A choline-releasing glycerophosphodiesterase essential for phosphatidylcholine biosynthesis and blood stage development in the malaria parasite
Figures
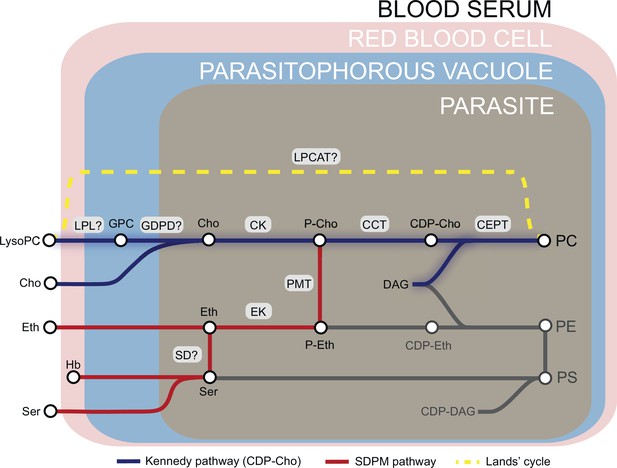
Phosphatidylcholine (PC) synthesis in malaria parasites.
The CDP-choline dependent Kennedy pathway, the SDPM pathway and Lands’ cycle produce PC from the metabolic precursors lysoPC, choline (Cho), ethanolamine (Eth), serine (Ser, including that obtained from digestion of haemoglobin, Hb), and fatty acids, all salvaged from the host milieu. PC is primarily produced through the Kennedy pathway using Cho sourced mainly from serum lysoPC. Breakdown of lysoPC into choline is thought to occur in the parasitophorous vacuole via a two-step hydrolysis process involving an unidentified lysophospholipase (LPL) and a glycerophosphodiesterase (GDPD; PF3D7_1406300) (this work). Other abbreviations: CCT, CTP:phosphocholine cytidyltransferase; CEPT, choline/ethanolamine-phosphotransferase; CK, choline kinase; DAG, diacylglycerol; EK, ethanolamine kinase; GPC, glycerophosphocholine; LPCAT, lysophosphatidylcholine acyltransferase; PMT, phosphoethanolamine methyltransferase; SD, serine decarboxylase. ‘?’ indicates parasite enzymes not yet identified.
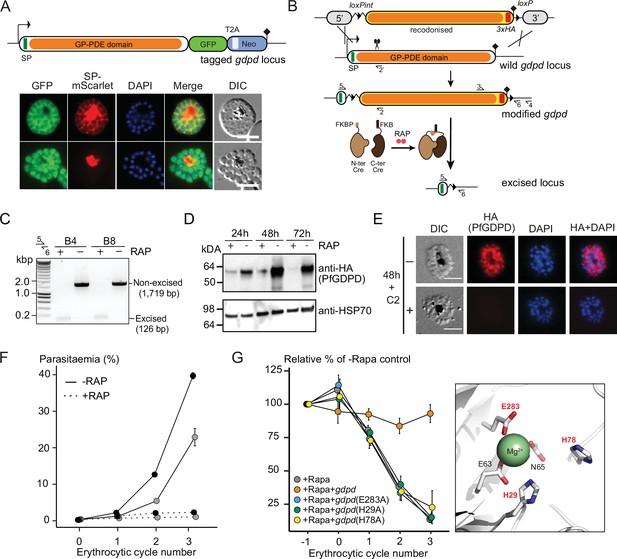
Subcellular localization and conditional ablation of PfGDPD.
(A) Endogenous PfGDPD tagged with GFP shows dual localization in the cytosol and PV. GDPD colocalization with soluble PV marker, SP-mScarlet (Mesén-Ramírez et al., 2019), expressed episomally in the GDPD-GFP line is shown in mature schizonts (top) and free merozoites (bottom). Scale bars, 5 µm. (B) Strategy used for conditional disruption of PfGDPD in parasite line GDPD:HA:loxPint. The predicted catalytic domain (GP-PDE, glycerophosphodiester phosphodiesterase; orange) was floxed by introducing an upstream loxPint and a loxP site following the translational stop site. Sites of targeted Cas9-mediated double-stranded DNA break (scissors), left and right homology arms for homology-directed repair (5’ and 3’), introduced loxP sites (arrow heads), secretory signal peptide (green), recodonized sequences (yellow), 3xHA epitope (red) and diagnostic PCR primers (half arrows 1–4) are indicated. RAP-induced DiCre-mediated excision results in removal of the catalytic domain. (C) Diagnostic PCR 12 hr following mock- or RAP-treatment of ring-stage GDPD:HA:loxPint parasites (representative of three independent experiments) confirms efficient gene excision. Expected amplicon sizes are indicated. (D) Western blots (representative of two independent experiments) showing successful RAP-induced ablation of PfGDPD expression in cycle 0 GDPD:HA:loxPint parasites sampled at 24 hr and 48 hr post invasion and cycle 1 trophozoites (72 hr). HSP70 was probed as loading control. (E) IFA of RAP-treated (+) and mock-treated (-) mature GDPD:HA:loxPint cycle 0 schizonts following mock- (-) or RAP-treatment (+) at ring-stage, showing that expression of PfGDPD-HA is lost following RAP treatment. Scale bar, 5 µm. (F) RAP-treatment results in loss of replication in two clonal lines, B4 (black) and B8 (grey), of GDPD:HA:loxPint parasites (error bars, ± SD). Data shown are averages from triplicate biological replicates using different blood sources. (G) Genetic complementation with an episomal, constitutively expressed mCherry-tagged PfGDPD fully restores growth of Rapa-treated GDPD:loxPint:HA:Neo-R parasites. In contrast, mutant PfGDPD alleles carrying Ala substitutions of the catalytic H29 and H78 residues or the metal-binding residue E283 do not complement. Inset, zoomed AlphaFold model of PfGDPD catalytic groove and coordinated Mg2+ ion, with relevant residues highlighted in red. The erythrocytic cycle when rapalog was added has been designated as cycle 0.
