of a site 3a, p28 to gp130, interaction, and a site 3b, Ebi3 to IL-27Rα, interaction. All proteins are colored as in (A). (F) Structural overlay of site 3 interacting domains from IL-27 complex, IL-6 complex (PDB 1P9M), and viral IL-6 (vIL-6) complex (PDB 1L1R). IL-27 quaternary complex colored as in (A), IL-6 ternary complex in orange (IL-6) and yellow (gp130), and vIL-6 binary complex in green (vIL-6) and teal (gp130).
Opposing the Ebi3-binding site, p28 engages the hinge region of IL-27Rα (Figure 2C, site 2 a). This interaction is stabilized by stem-stem contacts between the D2 domains of IL-27Rα and Ebi3 (Figure 2C, site 2b). Alpha helices A and C of p28 form the bulk of the site 2a interaction. In contrast to the site 1 interaction which forms the holo-cytokine, the site 2a interaction is more limited both in terms of hydrophobicity and buried surface area (Site 1: 1,339.4 Å2, Site 2a: 810.4 Å2) (Krissinel and Henrick, 2007). One notable feature of this interface is the contribution of a 13 amino acid polyglutamic acid region in p28. This region is not well resolved in the cryoEM density, but may form an alpha helix (polyE helix) and contact R74 and K77 in the IL-27Rɑ D1 domain. Cytokine binding is stabilized by ‘stem-stem’ interactions between D2 FNIII domains of IL-27Rα and Ebi3. This site 2b interaction is characterized by a high degree of shape and charge complementarity with an arginine-rich patch of Ebi3 (R143, R171, R194) packing against the positively charged base of IL-27Rɑ D2 domain (D138, D142, E146). The composite nature of IL-27Rɑ binding explains, in part, the inability of p28 to mediate IL-27 signaling in the absence of Ebi3 (Stumhofer et al., 2010).
On the posterior face of the cytokine, a classical site 3 interface is formed through the interaction of the D1 Ig domain of gp130 associating with both p28 (site 3a) and Ebi3 (site 3b) (Figure 2D, E). Similar to IL-12 family members, IL-23 and IL-12 (Glassman et al., 2021a), and the gp130 family members IL-6 (Boulanger et al., 2003) and viral IL-6 (Chow et al., 2001), this interaction is anchored by the conserved W197 on p28 which packs tightly against the base of the D1 Ig domain of gp130 (Figure 2F). The site 3 interaction is extended by the AB loop of p28 which contacts both gp130 and Ebi3. We do not observe an interaction between the N-terminus of gp130 and helix D of the four-helix bundle as observed for other site 3 interactions; however, this may be due to the use of a flexible GS linker which connects p28 to gp130. The tip of gp130 D1 packs against the top of Ebi3 D1 in a limited interface centered around Ebi3 F118.
The heterodimeric nature of IL-27 has led some to classify it as an IL-12 family cytokine (Trinchieri et al., 2003). However, comparison of the IL-27 receptor complex with that of IL-23 and IL-12 reveals striking differences (Figure 3A–C). In IL-27, the central p28 subunit engages all receptor components. In contrast, each subunit of IL-23 and IL-12 engages a different receptor in a modular interaction mechanism (Glassman et al., 2021a). The assembly of IL-27 more closely resembles that of IL-6, in which the central four-helix bundle encodes binding sites for all receptor components (Figure 3D). However, in the case of IL-6, this motif is duplicated through a C2 symmetry axis due to a dual role of gp130 at site 2 and site 3. The similarity between IL-27 and IL-6 is observed not only in overall architecture but also in molecular detail, where IL-6, vIL-6, and IL-27 all engage gp130 using a highly convergent interface in which a tryptophan from helix D of the cytokine (W197 in p28, W185 in IL-6, W166 in vIL-6) forms an aromatic anchor that is capped by Y116 of gp130 (Figures 2F and 3D, E). Alignment of gp130 D1–(v)IL-6 pairs to the IL-27 structure reveals a tight correlation in gp130 binding pose with Cα RMSDs of 1.039 Å (IL-6) and 1.015 Å (vIL-6) across 149 and 124 trimmed residue pairs. This striking convergence of gp130 binding modes helps to explain the ability of p28 to antagonize IL-6 signaling (Stumhofer et al., 2010) and contextualizes its place in respect to both gp130 and IL-12 family cytokines.
