Novel fast pathogen diagnosis method for severe pneumonia patients in the intensive care unit: randomized clinical trial
Figures
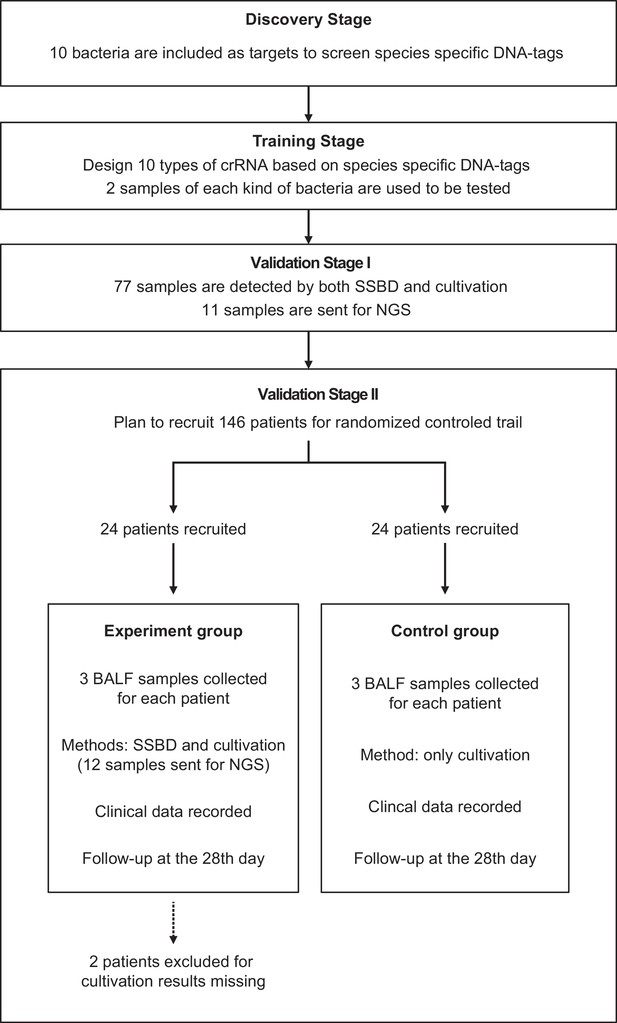
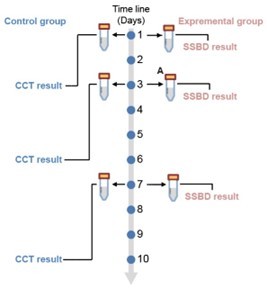
Study design.
This study contained four stages: discovery stage, training stage, validation stage I and validation stage II. All patients were from the Department of Critical Care Medicine, Nanjing Drum Tower Hospital. Patients were randomly divided into two groups for the clinical trial.
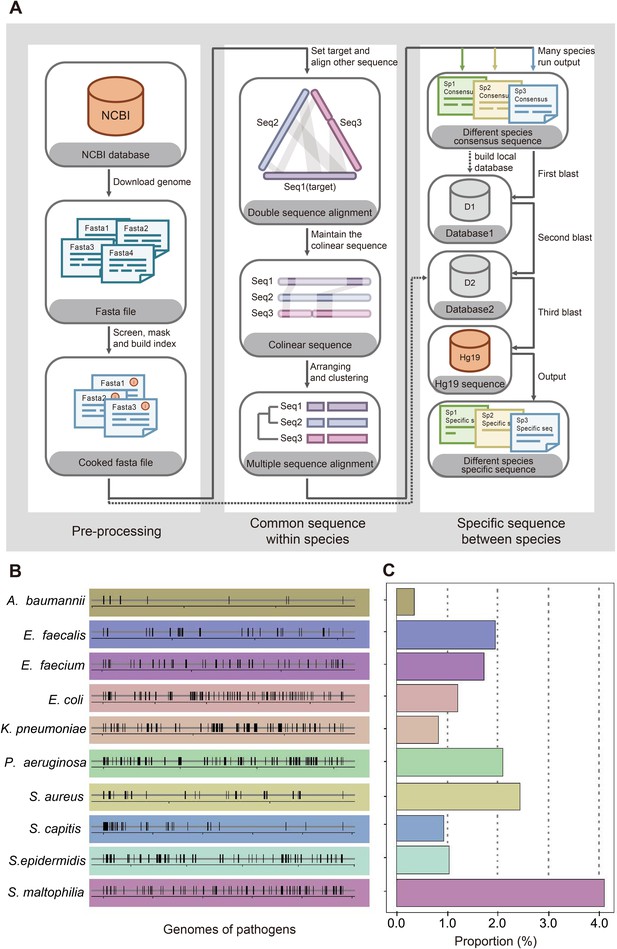
Screening workflow and statistics of species-specific DNA tags.
(A) Schematic diagram of screening species-specific DNA tags. (B) Genomic distribution of species-specific DNA tags in 10 bacteria. (C) Genomic proportion of species-specific DNA tags in 10 bacteria.
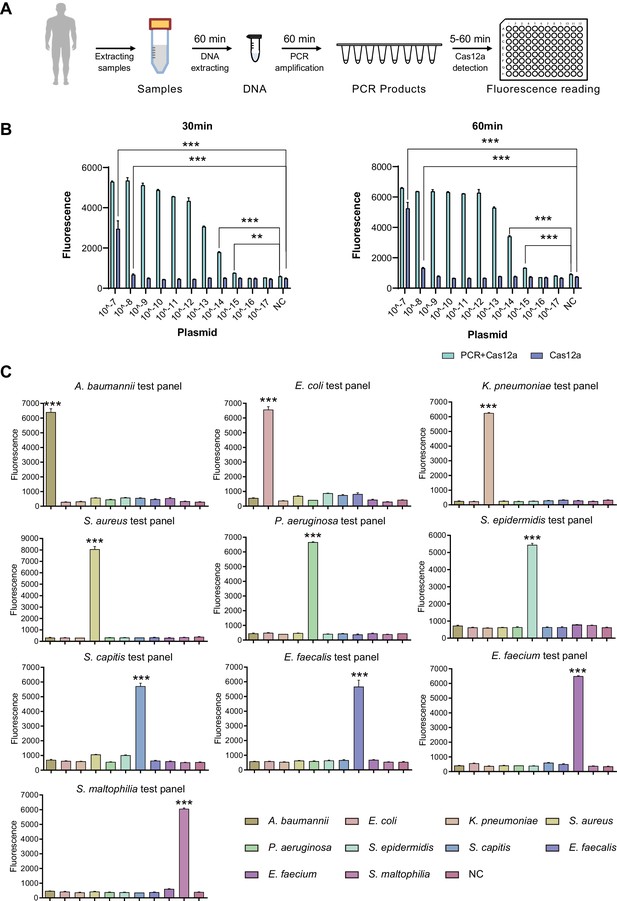
SSBD development and effectiveness validation.