-
Figure 2—source data 1
Source data for plotted graphs and the full raw unedited versions of the gel and blot images shown in Figure 2 and Figure 2—figure supplements 1–3.
- https://cdn.elifesciences.org/articles/82207/elife-82207-fig2-data1-v2.zip
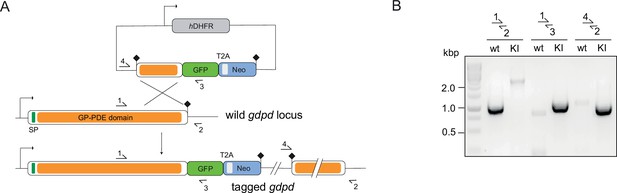
Endogenous tagging of PfGDPD.
(A) Schematic of SLI-based endogenous tagging of PfGDPD. GFP, green fluorescent protein; T2A, skip peptide; Neo-R, neomycin-resistance gene; hDHFR, human dihydrofolate reductase; asterisks, stop codons; arrows, promoters. (B) Diagnostic PCR showing correct integration of the GFP-tagging construct in the GDPD locus.
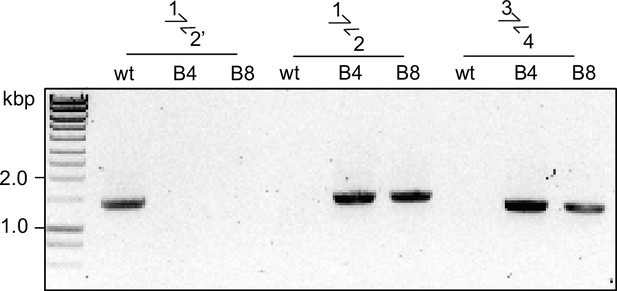
Diagnostic PCR for successful integration in GDPD:loxPint:HA line.
Diagnostic PCR showing correct integration of the pREP-GDPD modification plasmid in the PfGDPD locus in GDPD:loxPint:HA parasites. Primers used are denoted in Figure 2B.
Conditional knockout of PfGDPD expression using the SLI system.
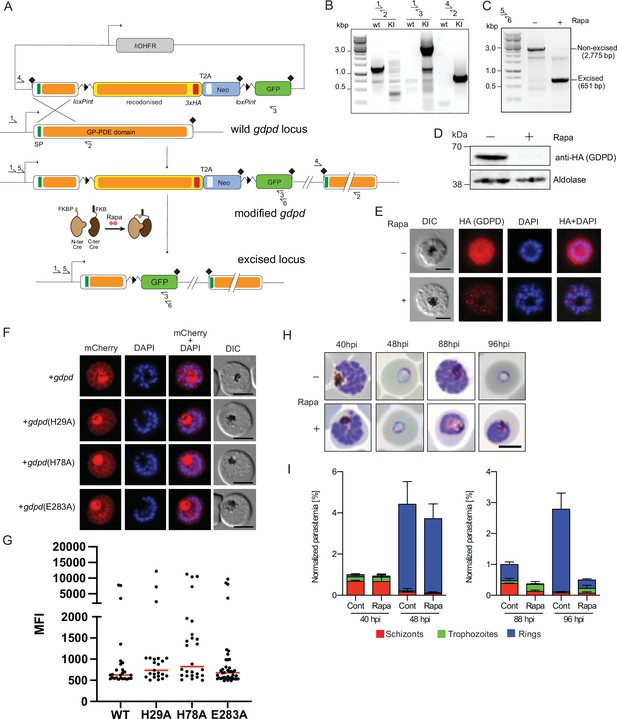
(A) Schematic of the SLI-based DiCre-mediated conditional knockout strategy. (B) Diagnostic PCR showing correct integration of the SLI modification plasmid in the GDPD locus in the GDPD:loxPint:HA:Neo-R line. (C) Diagnostic PCR 36 hr following mock- or Rapa- treatment confirms efficient gene excision. Expected amplicon sizes are indicated. (D) Western blot of 3xHA-tagged PfGDPD in control and Rapa-treated parasites 48 hr post-Rapa-treatment. (E) IFA of 3xHA-tagged PfGDPD in control and Rapa-treated parasites 48 hr post-Rapa treatment. Scale bars 5 µm. (F) Mutagenesis of several key functional residues does not affect localization of PfGDPD. Live-cell microscopy of C2-arrested GDPD:loxPint:HA:Neo-R schizonts expressing the non-mutant (WT) and mutant PfGDPD coding sequence C-terminally fused to mCherry. Nuclei were stained with DAPI (blue). Scale bars 5 µm. (G) PfGDPD expression levels in PfGDPD complementation cells lines. Late trophozoite and schizont/segmenter stage parasites episomally expressing WT or mutated PfGDPD-mCherry were analyzed by live cell microscopy using the same imaging settings and their mean fluorescence intensity (MFI) was determined. Shown are individual values and medians (red) of 23–40 imaged parasites per line. No statistically significant differences in expression levels between WT and mutated GDPDs were observed (One-way ANOVA, p=0.4219). (H) Light microscopic images of Giemsa-stained parasites following mock- or Rapa-treatment at ring stages. (I) Life stage quantification of parasites at selected time points after Rapa-treatment (error bars, ± SD, triplicate Rapa treatments).
PfGDPD is essential for asexual blood stage development.