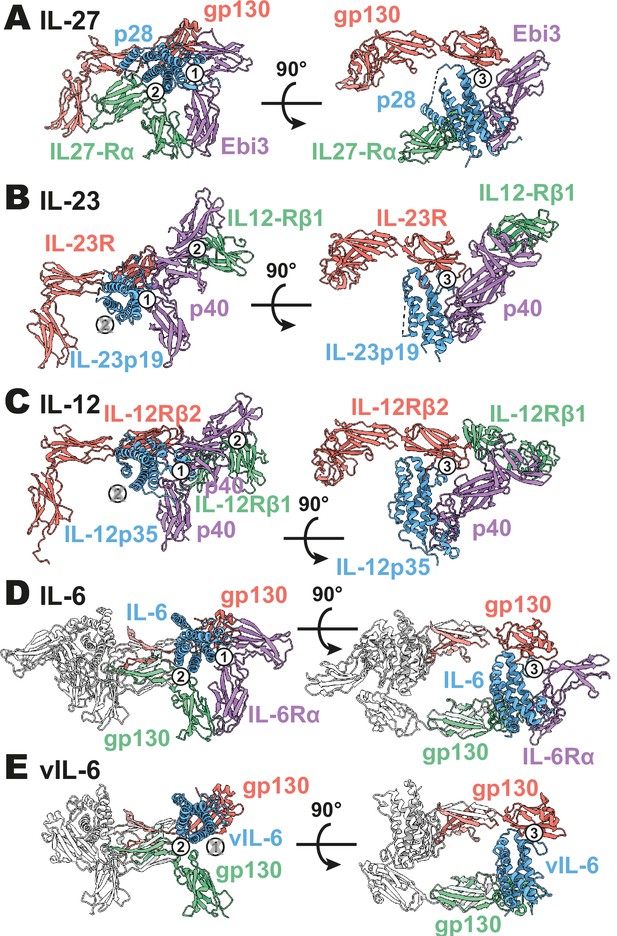
Comparison of IL-27 to IL-12 and IL-6 family complexes.
(A) Ribbon representation of the IL-27 signaling complex, containing gp130 (red), p28 (blue), Epstein-Barr Virus-Induced 3 (Ebi3; purple), and IL-27Rα (green). Sites 1, 2, and 3 are noted with a numbered circle. (B) Ribbon representation of the IL-23 signaling complex (IL-12 family), containing IL-23R (red), IL-23p19 (blue), p40 (purple), and IL-12Rβ1 (green) (PDB 6WDQ). Sites 1 and 3 are noted as in (A), with the unoccupied site 2 marked with a gray ‘X’ and the distinct IL-12/IL-23 site 2 marked with a circled number 2. (C) Ribbon representation of a model of the IL-12 signaling complex (IL-12 family), containing IL-12Rβ2 (red), IL-12p35 (blue), p40 (purple), and IL-12Rβ1 (green) (Baek et al., 2021; Glassman et al., 2021a). Sites 1 and 3 are noted as in (A), with the unoccupied site 2 marked with a gray ‘X’ and the distinct IL-12/IL-23 site 2 marked with a circled number 2. (D) Ribbon representation of the IL-6 signaling complex (IL-6 family), containing gp130 (red and green), IL-6 (blue), and IL-6Rα (purple) (PDB 1P9M). Secondary copies of each protein in the complex are colored in white. Sites 1, 2, and 3 are noted as in (A). (E) Ribbon representation of the viral IL-6 signaling complex (IL-6 family), containing gp130 (red and green) and (blue) (PDB 1I1R). Secondary copies of each protein in the complex are colored in white. Sites 2 and 3 are noted as in (A), with the unoccupied site 1 marked with a gray ‘X’.