(A) SSBD workflow for clinical validation stages. (B) Cas12a and Cas12a-after-PCR detection of different concentrations and reaction times including 30 min (left) and 60 min (right). Blue bars indicated the Cas12a-after-PCR test. Brown bars indicated Cas12a test only. The concentration gradient of pGL3 plasmid from 10–17 M-10–7M was established as the test group. NC stood for the fluorescence values of PCR products of using DEPC-H2O as input. Each group had three repeats. Error bars indicated mean ± SEM of fluorescence value. ** indicated p-value <0.01 and *** indicated p-value <0.001 of unpaired t-test. (C) SSBD results of 10 pathogenic bacteria. Every test panel for each of 10 bacteria was used to detect genome DNA samples of 10 bacteria by SSBD. NC stood for the fluorescence values of PCR products of using DEPC-H2O as input. Each group had three repeats. Error bars indicated mean ± SEM of fluorescence value. *** indicated p-value <0.001 of unpaired t-test.
-
Figure 3—source data 1
The reaction condition test of Cas12a detection.
- https://cdn.elifesciences.org/articles/79014/elife-79014-fig3-data1-v2.xlsx
-
Figure 3—source data 2
The cross-validation of 10 selected bacteria using SSBD.
- https://cdn.elifesciences.org/articles/79014/elife-79014-fig3-data2-v2.xlsx
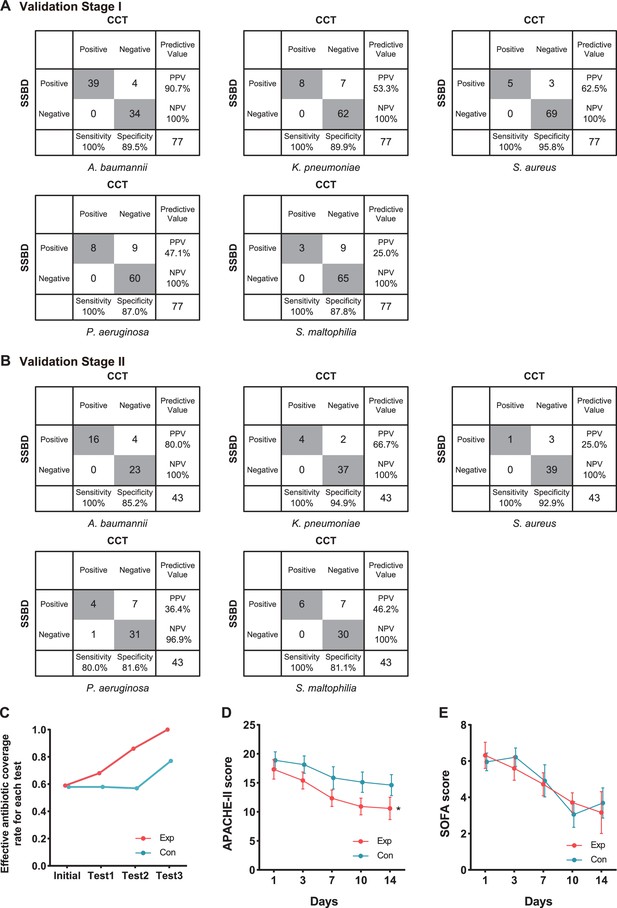
Statistical analysis of test results and clinical outcomes in the two validation stages.
(A) Cross-tables for 5 of 10 bacteria by both SSBD and CCT in the validation stage I. (B) Cross-tables for 5 of 10 bacteria by both SSBD and CCT in the validation stage II. (C) Antibiotics coverage rate of each test in the two groups. Exp meant the experimental group, and Con meant the control group. Test 1: Day 1. Test 2: Day 3–5. Test 3: Day 7+. Raw antibiotics coverage results of each patient were available in Appendix 1—figure 4B. Detailed judging guidelines were shown in Appendix 1. (D and E) Line charts for APACHE II and SOFA scores, respectively. Error bars indicated mean ± SEM of scores of all the recorded patients. * indicated a significant difference between the two groups using two-way ANOVA.
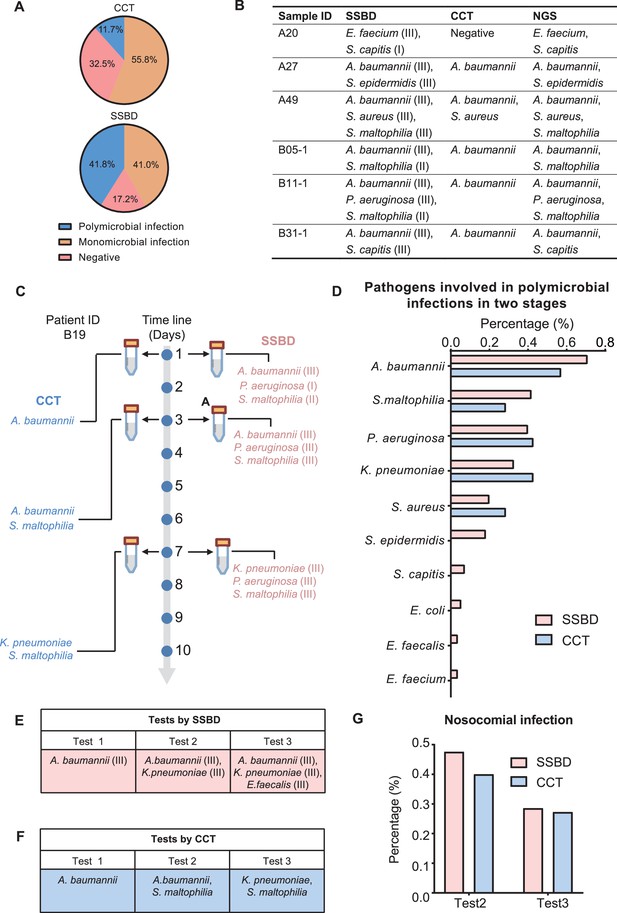
Statistical analysis of polymicrobial infection and nosocomial infection in the two validation stages.
(A) Statistics of pathogenic infection status of BALF samples in the two validation stages. (B) Verification from NGS results for 6 samples identified as polymicrobial infection by SSBD but not CCT or missed pathogens by CCT. (C) Case study of polymicrobial infection detected by SSBD and CCT. (D) Statistics of pathogens involved in polymicrobial infections in the two stages. (E) Case study of nosocomial infection identified by SSBD. (F) Case study of nosocomial infection identified by CCT. (G) Percentage of nosocomial infection identified by SSBD and CCT.