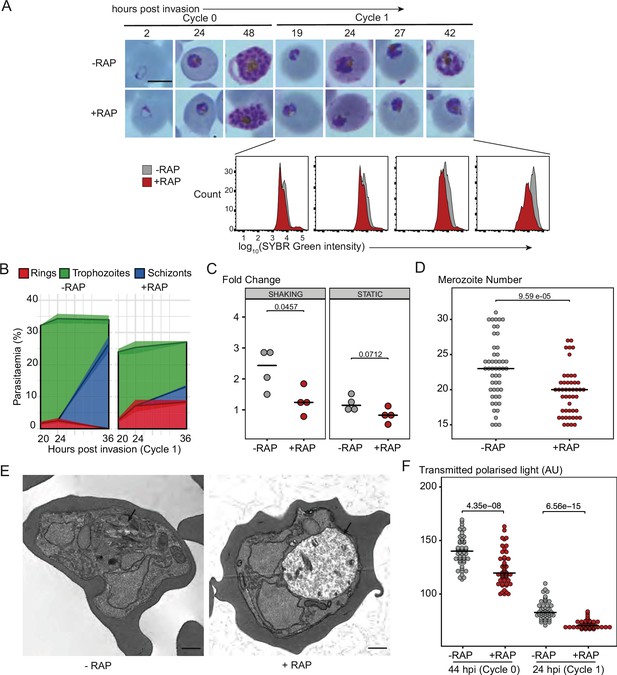
(A) Light microscopic images of Giemsa-stained cycle 0 and 1 GDPD:HA:loxPint parasites following mock- or RAP-treatment at ring stage in cycle 0 (representative of 2 independent experiments). PfGDPD-null parasites began to exhibit defective development at around 19 hr post-invasion (19 hpi) in cycle 1, producing abnormal trophozoites. The growth defect was confirmed and quantified using flow cytometry to measure parasite DNA content. Fluorescence intensity of the SYBR Green-stained RAP-treated population (red) was detectably lower than that of the control population (grey) from 19 hr into cycle 1. Scale bar, 5 µm. (B) Life stage quantification of GDPD:HA:loxPint parasites at selected time points in cycle 1 (error bars, ± SD, triplicate RAP treatments) following RAP treatment of rings in cycle 0. Mock-treated parasites (DMSO) transitioned normally from trophozoite to schizont stage while RAP-treated parasites showed accumulation of abnormal ring and trophozoite forms. (C) PfGDPD-null parasites exhibit an invasion defect. Fold change in parasitaemia after 4 hr invasion of mock-treated (-) and RAP-treated (+) GDPD:HA:loxPint schizonts under shaking and static conditions (crossbar represents median fold change in four replicate RAP treatments with different blood sources; individual points represent a single replicate). (D) Numbers of merozoites in highly mature cycle 0 schizonts (obtained by arresting egress using the reversible egress inhibitor C2) following mock (-) or RAP-treatment (+) at ring stage. Merozoite numbers were slightly but significantly lower in PfGDPD-null parasites (crossbar represents median; n=50; Student t-test with Bonferroni adjusted p-value). (E) TEM micrographs of control and RAP-treated GDPD:HA:loxPint parasites allowed to mature for ∼40 hr in cycle 1 in order to maximise proportions of abnormal forms. Less haemozoin formation was evident in the digestive vacuole (arrowed) of the PfGDPD-null mutants compared to mock-treated controls. Scale bar, 500 nm. (F) Haemozoin content of individual parasites measured as transmitted polarized light at 44 hpi in cycle 0 and 24 hpi in cycle 1. (crossbar represents median; n=50; Student t-test with Bonferroni adjusted p-value).
-
Figure 3—source data 1
Source data for plotted graphs in Figure 3.
- https://cdn.elifesciences.org/articles/82207/elife-82207-fig3-data1-v2.zip
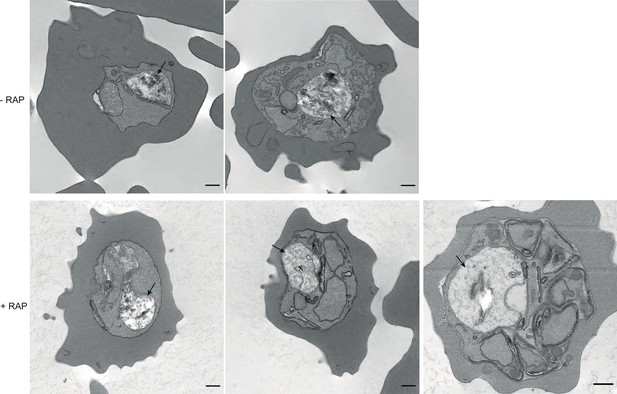
TEM images of mock and RAP-treated GDPD:loxPint:HA parasites.
TEM images of mock- and RAP-treated GDPD:loxPint:HA parasites at different stages of development – (from left to right) young trophozoites, late trophozoite (with double nuclei) and a partially segmented schizont. Less haemozoin formation was evident in the digestive vacuole (arrowed) at all stages of development in PfGDPD-null parasites. Scale bar, 500 nm.
Choline supplementation rescues growth of PfGDPD-null parasites.