Discussion
A key paradigm in cytokine structural biology is the use of shared receptor components that mediate diverse signaling outputs. IL-27 extends this, given that it shares the gp130 subunit with IL-6 but mediates distinct and often countervailing functional effects. Here we find that IL-27 assembles a receptor complex reminiscent of IL-6 but distinct from other heterodimeric IL-12/23 class cytokines. Interestingly, the soluble form of IL-6Rα (sIL-6Rα) generated through alternative splicing or proteolytic cleavage can complex with IL-6 to potentiate signaling in gp130-expressing cells (Kishimoto and Kang, 2022). This IL-6/sIL-6Rα complex thus may be functionally analogous to heterodimeric cytokine IL-27, albeit with gp130 playing the role of both site 2 and 3 receptors in the case of IL-6/sIL-6Rα signalling, allowing for broader range of cells which can be stimulated.
In addition to their roles in IL-27 signaling, p28 and Ebi3 have been implicated in the assembly of alternate cytokine complexes. Ebi3 has been reported to complex with the p35 subunit of IL-12 to form IL-35 (Collison et al., 2007) and the p19 subunit of IL-23 to form IL-39 (Wang et al., 2016), while p28 has been proposed to interact with IL-6Rα (Garbers et al., 2013) and the p40 subunit of IL-12/23 (Wang et al., 2012). These results suggest structural plasticity among heterodimeric cytokines; however, additional work is needed to characterize the physiological relevance and biochemical basis for these cytokines.
As in other types of cytokine signaling, there is context-dependent therapeutic potential in both the inhibition and potentiation of IL-27 signaling. The targeting of the heterodimeric cytokines of the IL-12 family has been well explored, with various clinically approved inhibitors, targeting either IL-23 signaling via p19 (Risankizumab, Guselkumab, Tildrakizumab) (Tait Wojno et al., 2019) or IL-12 and IL-23 signaling via the shared p40 (Ustekinumab) (Luo et al., 2010). In these cases, the inhibition of IL-12 family members antagonizes the pro-inflammatory effects of these cytokines. Due to the role of IL-27 in the regulation T cell-mediated inflammation (Tait Wojno et al., 2019), inhibition of IL-27 is currently being explored for its use in the inhibition of aberrant IL-27 signaling in cancer (Patnaik et al., 2021). The structure of the IL-27 quaternary signaling complex provides ample opportunities for the design of IL-27 inhibitors targeting different steps in receptor assembly to better regulate IL-27 signaling.
On the other end of the spectrum, IL-27 agonism has been explored for its therapeutic use in inflammatory autoimmune dysregulation such as experimental autoimmune encephalomyelitis (Fitzgerald et al., 2013) and colitis (Hanson et al., 2014). Despite the promise of IL-27 as a therapeutic, the bipartite nature of IL-27 limits its usefulness in the clinic. Using the interfacial information provided in the structure of the IL-27 quaternary complex, structure-guided protein engineering techniques can now be used to improve the therapeutic potential of IL-27. One such application may be to generate single agent IL-27 agonists by affinity maturing the interactions between p28 and its receptor subunit, as has been done recently to generate IL-6 which do not require IL-6Rα binding for efficient signaling (Martinez-Fabregas et al., 2019). Conversely, attenuating the affinity of p28 for IL-27Rα might be used to generate cell-type biased partial agonists with selective activity based on differences in IL-27Rα expression across cell-type and activation state (Villarino et al., 2005) as has been done for IL-2 (Glassman et al., 2021b), IL-10 (Saxton et al., 2021), and IL-12 (Glassman et al., 2021a).