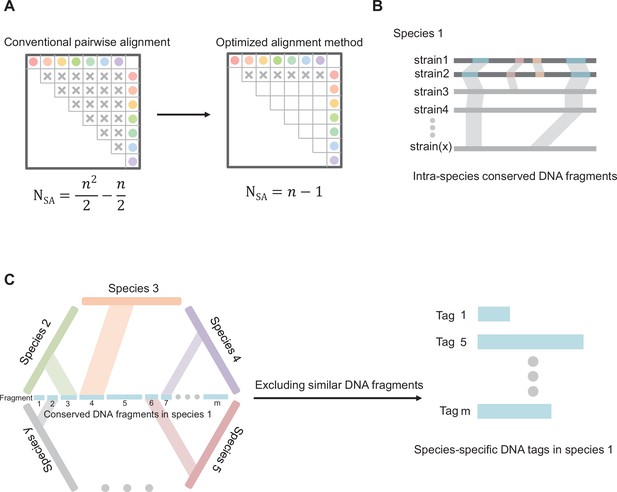
Diagram of core principles for screening species-specific DNA-tags.
(A) Optimizing the algorithm of sequence alignment. Abbreviations: SA, sequence alignment; N, number of double sequence alignment; n, number of sequences. (B) Schematic map of screening intra-species conserved DNA fragments. (C) Schematic map of screening species-specific DNA tags.
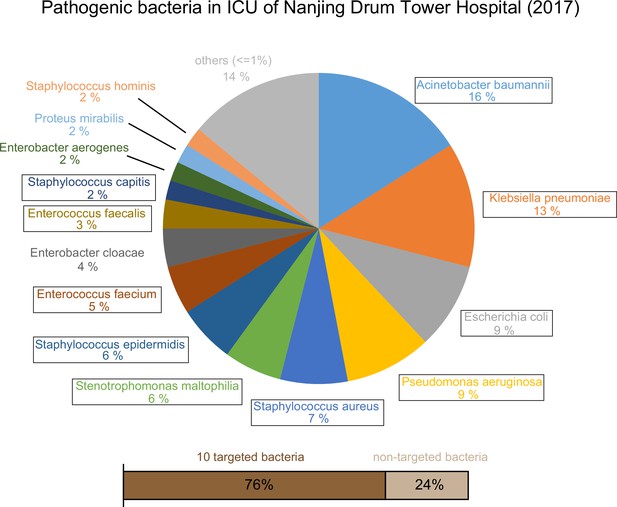
Epidemic data of pathogens in the Nanjing Drum Tower Hospital ICU in 2017.
10 targeted bacteria were indicated with the box.
-
Appendix 1—figure 2—source data 1
Epidemic data of pathogens in the ICU of Nanjing Drum Tower Hospital in 2017.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig2-data1-v2.xlsx
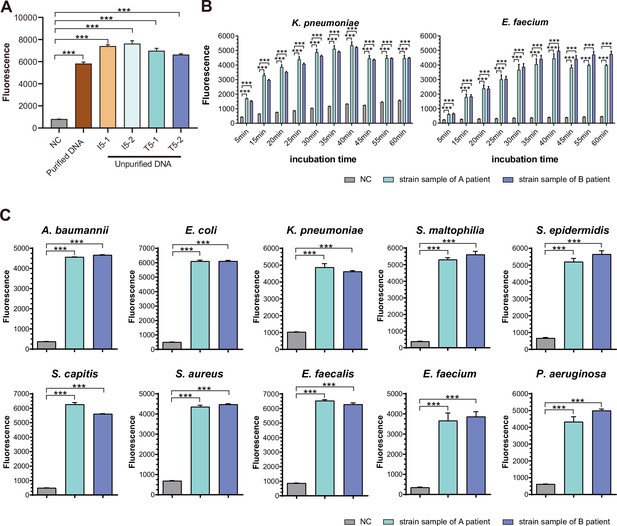
SSBD development and effectiveness validation.
(A) SSBD results of purified and unpurified DNA. NC, namely the fluorescence values of PCR products of using DEPC-H2O as input. Each group had three repeats. Error bars indicated mean ± SEM of fluorescence value. *** indicated p-value <0.001 of unpaired t-test. (B) SSBD results of reaction time gradient with Cas12a. Fluorescence values of K. pneumoniae and E. faecium by Cas12a through different incubation times after PCR. Gray represented NC, namely the fluorescence values of PCR products of using DEPC-H2O as input. Green and blue represented the fluorescence values of bacteria strains from different patients. Each group had three repeats. Error bars indicated mean ± SEM of fluorescence value. ** indicated p-value <0.01 and *** indicated p-value <0.001 of unpaired t-test. (C) SSBD results of 10 pathogenic bacteria with Cas12a. Gray represented NC, namely the fluorescence values of PCR products of using DEPC-H2O as input. Green and blue represented the fluorescence values of bacteria strains from different patients. Each group had three repeats. Error bars indicated mean ± SEM of fluorescence value. *** indicated p-value <0.001 of unpaired t-test.
-
Appendix 1—figure 3—source data 1
The test of SSBD with or without DNA purification.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig3-data1-v2.xlsx
-
Appendix 1—figure 3—source data 2
The test of SSBD with different incubation times.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig3-data2-v2.xlsx
-
Appendix 1—figure 3—source data 3
The validation of SSBD with clinical strains.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig3-data3-v2.xlsx
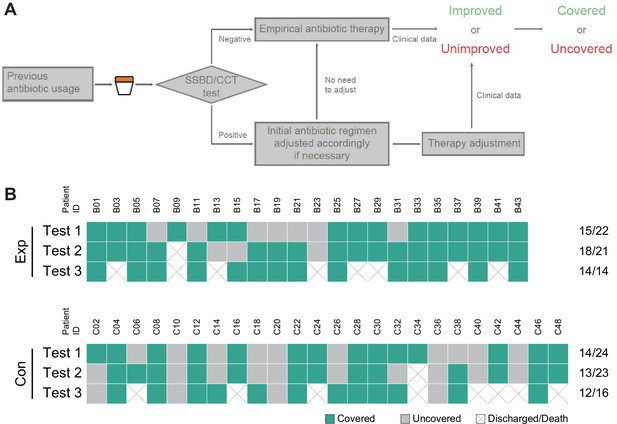
Judgment process and results of antibiotics coverage.
(A) Judgment process of antibiotics coverage. (B) The raw results of antibiotics coverage in two groups. Exp meant the experimental group, and Con meant the control group.
-
Appendix 1—figure 4—source data 1
Classification of antibiotic coverage for each test in two groups.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig4-data1-v2.xlsx
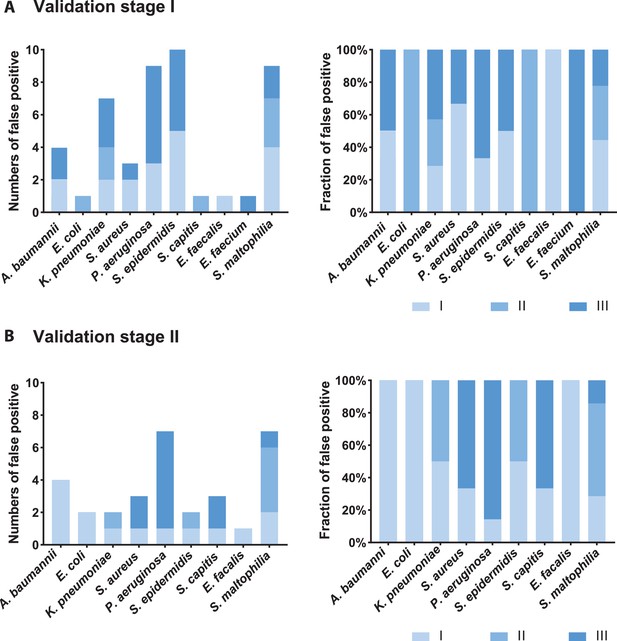
Analysis of false-positive samples.