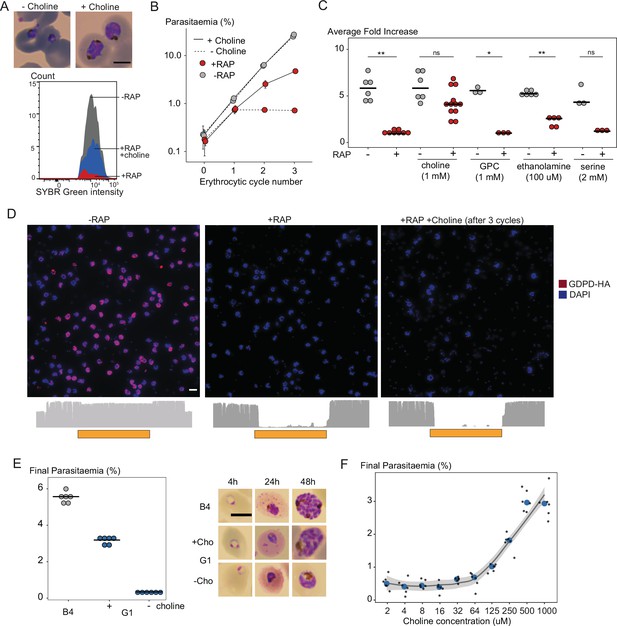
(A) Morphology of PfGDPD-null trophozoites at 32 hr in cycle 1 following RAP treatment of rings in cycle 0 in the presence or absence of choline. Fluorescence intensity of SYBR Green-stained populations at the same timepoint show choline-supplemented PfGDPD-null trophozoites (blue) can surpass the developmental arrest in non-supplemented parasites. Scale bar, 5 µm. (B) Replication of mock- (grey) and RAP-treated (red) GDPD:HA:loxPint parasites in the presence (solid line) or absence (dashed line) of choline (error bars, ± SD, triplicate experiments with different blood sources). (C) Effects of supplementation with different metabolic precursors on the replication of mock- (grey) or RAP-treated (red) GDPD:HA:loxPint parasites. Mean average fold increase in parasitaemia over two erythrocytic cycles was increased by 1 mM choline to close to wild type levels (grey). In contrast, 100 µM ethanolamine effected only a marginal improvement in the replication rate while 1 mM glycerophosphocholine (GPC) and 2 mM serine had no effect. (D) Continuous culture of PfGDPD-null parasites enabled by choline supplementation. Top, IFA showing absence of PfGDPD-HA expression in the emergent parasite population after three erythrocytic cycles of growth in choline-supplemented medium (right). For comparison, parasite populations in cycle 0 following treatment are shown (left and middle). Below, genome sequencing showing RAP-induced excision of the PfGDPD gene and no evidence of the non-excised locus in the choline-supplemented emergent RAP-treated parasite population. Scale bar, 10 µm. (E) Confirmation of the choline dependency of the PfGDPD-null parasite clone G1. Left, parasite cultures (starting parasitaemia 0.1%) were maintained with or without 1 mM choline for two erythrocytic cycles before measuring final parasitaemia (n=6). Right, effects of choline removal on intra-erythrocytic parasite development, assessed at different time points. In all cases results are shown compared to the parental GDPD:HA:loxPint line (B4) without choline supplementation. Scale bar, 5 µm. (F) Concentration-dependence of choline supplementation on replication of the choline-dependent PfGDPD-null parasite clone G1. Parasite cultures (starting parasitaemia 0.1%) were maintained for two erythrocytic cycles in the presence of a range of choline concentrations, before final parasitaemia quantified (n=6). Black dots, individual replicates. Blue dots, mean values. Grey band, dose-dependency curve ± SD.
-
Figure 4—source data 1
Source data for plotted graphs in Figure 4 and a representative microscopy image of choline-supplemented G1 line (G1+ in Figure 4E).
- https://cdn.elifesciences.org/articles/82207/elife-82207-fig4-data1-v2.zip
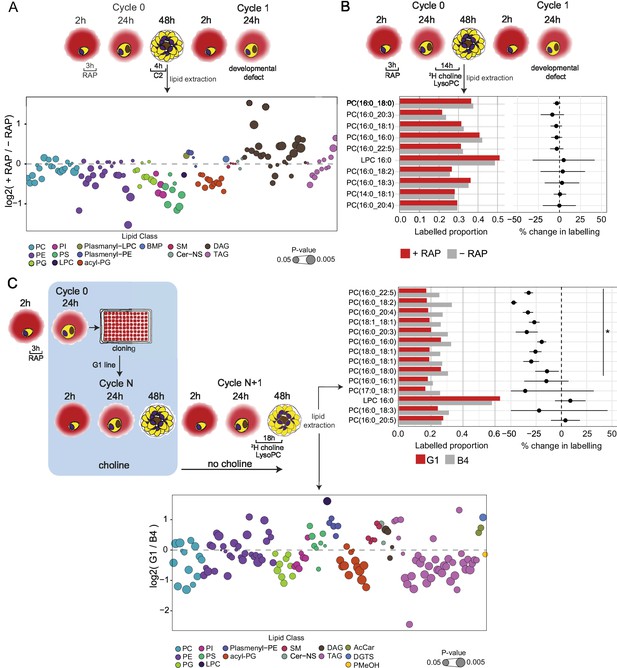
Lipidomic profiling and metabolic labelling of PfGDPD-null parasites show disruption in PC biosynthesis and choline uptake from lysoPC.
(A) Lipidome analysis of mature cycle 0 GDPD:loxPint:HA schizonts following mock-or RAP-treatment at ring stage. The bubble plot shows the fold change in levels of various lipid species in PfGDPD-null schizonts compared to controls (3 independent biological replicates). (B) Metabolic labelling of mock- and RAP-treated GDPD:loxPint:HA parasites by a 14 hr incubation with 2H choline-labelled lysoPC 16:0 during trophozoite development. Dotplots depict percentage change in mean labelled proportions in each PC or lysoPC species (shown as bar graphs) in PfGDPD-null schizonts compared to controls across three independent biological replicates. (C) Metabolic labelling (top panel) and lipidome analysis (bottom panel) of PfGDPD-expressing GDPD:loxPint:HA (B4) and PfGDPD-null parasites (clone G1) by treatment for 18 hr with 2H choline-labelled lysoPC 16:0 during trophozoite development. Choline was removed from the culture medium 24 hr prior to labelling.
-
Figure 5—source data 1
Source data for the plotted graphs in Figure 5 and the PRM inclusion list (Figure5C-sourcedata3) used for identification of DGTS species in Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/82207/elife-82207-fig5-data1-v2.zip
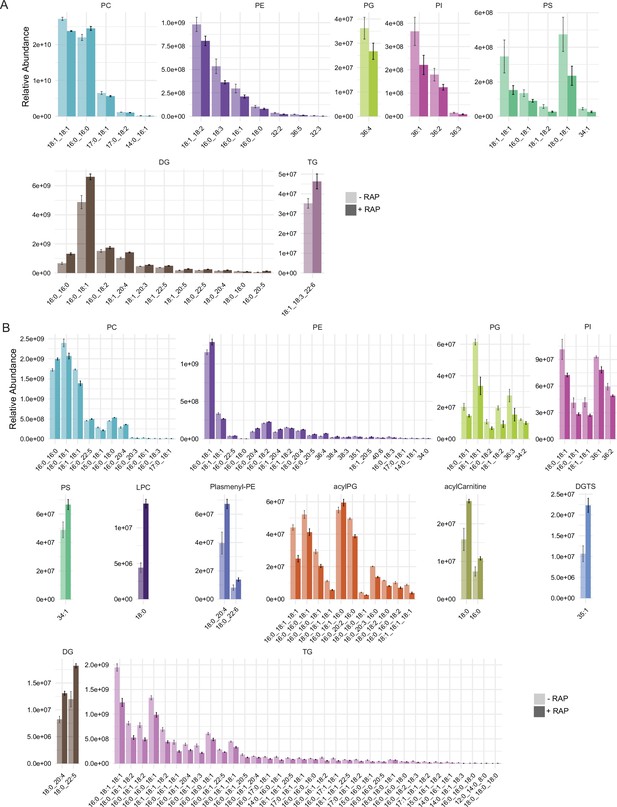
Relative peak intensities of the significantly altered lipid species.