Resolution of the complete IL-27 receptor complex reveals how this therapeutically important cytokine engages its receptor subunits. The receptor assembly mechanism bears striking resemblance to that of IL-6 but is distinct from IL-12 and IL-23, indicating divergent receptor binding modes for heterodimeric cytokines. This structural insight further paves the way for the continued development of therapeutics that modulate IL-27 signaling.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Human embryonic kidney cells | GIBCO | Expi293 | |
| Recombinant DNA reagent | pD649-IL-27Rα (plasmid) | This paper | See: Methods - Cloning and protein expression | |
| Recombinant DNA reagent | pD649-Ebi3 (plasmid) | This paper | See: Methods - Cloning and protein expression | |
| Recombinant DNA reagent | pD649-p28-gp130 (plasmid) | This paper | See: Methods - Cloning and protein expression | |
| Software, algorithm | Data collection software | SerialEM | SerialEM | |
| Software, algorithm | Data processing software | Structura Biotechnology Inc. | cryoSPARC | |
| Software, algorithm | Data sharpening software | Sanchez-Garcia et al., 2021 | DeepEMhancer | |
| Software, algorithm | Initial modeling software | Jumper et al., 2021 | AlphaFold | |
| Software, algorithm | Graphics software | Pettersen et al., 2021 | UCSF ChimeraX | |
| Software, algorithm | Modeling and refinement software | Adams et al., 2010 | Phenix | |
| Software, algorithm | Modeling and refinement software | Emsley and Cowtan, 2004 | Coot | |
| Software, algorithm | Model validation software | Barad et al., 2015 | EMRinger | |
| Software, algorithm | Model validation software | Chen et al., 2010 | MolProbity |
Cloning and protein expression
Human IL-27Rα (D1-D2, residues 36–231), Ebi3 (residues 21–228), and p28 (residues 29–243)-(GGGGS)4-gp130 (D1-D3, residues 23–321) were cloned into the expression vector pD649. All components were expressed with signal sequence of influenza hemagglutinin and IL-27Rα and Ebi3 contained C-terminal 6-His tags for affinity purification. Proteins were co-expressed in Expi293F cells (GIBCO) maintained in Expi293 Expression Media (GIBCO) at 37 °C with 5% CO2 and gentle agitation.
Material availability
Request a detailed protocolThe expression plasmids for the IL-27 complex are available from KCG (https://kcgarcia@stanford.edu) by request.
Protein purification
Request a detailed protocolIL-27 quaternary complex was purified by Ni-NTA chromatography followed by size exclusion chromatography with a Superdex 200 column (GE Lifesciences). Fractions containing pure IL-27 complex were pooled and stored at 4°C until vitrification.
Cryo-electron microscopy
Request a detailed protocolAliquots of 3 μL of IL-27 quaternary complex were applied to glow-discharged Quantifoil (1.2/1.3) grids. The grids were blotted for 3 s at 100% humidity with an offset of 3 and plunge frozen into liquid ethane using a Vitrobot Mark IV (Thermo Fisher). Grid screening and preliminary dataset collection occurred at Stanford cEMc. Final grids were imaged on a 300 kV FEI Titan Krios microscope (Thermo Fisher) located at the HHMI Janelia Research Campus and equipped with a K3 camera and energy filter (Gatan). Movies were collected at a magnification of ×105,000, corresponding to a 0.839 Å per physical pixel. The dose was set to a total of 50 electrons per Å2 over an exposure of 50 frames. Automated data collection was carried out using SerialEM with a nominal defocus range set from –0.8 to –2.0 μM. 18,168 movies were collected.
Image processing
Request a detailed protocolAll processing was performed in cryoSPARC (Punjani et al., 2017) unless otherwise noted (Figure 1—figure supplement 1). The 18,168 movies were motion corrected using patch motion correction and micrographs were binned to 0.839 Å per pixel. The contrast transfer functions (CTFs) of the flattened micrographs were determined using patch CTF and 6,387,370 particles were picked using blob picking and subsequently template picking. A subset of 2,008,987 particles were used in reference-free 2D classification. A particle stack containing 109,536 2D cleaned particles was used to generate three ab initio models, one of which resembled a complete complex. This ab initio model was then used against two junk classes in six rounds of iterative heterogenous refinement to reduce the full particle stack to 548,147 particles. These particles were refined using cryoSPARC non-uniform refinement (Punjani et al., 2020) followed by local refinement with a mask excluding gp130 D3 domain to achieve a resolution of 3.47 Å. Resolution was determined at a criterion of 0.143 Fourier shell correlation gold-standard refinement procedure. The final map was sharpened using deepEMhancer (Sanchez-Garcia et al., 2021).