Numbers and fractions of different strength levels among all false-positive samples of each bacteria species in the validation stage I (A) and II (B). Strength could be seen roughly as bacterial amounts (level I-level III, the definition could be seen in the Appendix 1). False-positive situations meant pathogenic bacteria detected by SSBD but not by CCT in a given BALF sample.
Tables
Demographic and baseline characteristics of the patients in the validation stage II.
| Experimental group (n=22) | Control group(n=24) | p value | |
|---|---|---|---|
| Women | 9 (40.9%) | 11 (45.8%) | 0.774 |
| Men | 13 (59.1%) | 13 (54.2%) | 0.774 |
| Age, years (SD) | 58 (17.4) | 68 (9.5) | 0.015* |
| Patients' numbers of chronic comorbidities | |||
| Hypertension | 9 (40.9%) | 17 (70.8%) | 0.073 |
| Coronary artery disease | 1 (4.5%) | 3 (12.5%) | 0.609 |
| Chronic pulmonary disease | 2 (9.1%) | 4 (16.7%) | 0.667 |
| Chronic kidney disease | 2 (9.1%) | 6 (25.0%) | 0.247 |
| Diabetes | 5 (22.7%) | 12 (50.0%) | 0.072 |
| Malignancy | 0 (0.0%) | 2 (8.3%) | 0.490 |
| Stroke | 3 (13.6%) | 8 (33.3%) | 0.171 |
| Immunodeficiency/immune suppressive therapy | 5 (22.7%) | 3 (12.5%) | 0.451 |
| Recent surgery | 4 (18.2%) | 3 (12.5%) | 0.694 |
| Hemodynamic support (using vasoactive drugs) | 7 (31.8%) | 7 (29.2%) | 1.000 |
| Norepinephrine ≤0.1 μg/(kg•min) | 2 | 3 | |
| Norepinephrine >0.1 μg/(kg•min) | 1 | 1 | |
| Dopamine ≤5 μg/(kg•min) | 3 | 2 | |
| Dopamine >5 μg/(kg•min) | 0 | 1 | |
| Dobutamine ≤5 μg/(kg•min) | 1 | 0 | |
| Dobutamine >5 μg/(kg•min) | 0 | 0 | |
| Status at randomization (D1) | |||
| Temperature, °C | 38.4 (0.6) | 38.3 (0.7) | 0.345 |
| Coma | 6 (27.3%) | 6 (25.0%) | 1.000 |
| Systolic blood pressure, mmHg | 112.2 (19.3) | 121.6 (17.8) | 0.057 |
| Invasive mechanical ventilation | 20 (90.9%) | 24 (100.0%) | 0.223 |
| Renal replacement therapy | 0 (0.0%) | 2 (8.3%) | 0.493 |
| SOFA score | 6.3 (0.7) | 6.0 (0.5) | 0.935 |
| APACHE II score | 17.3 (1.6) | 18.9 (1.5) | 0.422 |
| Albumin, g/L | 32.1 (5.1) | 31.0 (3.7) | 0.442 |
| Globulin, g/L | 21.8 (3.9) | 23.1 (5.9) | 0.489 |
| Absolute lymphocyte count, 109 /L | 0.9 (0.7) | 0.7 (0.3) | 0.909 |
| White blood cells, 109 /L | 11.0 (5.7) | 12.7 (6.3) | 0.210 |
| CRP, mg/L | 94.7 (101.2) | 108.3 (84.0) | 0.424 |
-
SOFA score and APACHE II score are mean (SEM), other data are mean (SD), n (%). Mean (SEM/SD) is compared using Mann-Whitney test, and n (%) is compared using Fisher’s exact test. * indicated p-value <0.05.
Primers used in experiments.
| Name | Sequence (5' ->3') | Target | Product length (bp) |
|---|---|---|---|
| pGL3-amplify-F | GAAGATGGAACCGCTGGAGA | pGL3 | 597 |
| pGL3-amplify-R | GCAGGCAGTTCTATGAGGCA | ||
| Aba-amplify-F | CACAGCGTTTAACCCATGCC | A. baumannii | 564 |
| Aba-amplify-R | TATCGCCACCTGCACAGAAG | ||
| Eco-amplify-F | GTTCCTGACTATCTGGCGGG | E. coli | 371 |
| Eco-amplify-R | GCTTCCTGACTCCAGACACC | ||
| Kpn-amplify-F | CATGGGCATATCGACGGTCA | K. pneumoniae | 740 |
| Kpn-amplify-R | CCTGCAACATAGGCCAGTGA | ||
| Sau-amplify-F | AGGTGCAGTAGACGCATAGC | S. aureus | 563 |
| Sau-amplify-R | CATTCGCTGCGCCAATACAA | ||
| Pae-amplify-F | TCTCTCTATCACGCCGGTCA | P. aeruginosa | 467 |
| Pae-amplify-R | TCGCATCGAGGTATTCCAGC | ||
| Sep-amplify-F | CACGCATGGCACTAGGTACA | S. epidermidis | 383 |
| Sep-amplify-R | CGAAAAAGAGTTGTCCTTGTTGA | ||
| Sca-amplify-F | GGTTCAGTCATCCCCACGTT | S. capitis | 591 |
| Sca-amplify-R | CAGCTGCGACAACTGCTTAC | ||
| Efa-amplify-F | CGGCAAGTTTGGAAGCAGAC | E. faecalis | 627 |
| Efa-amplify-R | CAGCGCCTAGTCCTTGTGAT | ||
| Efm-amplify-F | ATCGGAAATCGGTGTGGCTT | E. faecium | 507 |
| Efm-amplify-R | TCAAATGCATCCCTGTGCCT | ||
| Sma-amplify-F | CGCCTCCCGTTTACAGATTA | S. maltophilia | 356 |
| Sma-amplify-R | TCGGCTCCACCACATACAC |
Oligonucleotide templates for synthesis of crRNAs.