Relative peak intensities of the significantly altered lipid species in (A) comparison between mock- or RAP-treated GDPD:loxPint:HA mature schizonts from cycle 0 and (B) comparison between choline-starved GDPD:loxPint:HA (B4) and PfGDPD-null (clone G1) parasites.
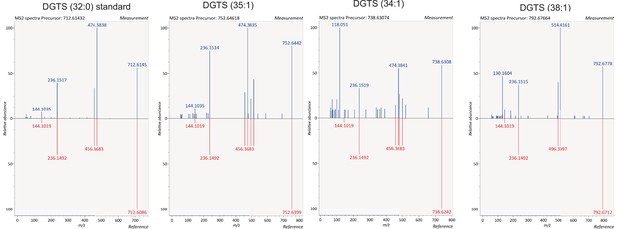
Identification of DGTS species.
Identification of DGTS species in lipids extracted from B4 and PfGDPD-null G1 parasites by comparing fragmentation spectra with a commercially available DGTS (32:0) standard.
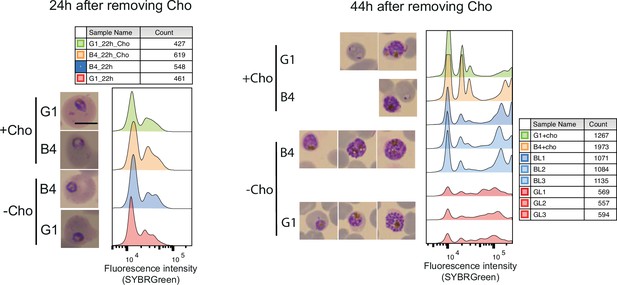
DNA content-based assessment of parasite development.
DNA content-based assessment of parasite development in choline-starved PfGDPD-null G1 and parental B4 parasites before lipid extraction. No difference in growth was observed between choline-supplemented cultures and cultures 24 hr after removing choline. However, a significant lag in development was observed in labelled G1 parasites (three replicates GL1, GL2, GL3) at 44 hr compared to B4 and choline-supplemented controls.
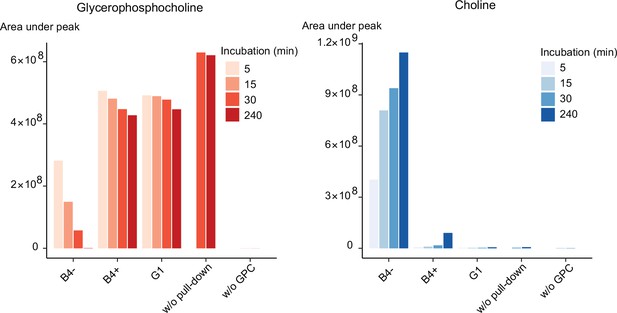
Purified PfGDPD releases choline from GPC.
GPC and choline content in enzymatic reactions set up with affinity-purified GDPD-HA from similar numbers of mock- (B4-) and RAP-treated (B4+) GDPD:loxPint:HA parasites or the GDPD-null clonal parasite line (G1). Pulled-down samples were incubated with 10 mM GPC in a reaction buffer containing 10 mM MgCl2 for different durations at 37°C. Reactions without pulled-down fraction or GPC substrate were included as controls.
-
Figure 6—source data 1
Source data for plotted graph in Figure 6 and full raw unedited versions of the blot images shown in Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/82207/elife-82207-fig6-data1-v2.zip
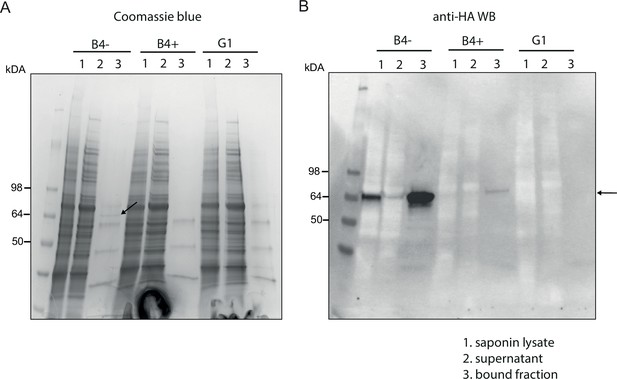
Affinity purification of PfGDPD-HA.
Affinity purification of PfGDPD-HA from GDPD:loxPint:HA and GDPD-null parasites. (A) SDS-PAGE stained with Coomassie blue showing saponin lysate, the bound and supernatant fractions. Arrow points to the band only present in mock-treated GDPD:loxPint:HA parasites. (B) Western blot with anti-HA antibody showing abundance of GDPD-HA in mock-treated GDPD:loxPint:HA, residual levels in RAP-treated GDPD:loxPint:HA and absence in GDPD-null clonal parasites.