Model building and refinement
Request a detailed protocolAlphaFold models (Jumper et al., 2021) of IL-27Rα, Ebi3, p28, and gp130 were docked into the map using UCSF Chimera X (Pettersen et al., 2021). The resultant model was then refined using Phenix real space refine (Adams et al., 2010) and manual building in Coot (Emsley and Cowtan, 2004). The final model fit the map well (EMRinger [Barad et al., 2015] score 2.33) and produced a favorable MolProbity score of 1.60 (Chen et al., 2010) with side-chain rotamers occupying 97.37% Ramachandran favored and 0.00% outliers (Table 1).
Data availability
CryoEM maps and atomic coordinates for human IL-27 quaternary complex have been deposited in the EMDB (EMD- 26382) and PDB (7U7N) respectively.
-
RCSB Protein Data BankID 7U7N. IL-27 quaternary receptor signaling complex.
-
Electron Microscopy Data BankID EMD-26382. IL-27 quaternary receptor signaling complex.
References
-
PHENIX: a comprehensive Python-based system for macromolecular structure solutionActa Crystallographica. Section D, Biological Crystallography 66:213–221.https://doi.org/10.1107/S0907444909052925
-
Accurate prediction of protein structures and interactions using a three-track neural networkScience (New York, N.Y.) 373:871–876.https://doi.org/10.1126/science.abj8754
-
Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complexScience (New York, N.Y.) 300:2101–2104.https://doi.org/10.1126/science.1083901
-
MolProbity: all-atom structure validation for macromolecular crystallographyActa Crystallographica. Section D, Biological Crystallography 66:12–21.https://doi.org/10.1107/S0907444909042073
-
Structure of an extracellular gp130 cytokine receptor signaling complexScience (New York, N.Y.) 291:2150–2155.https://doi.org/10.1126/science.1058308
-
Coot: model-building tools for molecular graphicsActa Crystallographica. Section D, Biological Crystallography 60:2126–2132.https://doi.org/10.1107/S0907444904019158
-
The Structural Basis for Class II Cytokine Receptor Recognition by JAK1Structure (London, England 24:897–905.https://doi.org/10.1016/j.str.2016.03.023
-
The Immunological Genome Project: networks of gene expression in immune cellsNature Immunology 9:1091–1094.https://doi.org/10.1038/ni1008-1091
-
IL-6 Revisited: From Rheumatoid Arthritis to CAR T Cell Therapy and COVID-19Annual Review of Immunology 40:323–348.https://doi.org/10.1146/annurev-immunol-101220-023458
-
Inference of macromolecular assemblies from crystalline stateJournal of Molecular Biology 372:774–797.https://doi.org/10.1016/j.jmb.2007.05.022
-
Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumabJournal of Molecular Biology 402:797–812.https://doi.org/10.1016/j.jmb.2010.07.046
-
WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27Journal of Immunology (Baltimore, Md 172:2225–2231.https://doi.org/10.4049/jimmunol.172.4.2225
-
Activation of JAK2-V617F by components of heterodimeric cytokine receptorsThe Journal of Biological Chemistry 285:16651–16663.https://doi.org/10.1074/jbc.M109.071191
-
Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10Science (New York, N.Y.) 371:eabc8433.https://doi.org/10.1126/science.abc8433
-
Insights into cytokine-receptor interactions from cytokine engineeringAnnual Review of Immunology 33:139–167.https://doi.org/10.1146/annurev-immunol-032713-120211
-
Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10Nature Immunology 8:1363–1371.https://doi.org/10.1038/ni1537
-
A role for IL-27p28 as an antagonist of gp130-mediated signalingNature Immunology 11:1119–1126.https://doi.org/10.1038/ni.1957
-
Positive and negative regulation of the IL-27 receptor during lymphoid cell activationJournal of Immunology (Baltimore, Md 174:7684–7691.https://doi.org/10.4049/jimmunol.174.12.7684
-