| Name | Sequence (5′->3′) | Target |
|---|---|---|
| T7-Forward | TAATACGACTCACTATAGGT | |
| LbcrRNA-Reverse-PGL3 | ATGTGACGAACGTGTACATCGACTATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | PGL3 |
| LbcrRNA-Reverse-Aba | TTCAAGTAATTCTTCTTTACATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | A. baumannii |
| LbcrRNA-Reverse-Eco | CTTGCCATCATAGCGACCGTATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | E. coli |
| LbcrRNA-Reverse-Kpn | AAGATGGCGATTACCGCAGTATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | K. pneumoniae |
| LbcrRNA-Reverse-Sau | CCTCAGCAAGTTCACGTTGTATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | S. aureus |
| LbcrRNA-Reverse-Pae | CTCCTCATGTGTGTTTACAAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | P. aeruginosa |
| LbcrRNA-Reverse-Sep | TCTCTAATTGATGAATATTAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | S. epidermidis |
| LbcrRNA-Reverse-Sca | TATTGATTAATAAGGTGATTATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | S. capitis |
| LbcrRNA-Reverse-Efa | TCAGCTGTGTTATTTGGTGCATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | E. faecalis |
| LbcrRNA-Reverse-Efm | TGTATATAAGTTCAGGTAGTATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | E. faecium |
| LbcrRNA-Reverse-Sma | CCTGGTCGCAGGTGTCATGCATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTA | S. maltophilia |
SSBD and CCT results of BALF samples in the validation stage I.
| Sample ID | SSBD Results | CCT Results | NGS Results |
|---|---|---|---|
| A01 | S. aureus (I) | N | S. aureus |
| A02 | A. baumannii (III) | A. baumannii | |
| A03 | A. baumannii (III) S. aureus (I) | A. baumannii S. aureus | A. baumannii S. aureus |
| A04 | A. baumannii (III) K. pneumoniae (II) S. maltophilia (II) | N | |
| A05 | A. baumannii (III) K. pneumoniae (III) P. aeruginosa (III) | K. pneumoniae P. aeruginosa | |
| A06 | A. baumannii (III) | A. baumannii | |
| A07 | S. maltophilia (II) | S. maltophilia | |
| A08 | A. baumannii (III) | A. baumannii | |
| A09 | A. baumannii (II) S. aureus (I) | A. baumannii | |
| A10 | A. baumannii (III) K. pneumoniae (III) P. aeruginosa (I) | A. baumannii K. pneumoniae | |
| A11 | A. baumannii (III) K. pneumoniae (I) | A. baumannii | |
| A12 | N | N | |
| A13 | A. baumannii (II) | A. baumannii | |
| A14 | N | N | |
| A15 | K. pneumoniae (III) | K. pneumoniae | |
| A16 | K. pneumoniae (III) P. aeruginosa (III) | K. pneumoniae | |
| A17 | N | N | |
| A18 | K. pneumoniae (I) P. aeruginosa (III) | K. pneumoniae P. aeruginosa | |
| A19 | P. aeruginosa (I) | P. aeruginosa | |
| A20 | E. faecium (III) S. capitis (II) | N | E. faecium S. capitis |
| A21 | A. baumannii (III) K. pneumoniae (III) S. aureus (I) | A. baumannii S. aureus | |
| A22 | A. baumannii (III) S. epidermidis (III) | A. baumannii | |
| A23 | N | N | |
| A24 | A. baumannii (III) | A. baumannii | |
| A25 | N | N | N |
| A26 | P. aeruginosa (III) | P. aeruginosa | |
| A27 | A. baumannii (III) S. epidermidis (III) | A. baumannii | A. baumannii S. epidermidis |
| A28 | A. baumannii (II) P. aeruginosa (III) | A. baumannii | |
| A29 | S. epidermidis (I) S. maltophilia (I) | N | |
| A30 | A. baumannii (III) S. aureus (III) S. epidermidis (I) | A. baumannii | |
| A31 | K. pneumoniae (I) P. aeruginosa (I) S. epidermidis (I) | N | K. pneumoniae |
| A32 | P. aeruginosa (I) S. epidermidis (I) | N | |
| A33 | S. aureus (I) S. maltophilia (II) | S. aureus | |
| A34 | N | N | A. baumannii |
| A35 | A. baumannii (III) | A. baumannii | |
| A36 | A. baumannii (III) S. maltophilia (I) | A. baumannii | |
| A37 | A. baumannii (III) S. epidermidis (III) | A. baumannii | |
| A38 | A. baumannii (III) | A. baumannii | |
| A39 | A. baumannii (III) | A. baumannii | |
| A40 | N | N | |
| A41 | N | N | |
| A42 | A. baumannii (I) | N | |
| A43 | A. baumannii (III) | A. baumannii | |
| A44 | A. baumannii (III) | A. baumannii | A. baumannii |
| A45 | A. baumannii (III) K. pneumoniae (II) | A. baumannii | |
| A46 | A. baumannii (III) | A. baumannii | |
| A47 | A. baumannii (I) | A. baumannii | |
| A48 | A. baumannii (III) | A. baumannii | |
| A49 | A. baumannii (III) S. aureus (III) S. maltophilia (III) | A. baumannii S. aureus | A. baumannii S. aureus S. maltophilia |
| A50 | S. epidermidis (III) | N | |
| A51 | N | N | |
| A52 | N | N | |
| A53 | K. pneumoniae (III) | K. pneumoniae | |
| A54 | N | N | |
| A55 | A. baumannii (III) E. coli (II) K. pneumoniae (III) S. maltophilia (III) | A. baumannii | |
| A56 | S. epidermidis (III) E. faecalis (I) | N | |
| A57 | N | N | |
| A58 | P. aeruginosa (III) | N | |
| A59 | S. maltophilia (I) | S. maltophilia | S. aureus S. maltophilia |
| A60 | A. baumannii (I) P. aeruginosa (III) | A. baumannii P. aeruginosa | |
| A61 | A. baumannii (II) | A. baumannii | |
| A62 | K. pneumoniae (II) S. maltophilia (III) | K. pneumoniae S. maltophilia | |
| A63 | A. baumannii (III) | A. baumannii | |
| A64 | A. baumannii (III) | A. baumannii | |
| A65 | A. baumannii (III) P. aeruginosa (I) S. maltophilia (II) | A. baumannii P. aeruginosa | |
| A66 | K. pneumoniae (II) | K. pneumoniae | |
| A67 | S. aureus (III) | S. aureus | |
| A68 | A. baumannii (III) P. aeruginosa (III) | A. baumannii | |
| A69 | A. baumannii (I) P. aeruginosa (III) | A. baumannii | P. aeruginosa |
| A70 | A. baumannii (I) | A. baumannii | |
| A71 | N | N | |
| A72 | A. baumannii (III) S. maltophilia (I) | A. baumannii | |
| A73 | A. baumannii (III) K. pneumoniae (III) P. aeruginosa (III) | A. baumannii P. aeruginosa | |
| A74 | A. baumannii (III) | A. baumannii | |
| A75 | A. baumannii (III) P. aeruginosa (III) | A. baumannii | |
| A76 | A. baumannii (I) P. aeruginosa (III) S. maltophilia (I) | P. aeruginosa | |
| A77 | S. epidermidis (I) | N |
Comparative analysis of test results by SSBD, CCT and NGS in the validation stage I.