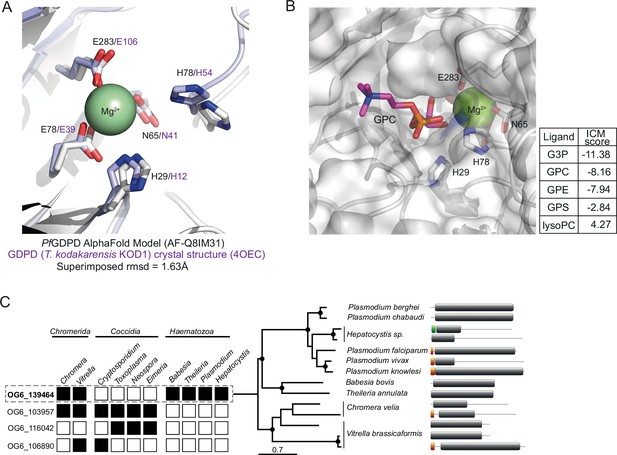
In silico substrate docking in PfGDPD model.
(A) Structural conservation of PfGDPD active site residues and the Mg2+ binding site. The AlphaFold model of PfGDPD (AF-Q8IM31; shown as a light grey cartoon) indicates structural conservation of the active site residues and the metal ion binding site (coloured sticks) when superimposed onto its closest structural analogue, the magnesium-dependent marine phosphodiesterases KOD1 from Thermococcus kodarensis (4OEC, rmsd = 1.63 Å; tinted purple cartoon). (B) In silico docking and simulation of substrate specificity of PfGDPD. GPC is shown as a stick (C in pink, N in blue, P in orange, O in red), docked into the active site of PfGDPD (unitless ICM-Pro score –10). The GPC phosphate group is found in the vicinity of the active site residues His29 and His78 and the Mg2+ metal ion. ICM docking scores were low (around –10) possibly due to non-optimum side chain conformations in the active site pocket residues of the rigid PfGDPD receptor AlphaFold model. Docking of G3P, GPC, glycerophosphoethanolamine (GPE) and glycerophosphoserine (GPS) were successful with a preference for G3P, GPC, and GPE. As expected, docking with lysoPC (16:0) did not perform well suggesting a low preference for PfGDPD. These results suggest that PfGDPD has a substrate preference for GPC and GPE, but activity against GPS cannot be ruled out. (C) Orthology analysis of GDPD catalytic domain-containing proteins across all apicomplexan parasites and their chromerid ancestors reveals four distinct ortholog groups. Four ortholog groups can be identified within apicomplexan parasites and their algal ancestors (Chromera and Vitrella) with member orthologous genes present (black box) or absent (white box) in some organisms. Orthologs of PfGDPD (PF3D7_1406300) (OG6_139464) are found only the Haematozoan group of apicomplexan parasites (i.e. those possessing an intra-erythrocytic life cycle). Maximum likelihood phylogeny inferred from multiple sequence alignment and the protein domain information for the GDPD orthologs are shown (nodes with bootstrap values >80 are marked; domains: grey, GP-PDE domain; orange, signal peptide; green, transmembrane domain).
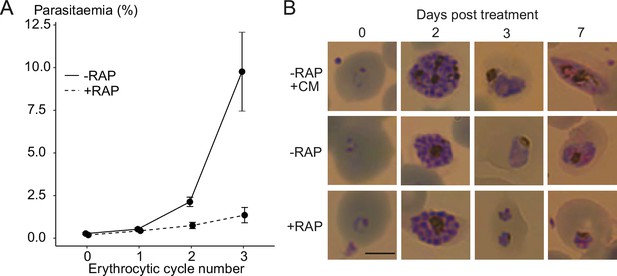
Ablation of GDPD expression does not induce sexual differentiation.
(A) Replication of mock- (solid line) and RAP-treated (dashed line) clonal line of GDPD:loxPint:HANF54 parasites over three erythrocytic cycles (error bars, ± SD). Data shown are averages from triplicate biological replicates using different blood sources. (B) Light microscopic images of Giemsa-stained GDPD:loxPint:HANF54 parasites at days 0, 2, 3 and 7 post treatment with conditioned media (-RAP+CM, known to induce sexual commitment), DMSO (-RAP) or rapamycin (+RAP). Gametocyte stages were apparent from day 6–7 in cultures treated with conditioned media while DMSO-treated cultures showed normal asexual stage progression and RAP-treated cultures showed development-stalled trophozoite stages from day 3. Images are representative of three independent treatments. Scale bar, 5 µm.
-
Figure 7—source data 1
Source data for plotted graph in Figure 7A and full raw unedited versions of the gel image shown in Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/82207/elife-82207-fig7-data1-v2.zip
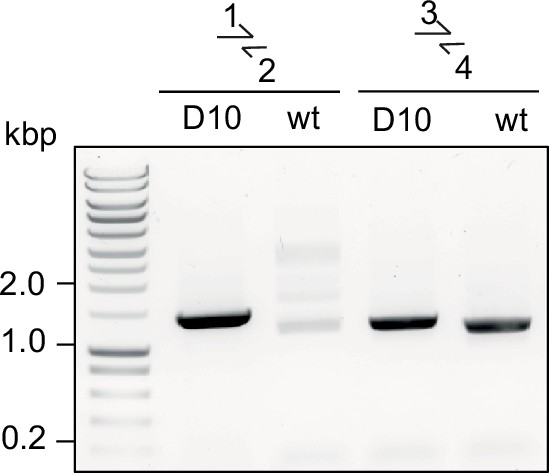
Diagnostic PCR for successful integration in GDPD:loxPint:HANF54 line.