Novel IL27p28/IL12p40 cytokine suppressed experimental autoimmune uveitis by inhibiting autoreactive Th1/Th17 cells and promoting expansion of regulatory T cellsThe Journal of Biological Chemistry 287:36012–36021.https://doi.org/10.1074/jbc.M112.390625
-
A novel IL-23p19/Ebi3 (IL-39) cytokine mediates inflammation in Lupus-like miceEuropean Journal of Immunology 46:1343–1350.https://doi.org/10.1002/eji.201546095
Article and author information
Author details
Funding
Canadian Institutes of Health Research
- Nathanael A Caveney
Howard Hughes Medical Institute
- K Christopher Garcia
National Institute of Allergy and Infectious Diseases (R01-AI51321)
- K Christopher Garcia
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Rui Yan at the HHMI Janelia CryoEM Facility for help in microscope operation and final data collection. We thank Liz Montabana and Stanford cEMc for microscope access for preliminary data collection. NAC is a CIHR postdoctoral fellow. KCG is an investigator with the Howard Hughes Medical Institute. KCG is supported by National Institutes of Health grant R01-AI51321, the Mathers Foundation, and the Ludwig Foundation.
Copyright
© 2022, Caveney, Glassman et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 3,562
- views
-
- 724
- downloads
-
- 28
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Immunology and Inflammation
The incidence of metabolic dysfunction-associated steatotic liver disease (MASLD) has been increasing worldwide. Since gut-derived bacterial lipopolysaccharides (LPS) can travel via the portal vein to the liver and play an important role in producing hepatic pathology, it seemed possible that (1) LPS stimulates hepatic cells to accumulate lipid, and (2) inactivating LPS can be preventive. Acyloxyacyl hydrolase (AOAH), the eukaryotic lipase that inactivates LPS and oxidized phospholipids, is produced in the intestine, liver, and other organs. We fed mice either normal chow or a high-fat diet for 28 weeks and found that Aoah-/- mice accumulated more hepatic lipid than did Aoah+/+ mice. In young mice, before increased hepatic fat accumulation was observed, Aoah-/- mouse livers increased their abundance of sterol regulatory element-binding protein 1, and the expression of its target genes that promote fatty acid synthesis. Aoah-/- mice also increased hepatic expression of Cd36 and Fabp3, which mediate fatty acid uptake, and decreased expression of fatty acid-oxidation-related genes Acot2 and Ppara. Our results provide evidence that increasing AOAH abundance in the gut, bloodstream, and/or liver may be an effective strategy for preventing or treating MASLD.
-
- Immunology and Inflammation
- Microbiology and Infectious Disease
Pseudomonas aeruginosa (PA) is an opportunistic, frequently multidrug-resistant pathogen that can cause severe infections in hospitalized patients. Antibodies against the PA virulence factor, PcrV, protect from death and disease in a variety of animal models. However, clinical trials of PcrV-binding antibody-based products have thus far failed to demonstrate benefit. Prior candidates were derivations of antibodies identified using protein-immunized animal systems and required extensive engineering to optimize binding and/or reduce immunogenicity. Of note, PA infections are common in people with cystic fibrosis (pwCF), who are generally believed to mount normal adaptive immune responses. Here, we utilized a tetramer reagent to detect and isolate PcrV-specific B cells in pwCF and, via single-cell sorting and paired-chain sequencing, identified the B cell receptor (BCR) variable region sequences that confer PcrV-specificity. We derived multiple high affinity anti-PcrV monoclonal antibodies (mAbs) from PcrV-specific B cells across three donors, including mAbs that exhibit potent anti-PA activity in a murine pneumonia model. This robust strategy for mAb discovery expands what is known about PA-specific B cells in pwCF and yields novel mAbs with potential for future clinical use.