| Sample ID | SSBD Results | CCT Results | NGS Results |
|---|---|---|---|
| A01 | S. aureus (I) | N | S. aureus |
| A03 | A. baumannii (III) S. aureus (I) | A. baumannii S. aureus | A. baumannii S. aureus |
| A20 | E. faecium (III) S. capitis (II) | N | E. faecium S. capitis |
| A25 | N | N | N |
| A27 | A. baumannii (III) S. epidermidis (III) | A. baumannii | A. baumannii S. epidermidis |
| A31 | K. pneumoniae (I) P. aeruginosa (I) S. epidermidis (I) | N | K. pneumoniae |
| A34 | N | N | A. baumannii |
| A44 | A. baumannii (III) | A. baumannii | A. baumannii |
| A49 | A. baumannii (III) S. aureus (III) S. maltophilia (III) | A. baumannii S. aureus | A. baumannii S. aureus S. maltophilia |
| A59 | S. maltophilia (I) | S. maltophilia | S. maltophilia S. aureus |
| A69 | A. baumannii (I) P. aeruginosa (III) | A. baumannii | P. aeruginosa |
SSBD and CCT results of BALF samples from experiment group (n=22) and control group (n=24) during the validation stage II.
| Patient ID | Sample ID | Test No. | SSBD Results | CCT Results | NGS Results |
|---|---|---|---|---|---|
| Exp. | |||||
| B01 | B01-1 | Test 1 | N | N | |
| B01-2 | Test 2 | N | N | N | |
| B01-3 | Test 3 | A. baumannii (III) | A. baumannii | ||
| B03 | B03-1 | Test 1 | N | N | N |
| B03-2 | Test 2 | A. baumannii (III) | A. baumannii | ||
| B05 | B05-1 | Test 1 | A. baumannii (III) S. maltophilia (II) | A. baumannii | A. baumannii S. maltophilia |
| B05-2 | Test 2 | A. baumannii (III) S. maltophilia (I) | A. baumannii | ||
| B05-3 | Test 3 | A. baumannii (III) S. maltophilia (I) | |||
| B07 | B07-1 | Test 1 | P. aeruginosa (III) | P. aeruginosa | P. aeruginosa |
| B07-2 | Test 2 | P. aeruginosa (III) | P. aeruginosa | ||
| B07-3 | Test 3 | P. aeruginosa (III) | P. aeruginosa | ||
| B09 | B09-1 | Test 1 | A. baumannii (III) S. aureus (III) S. capitis (III) S. maltophilia (III) | A. baumannii | |
| B11 | B11-1 | Test 1 | A. baumannii (III) P. aeruginosa (III) S. maltophilia (II) | A. baumannii | A. baumannii P. aeruginosa S. maltophilia |
| B11-2 | Test 2 | A. baumannii (III) S. aureus (I) S. maltophilia (III) | |||
| B11-3 | Test 3 | P. aeruginosa (III) | N | ||
| B13 | B13-1 | Test 1 | S. epidermidis (I) | N | N |
| B13-2 | Test 2 | K. pneumoniae (I) | K. pneumoniae | ||
| B15 | B15-1 | Test 1 | A. baumannii (I) E. coli (I) K. pneumoniae (II) | N | |
| B15-2 | Test 2 | K. pneumoniae (I) E. faecium (I) | |||
| B15-3 | Test 3 | E. faecium (III) | |||
| B17 | B17-1 | Test 1 | A. baumannii (III) | ||
| B17-2 | Test 2 | A. baumannii (III) K. pneumoniae (III) | |||
| B17-3 | Test 3 | A. baumannii (I) K. pneumoniae (I) E. faecalis (I) | A. baumannii | ||
| B19 | B19-1 | Test 1 | A. baumannii (III) P. aeruginosa (I) S. maltophilia (II) | A. baumannii | |
| B19-2 | Test 2 | A. baumannii (III) P. aeruginosa (III) S. maltophilia (III) | A. baumannii S. maltophilia | ||
| B19-3 | Test 3 | K. pneumoniae (III) P. aeruginosa (III) S. maltophilia (III) | K. pneumoniae S. maltophilia | ||
| B21 | B21-1 | Test 1 | P. aeruginosa (III) S. aureus (III) S. maltophilia (III) | S. aureus S. maltophilia | |
| B21-2 | Test 2 | P. aeruginosa (III) S. aureus (III) S. maltophilia (III) | S. maltophilia | ||
| B21-3 | Test 3 | S. aureus (I) S. maltophilia (II) | N | ||
| B23 | B23-1 | Test 1 | S. capitis (I) S. maltophilia (III) | S. maltophilia | |
| B23-2 | Test 2 | S. maltophilia (III) | S. maltophilia | ||
| B25 | B25-1 | Test 1 | A. baumannii (II) | A. baumannii | |
| B25-2 | Test 2 | A. baumannii (III) | |||
| B25-3 | Test 3 | A. baumannii (III) | A. baumannii | ||
| B27 | B27-1 | Test 1 | S. maltophilia (I) | N | N |
| B27-2 | Test 2 | A. baumannii (III) | |||
| B29 | B29-1 | Test 1 | K. pneumoniae (III) | K. pneumoniae | K. pneumoniae |
| B29-2 | Test 2 | K. pneumoniae (III) | |||
| B31 | B31-1 | Test 1 | A. baumannii (III) S. capitis (III) | A. baumannii | A. baumannii S. capitis |
| B31-2 | Test 2 | A. baumannii (III) | |||
| B31-3 | Test 3 | A. baumannii (I) | N | ||
| B33 | B33-1 | Test 1 | N | N | |
| B33-2 | Test 2 | A. baumannii (I) S. epidermidis (II) | N | ||
| B33-3 | Test 3 | N | N | ||
| B35 | B35-1 | Test 1 | A. baumannii (III) | A. baumannii | A. baumannii |
| B35-2 | Test 2 | A. baumannii (III) | A. baumannii | ||
| B35-3 | Test 3 | A. baumannii (III) | A. baumannii | ||
| B37 | B37-1 | Test 1 | N | N | A. baumannii |
| B37-2 | Test 2 | P. aeruginosa (I) | |||
| B39 | B39-1 | Test 1 | N | P. aeruginosa | |
| B39-2 | Test 2 | A. baumannii (I) E. coli (I) K. pneumoniae (III) P. aeruginosa (III) | K. pneumoniae P. aeruginosa | ||
| B39-3 | Test 3 | K. pneumoniae (I) P. aeruginosa (III) | |||
| B41 | B41-1 | Test 1 | N | N | N |
| B41-2 | Test 2 | N | |||
| B43 | B43-1 | Test 1 | N | N | |
| B43-2 | Test 2 | A. baumannii (I) S. epidermidis (I) | |||
| B43-3 | Test 3 | A. baumannii (II) | A. baumannii | ||
| Con. | |||||
| C02 | Test 1 | A. baumannii | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C04 | Test 1 | N | |||
| Test 2 | N | ||||
| Test 3 | N | ||||
| C06 | Test 1 | N | |||
| Test 2 | A. baumannii | ||||
| C08 | Test 1 | E. coli | |||
| Test 2 | E. coli | ||||
| Test 3 | K. pneumoniae | ||||
| C10 | Test 1 | N | |||
| Test 2 | N | ||||
| Test 3 | A. baumannii | ||||
| C12 | Test 1 | N | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C14 | Test 1 | A. baumannii | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C16 | Test 1 | N | |||
| Test 2 | N | ||||
| C18 | Test 1 | A. baumannii | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C20 | Test 1 | N | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C22 | Test 1 | N | |||
| Test 2 | N | ||||
| Test 3 | A. baumannii | ||||
| C24 | Test 1 | N | |||
| Test 2 | N | ||||
| C26 | Test 1 | A. baumannii | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C28 | Test 1 | S. aureus | |||
| Test 2 | N | ||||
| Test 3 | A. baumannii | ||||
| C30 | Test 1 | N | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C32 | Test 1 | N | |||
| Test 2 | K. pneumoniae | ||||
| Test 3 | A. baumannii K. pneumoniae | ||||
| C34 | Test 1 | N | |||
| C36 | Test 1 | S. aureus | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C38 | Test 1 | A. baumannii | |||
| Test 2 | A. baumannii | ||||
| Test 3 | A. baumannii | ||||
| C40 | Test 1 | P. aeruginosa | |||
| Test 2 | P. aeruginosa | ||||
| C42 | Test 1 | N | |||
| Test 2 | N | ||||
| C44 | Test 1 | S. aureus | |||
| Test 2 | A. baumannii | ||||
| C46 | Test 1 | N | |||
| Test 2 | N | ||||
| Test 3 | A. baumannii | ||||
| C48 | Test 1 | A. baumannii | |||
| Test 2 | A. baumannii |
Comparative analysis of test results by SSBD, CCT and NGS in the validation stage II.