Diagnostic PCR showing correct integration of the pREP-GDPD modification plasmid in the PfGDPD locus in GDPD:loxPint:HANF54 parasites (clonal line D10). Primers used are denoted in Figure 2B.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Plasmodium falciparum) | PfGDPD | PlasmoDB (https://plasmodb.org) | PF3D7_1406300 | P. falciparum GDPD gene |
| Genetic reagent (P. falciparum) | GDPD:HA:loxPint | This paper | For inducible disruption of PfGDPD in B11 line. Line maintained in and available from Blackman lab, Francis Crick Institute. | |
| Genetic reagent (P. falciparum) | GDPD:HA:loxPintNF54 | This paper | For inducible disruption of PfGDPD in NF54::DiCre line. Line maintained in and available from Blackman lab. | |
| Genetic reagent (P. falciparum) | G1 | This paper | Clonal GDPD-null line supplemented with choline. Line maintained in and available from Blackman lab. | |
| Genetic reagent (P. falciparum) | GDPD:GFP | This paper | Endogenous tagging of PfGDPD with GFP. Line maintained in and available from Gilberger lab at Centre for Structural Systems Biology, Hamburg. | |
| Genetic reagent (P. falciparum) | GDPD:GFP +EpiSP-mScarlet | This paper | Endogenous tagging of PfGDPD with GFP. Episomal expression of PV marker protein SP-mScarlet. Line maintained in and available from Gilberger lab. | |
| Genetic reagent (P. falciparum) | GDPD:loxPint:HA:Neo-R | This paper | For inducible disruption of PfGDPD. Generated using SLI system. Line maintained in and available from Gilberger lab. | |
| Cell line (P. falciparum) | GDPD:loxPint:HA:Neo-R+EpiNMD3:GDPD-mCherry | This paper | For inducible disruption of PfGDPD. Episomal expression of GDPD-mCherry. Line maintained in and available from Gilberger lab | |
| Cell line (P. falciparum) | GDPD:loxPint:HA:Neo-R+EpiNMD3:GDPD(H29A)-mCherry | This paper | For inducible disruption of PfGDPD. Episomal expression of GDPD(H29A)-mCherry. Line maintained in and available from Gilberger lab | |
| Cell line (P. falciparum) | GDPD:loxPint:HA:Neo-R+EpiNMD3:GDPD(H78A)-mCherry | This paper | For inducible disruption of PfGDPD. Episomal expression of GDPD(H78A)-mCherry. Line maintained in and available from Gilberger lab | |
| Cell line (P. falciparum) | GDPD:loxPint:HA:Neo-R+EpiNMD3:GDPD(E283A)-mCherry | This paper | For inducible disruption of PfGDPD. Episomal expression of GDPD(E283A)-mCherry. Line maintained in and available from Gilberger lab | |
| Cell line (P. falciparum) | B11 | Perrin et al., 2018 | DiCre-expressing 3D7 parasite line. Maintained in and available from Blackman lab, Francis Crick Institute. | |
| Cell line (P. falciparum) | NF54::DiCre | Tibúrcio et al., 2019 | DiCre-expressing NF54 parasite line. Maintained in and available from Treeck lab, Francis Crick Institute. | |
| Transfected construct (P. falciparum) | pCas9_1406300_gRNA01 | This paper | Cas9-targeting plasmid for producing GDPD:loxPint:HA line. Available from Blackman lab. | |
| Transfected construct (P. falciparum) | pCas9_1406300_gRNA02 | This paper | Cas9-targeting plasmid for producing GDPD:loxPint:HA line. Available from Blackman lab. | |
| Transfected construct (P. falciparum) | pREP-GDPD | This paper | Repair plasmid for producing GDPD:loxPint:HA line. Available from Blackman lab. | |
| Transfected construct (P. falciparum) | pSLI-PF3D7_1406300-GFP-GlmS-WT | This paper | GFP-tagging construct for producing GDPD:GFP line. Available from Gilberger lab. | |
| Transfected construct (P. falciparum) | pSLI-PF3D7_1406300-TGD | This paper | SLI-based construct for testing essentiality of PfGDPD. Available from Gilberger lab. | |
| Transfected construct (P. falciparum) | pSLI- PF3D7_1406300-loxPint:HA:T2A:Neo | This paper | SLI-based construct for producing GDPD:loxPint:HA:Neo-R line. Available from Gilberger lab. | |
| Transfected construct (P. falciparum) | pSkipFlox | Birnbaum et al., 2017 | Plasmid for ectopic expression of DiCre in GDPD:loxPint:HA:Neo-R line. | |
| Transfected construct (P. falciparum) | pNMD3:PF3D7_1406300-mCherry-DHODH | This paper | Gene complementation vector for GDPD:loxPint:HA:Neo-R line leading to episomal expression of GDPD-mCherry. Available from Gilberger lab. | |
| Transfected construct (P. falciparum) | pNMD3:PF3D7_1406300(H29A)-mCherry-DHODH | This paper | Gene complementation vector for GDPD:loxPint:HA:Neo-R line leading to episomal expression of GDPD(H29A)-mCherry. Available from Gilberger lab. | |
| Transfected construct (P. falciparum) | pNMD3:PF3D7_1406300(H78A)-mCherry-DHODH | This paper | Gene complementation vector for GDPD:loxPint:HA:Neo-R line leading to episomal expression of GDPD(H78A)-mCherry. Available from Gilberger lab. | |
| Transfected construct (P. falciparum) | pNMD3:PF3D7_1406300(E283A)-mCherry-DHODH | This paper | Gene complementation vector for GDPD:loxPint:HA:Neo-R line leading to episomal expression of GDPD(E283A)-mCherry. Available from Gilberger lab. | |
| Transfected construct (P. falciparum) | SP-mScarlet | Mesén-Ramírez et al., 2019 | PV marker for GDPD:GFP line. | |
| biological sample (Homo sapiens) | Human red blood cells | UK NHS Blood and Transplant; University Medical Center Hamburg- Eppendorf (UKE), Germany | Provided anonymised. | |