| Patient ID | Sample ID | Test No. | SSBD Results | CCT Results | NGS Results |
|---|---|---|---|---|---|
| B01 | B01-2 | Test 2 | N | N | N |
| B03 | B03-1 | Test 1 | N | N | N |
| B05 | B05-1 | Test 1 | A. baumannii (III) S. maltophilia (II) | A. baumannii | A. baumannii S. maltophilia |
| B07 | B07-1 | Test 1 | P. aeruginosa (III) | P. aeruginosa | P. aeruginosa |
| B11 | B11-1 | Test 1 | A. baumannii (III) P. aeruginosa (III) S. maltophilia (II) | A. baumannii | A. baumannii P. aeruginosa S. maltophilia |
| B13 | B13-1 | Test 1 | S. epidermidis (I) | N | N |
| B27 | B27-1 | Test 1 | S. maltophilia (I) | N | N |
| B29 | B29-1 | Test 1 | K. pneumoniae (III) | K. pneumoniae | K. pneumoniae |
| B31 | B31-1 | Test 1 | A. baumannii (III) S. capitis (III) | A. baumannii | A. baumannii S. capitis |
| B35 | B35-1 | Test 1 | A. baumannii (III) | A. baumannii | A. baumannii |
| B37 | B37-1 | Test 1 | N | N | A. baumannii |
| B41 | B41-1 | Test 1 | N | N | N |
Antibiotic use of the patients prior to clinical trial in the experimental group and control group.
| Empirical antibiotic therapy | |
|---|---|
| Experimental group | |
| B01 | Piperacillin -tazobactam |
| B03 | Biapenem |
| B05 | Biapenem, Teicoplanin and Tigecycline |
| B07 | Biapenem |
| B09 | Biapenem and vancomycin |
| B11 | Cefoperazone-sulbactam and Tigecycline |
| B13 | Piperacillin-Tazobactam, trimethoprim-sulfamethoxazole and Teicoplanin |
| B15 | Imipenem-cilastatin and Linezolid |
| B17 | Biapenem |
| B19 | Biapenem |
| B21 | Ceftazidine-avibatam |
| B23 | Imipenem-Cilastatin |
| B25 | Imipenem-Cilastatin and Linezolid |
| B27 | Piperacillin-Tazobactam and trimethoprim-sulfamethoxazole |
| B29 | Imipenem-Cilastatin |
| B31 | Imipenem-Cilastatin and Teicoplanin |
| B33 | Piperacillin-tazobactam |
| B35 | Moxifloxacin and Piperacillin -tazobactam |
| B37 | Biapenem, trimethoprim-sulfamethoxazole and Linezolid |
| B39 | Piperacillin-tazobactam |
| B41 | Piperacillin-tazobactam |
| B43 | Meropenem and Linezolid |
| Control group | |
| C02 | Meropenem and vancomycin |
| C04 | Biapenem and vancomycin |
| C06 | Piperacillin-tazobactam |
| C08 | Imipenem-Cilastatin |
| C10 | Cefoperazone-sulbactam |
| C12 | Moxifloxacin |
| C14 | Biapenem and Linezolid |
| C16 | Cefoperazone-sulbactam |
| C18 | Moxifloxacin |
| C20 | Piperacillin-tazobactam |
| C22 | Piperacillin-tazobactam and vancomycin |
| C24 | Cefoperazone-sulbactam, Linezolid and trimethoprim-sulfamethoxazole |
| C26 | Cefoperazone-sulbactam and Moxifloxacin |
| C28 | Piperacillin-tazobactam |
| C30 | Piperacillin-tazobactam |
| C32 | Piperacillin-tazobactam and Moxifloxacin |
| C34 | Meropenem |
| C36 | Cefoperazone-sulbactam |
| C38 | Piperacillin-tazobactam |
| C40 | Imipenem-Cilastatin |
| C42 | Cefoperazone-sulbactam |
| C44 | Piperacillin-tazobactam |
| C46 | Cefoperazone-sulbactam |
| C48 | Piperacillin-tazobactam and Linezolid |
Patients' clinical outcomes.
| Experimental group (n=22) | Control group(n=24) | p | |
|---|---|---|---|
| Number of patients who have clinical indexes improved | |||
| Day 3 vs. Day 1 | |||
| Temperature, °C | 15 (68.2%) | 9 (37.5%) | 0.045* |
| WBC, 109 /L | 15 (68.2%) | 12 (50.0%) | 0.211 |
| PCT, ng/mL | 18 (81.8%) | 19 (82.6%) | 0.945 |
| Day 7 vs. Day 1 | |||
| Temperature, °C | 13 (72.2%) | 12 (54.5%) | 0.332 |
| WBC, 109 /L | 16 (84.2%) | 11 (50.0%) | 0.021* |
| PCT, ng/mL | 13 (68.4%) | 19 (86.4%) | 0.166 |
| Day 10 vs. Day 1 | |||
| Temperature, °C | 13 (82.6%) | 12 (70.6%) | 0.688 |
| WBC, 109 /L | 9 (56.3%) | 8 (47.1%) | 0.598 |
| PCT, ng/mL | 12 (75.0%) | 15 (88.2%) | 0.325 |
| Number of patients undergoing effective treatment | |||
| Day 3 | 13 (59.1%) | 11 (45.8%) | 0.395 |
| Day 7 | 16 (84.2%) | 11 (50.0%) | 0.046* |
| Day 10 | 13 (81.3%) | 10 (58.8%) | 0.259 |
| Clinical endpoint outcomes | |||
| Hospital stay duration, days | 21 (13.7) | 23.5 (17.2) | 0.987 |
| 28 days mortality | 8 (36.4%) | 8 (33.3%) | 1.000 |
| Mechanical ventilation from randomization to 28th day, days | 11.3 (7.7) | 11.5 (7.7) | 0.970 |
| Shock from randomization to 28th day, days | 3.1 (4.3) | 2.3 (3.6) | 0.456 |
| Numbers of antibiotic-associated diarrhea | 0 (0.0%) | 2 (8.3%) | 0.490 |
-
For those data are n (%), all p values are calculated using Fisher’s exact tests. For those data are mean (SD), all p values are calculated using Mann-Whitney tests. * indicated P-value <0.05.
Potential competitive analysis among bacteria.
| Sample ID | Bacteria detected by SSBD grow in CCT tests | Bacteria detected by SSBD could not grow in CCT tests | Probable relations among bacteria |
|---|---|---|---|
| A05 | K. pneumoniae (III) P. aeruginosa (III) | A. baumannii (III) | K. pneumoniae + P. aeruginosa > A. baumannii |
| A09 | A. baumannii (II) | S. aureus (I) | Strength: II > I |
| A10 | A. baumannii (III) K. pneumoniae (III) | P. aeruginosa (I) | Strength: III + III > I |
| A11 | A. baumannii (III) | K. pneumoniae (I) | Strength: III > I |
| A16 | K. pneumoniae (III) | P. aeruginosa (III) | K. pneumoniae > P. aeruginosa |
| A21 | A. baumannii (III) S. aureus (I) | K. pneumoniae (III) | Strength: III + I > III |
| A22 | A. baumannii (III) | S. epidermidis (III) | A. baumannii > S. epidermidis |
| A27 | A. baumannii (III) | S. epidermidis (III) | A. baumannii > S. epidermidis |
| A28 | A. baumannii (II) | P. aeruginosa (III) | A. baumannii > P. aeruginosa |
| A30 | A. baumannii (III) | S. aureus (III) S. epidermidis (I) | A. baumannii > S. aureus + S. epidermidis |
| A33 | S. aureus (I) | S. maltophilia (II) | S. aureus > S. maltophilia |
| A36 | A. baumannii (III) | S. maltophilia (I) | A. baumannii > S. maltophilia |
| A37 | A. baumannii (III) | S. epidermidis (III) | A. baumannii > S. epidermidis |
| A45 | A. baumannii (III) | K. pneumoniae (II) | A. baumannii > K. pneumoniae |
| A49 | A. baumannii (III) S. aureus (III) | S. maltophilia (III) | A. baumannii + S. aureus > S. maltophilia |
| A55 | A. baumannii (III) | E. coli (II) K. pneumoniae (III) S. maltophilia (III) | A. baumannii > E. coli. coli + K. pneumoniae + S. maltophilia |
| A65 | A. baumannii (III) P. aeruginosa (I) | S. maltophilia (II) | Strength: III + I > II |
| A68 | A. baumannii (III) | P. aeruginosa (III) | A. baumannii > P. aeruginosa |
| A69 | A. baumannii (I) | P. aeruginosa (III) | A. baumannii > P. aeruginosa |
| A72 | A. baumannii (III) | S. maltophilia (I) | A. baumannii > S. maltophilia |
| A73 | A. baumannii (III) P. aeruginosa (III) | K. pneumoniae (III) | A. baumannii + P. aeruginosa > K. pneumoniae |
| A75 | A. baumannii (III) | P. aeruginosa (III) | A. baumannii > P. aeruginosa |
| A76 | P. aeruginosa (III) | A. baumannii (I) S. maltophilia (I) | Strength: III > I + I |
| B05-1 | A. baumannii (III) | S. maltophilia (II) | A. baumannii > S. maltophilia |
| B05-2 | A. baumannii (III) | S. maltophilia (I) | A. baumannii > S. maltophilia |
| B09-1 | A. baumannii (III) | S. aureus (III) S. capitis (III) S. maltophilia (III) | A. baumannii > S. aureus + S. capitis + S. maltophilia |
| B11-1 | A. baumannii (III) | P. aeruginosa (III) S. maltophilia (II) | A. baumannii > P. aeruginosa + S. maltophilia |
| B17-3 | A. baumannii (I) | K. pneumoniae (I) E. faecalis (I) | A. baumannii > K. pneumoniae +E. faecalis |
| B19-1 | A. baumannii (III) | P. aeruginosa (I) S. maltophilia (II) | Strength: III >I + II |
| B19-2 | A. baumannii (III), S. maltophilia (III) | P. aeruginosa (III) | A. baumannii + S. maltophilia > P. aeruginosa |
| B19-3 | K. pneumoniae (III), S. maltophilia (III) | P. aeruginosa (III) | K. pneumoniae + S. maltophilia > P. aeruginosa |
| B21-1 | S. aureus (III) S. maltophilia (III) | P. aeruginosa (III) | S. maltophilia + S. aureus > P. aeruginosa |
| B21-2 | S. maltophilia (III) | S. aureu (III), P. aeruginosa (III) | S. maltophilia > S. aureus + P. aeruginosa |
| B23-1 | S. maltophilia (III) | S. capitis (I) | Strength: III > I |
| B31-1 | A. baumannii (III) | S. capitis (III) | A. baumannii > S. capitis |
| B39-2 | K. pneumoniae (III) P. aeruginosa (III) | A. baumannii (I) E. coli (I) | Strength: III + III > I+I |
Comparation of CCT, SSBD and NGS.
| CCT | SSBD | NGS | ||
|---|---|---|---|---|
| Turnover time | 2–5 days | Less than 4 hours | 2–3 days | |
| Cost | Low | Low | High | |
| Detection target | Culturable bacteria | Selected targets | All microorganisms in the sample | |
| Quantification | Semi | Relative | Semi | |
| Instrument requirement | Low | Low | High |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/79014/elife-79014-mdarchecklist1-v2.pdf
-
Appendix 1—figure 2—source data 1
Epidemic data of pathogens in the ICU of Nanjing Drum Tower Hospital in 2017.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig2-data1-v2.xlsx
-
Appendix 1—figure 3—source data 1
The test of SSBD with or without DNA purification.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig3-data1-v2.xlsx
-
Appendix 1—figure 3—source data 2
The test of SSBD with different incubation times.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig3-data2-v2.xlsx
-
Appendix 1—figure 3—source data 3
The validation of SSBD with clinical strains.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig3-data3-v2.xlsx
-
Appendix 1—figure 4—source data 1
Classification of antibiotic coverage for each test in two groups.
- https://cdn.elifesciences.org/articles/79014/elife-79014-app1-fig4-data1-v2.xlsx