| Antibody | 3 F10 High affinity anti-HA (rat monoclonal) | Roche | Cat# 11867423001, RRID:AB_390918 | IFA (1:500), western blot (1:1000) |
| Antibody | Biotinylated anti-rat (goat polyclonal) | Sigma-Aldrich | Cat# AP183B, RRID:AB_92595 | IFA (1:1000), western blot (1:8000) |
| Antibody | anti-aldolase (rabbit polyclonal) | Mesén-Ramírez et al., 2016 | IFA (1:2000) | |
| Antibody | goat-anti-rat-800CW (goat polyclonal) | LI-COR Biosciences | Cat# 925–32219, RRID:AB_2721932 | Western blot (1:10,000) |
| Antibody | goat-anti-rabbit-680RD (goat polyclonal) | LI-COR Biosciences | Cat# 925–68071, RRID:AB_2721181 | Western blot (1:10,000) |
| Antibody | Goat anti-rat-AlexaFluor 594 (goat polyclonal) | ThermoFisher | Cat# A-11007, RRID:AB_10561522 | IFA (1:2000) |
| Chemical compound, drug | AlexaFluor 594 conjugated Streptavidin | ThermoFisher | Cat# S32356 | |
| Chemical compound, drug | Streptavidin peroxidase | Sigma-Aldrich | Cat# S2438 | |
| Chemical compound, drug | WR99210 | Sigma-Aldrich | Cat# W1770 | |
| Chemical compound, drug | Rapamycin | Sigma-Aldrich | Cat# R0395-1MG | |
| Chemical compound, drug | Compound 2 (4-[7-dimethylamino)methyl]−2-(4-fluorphenyl)imidazo[1,2-α]pyridine-[3-yl]pyrimidin-2-amine | LifeArc (https://www.lifearc.org/) | Kindly provided by Dr. Simon A. Osborne (LifeArc). | |
| Chemical compound, drug | SYBR Green I | ThermoFisher | Cat# S7563 | |
| Chemical compound, drug | rapalog (AP21967) | Clontech | Cat# 635057 | |
| Chemical compound, drug | Neomycin/G418 | Sigma-Aldrich | Cat# G418-RO | 400 μg/ml |
| Chemical compound, drug | blasticidin S HCl | Invitrogen | Cat# R21001 | 2 μg/ml |
| Chemical compound, drug | DSM1 | BEI Resources | 0.9 μM | |
| Chemical compound, drug | choline chloride | Sigma-Aldrich | Cat# C7017 | 1 mM |
| Chemical compound, drug | ethanolamine | Sigma-Aldrich | Cat# E9508 | 100 μM |
| Chemical compound, drug | L-serine | Sigma-Aldrich | Cat# S4500 | 2 mM |
| Chemical compound, drug | sn-glycero-3-phosphocholine | Cayman chemical | Cat# 20736 | 1 mM |
| Chemical compound, drug | 2H choline-labelled lysoPC | Brancucci et al., 2017 | 20 μM. Kindly provided by Dr. Matthias Marti. | |
| Chemical compound, drug | DGTS 32:0 | Avanti Polar Lipids | Cat# 857464 | |
| Commercial assay, kit | Ligation Sequencing Kit | Oxford Nanopore Technologies | Cat# SQK-LSK109 | |
| Commercial assay, kit | Native Barcoding Expansion 1–12 | Oxford Nanopore Technologies | Cat# EXP-NBD104 | |
| Commercial assay, kit | MinION flow cell | Oxford Nanopore Technologies | Cat# R9.4.1 | |
| Software, algorithm | BD FACSDiva software | BD Bioscience | RRID:SCR_001456 | |
| Software, algorithm | FlowJo for Mac (version 10.3.0) software | Becton Dickinson Life Sciences | RRID:SCR_008520 | |
| Software, algorithm | Fiji (Image J version 2.0) software | Imagej.net | RRID:SCR_003070 | |
| Software, algorithm | Thermo Xcalibur v3.0.63 software | Thermo Scientific | RRID:SCR_014593 | |
| Software, algorithm | Free Style v1.5 | Thermo Scientific | RRID:SCR_022877 | |
| Software, algorithm | Progenesis QI | Nonlinear Dynamics | RRID:SCR_018923 | |
| Software, algorithm | LipidMatch | Koelmel et al., 2017 | ||
| Software, algorithm | TraceFinder v5.1 | Thermo Scientific | ||
| Software, algorithm | MS-Dial v4.80 | Tsugawa et al., 2015 | ||
| Software, algorithm | MinKNOW v20.10 | Oxford Nanopore Technologies; https://community.nanoporetech.com/downloads | ||
| Software, algorithm | Guppy v3.2.2 | Oxford Nanopore Technologies; https://community.nanoporetech.com/downloads | ||
| Software, algorithm | IGV v2.9.4 | Robinson et al., 2011; https://software.broadinstitute.org/software/igv/ | ||
| Software, algorithm | PDBeFold server | https://www.ebi.ac.uk/msd-srv/ssm/ | ||
| Software, algorithm | ICM-Pro v3.9–1 c/MacOSX | Molsoft LLC; https://www.molsoft.com/icm_pro.html | ||
| Software, algorithm | Muscle v3.8.31 | Edgar, 2004 | RRID:SCR_011812 | |
| Software, algorithm | trimAl v1.2 | Capella-Gutiérrez et al., 2009 | RRID:SCR_017334 | |
| Software, algorithm | RAxML | Stamatakis, 2014 | RRID:SCR_006086 | |
| Software, algorithm | the iTOL server | Letunic and Bork, 2021 | RRID:SCR_018174 | |
| Software, algorithm | InterProScan | Jones et al., 2014 | RRID:SCR_006695 | |
| Software, algorithm | myDomains | Hulo et al., 2008 | ||
| Software, algorithm | R v4.0.2 | http://www.r-project.org/ | RRID:SCR_001905 | |
| Software, algorithm | ggplot2 | https://cran.r-project.org/web/packages/ggplot2/index.html | RRID:SCR_014601 | |
| Software, algorithm | RStudio | http://www.rstudio.com/ | RRID:SCR_000432 | |
| Other | Pierce Anti-HA Magnetic Beads | Thermo Scientific | Cat# 88836 | Magnetic beads conjugated with highly specific anti-HA monoclonal antibody (clone 2–2.2.14). For immunoprecipiation of HA-tagged proteins. |
| Other | AMPure XP beads | Beckman Coulter | Cat# A63881 | Paramagnetic beads that selectively binds to DNA of length greater than 100 bp. Used for high recovery/purification of genomic or amplicons from other contaminants. |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/82207/elife-82207-mdarchecklist1-v2.pdf
-
Supplementary file 1
Sequences of all oligos and synthesized constructs used in this study.
- https://cdn.elifesciences.org/articles/82207/elife-82207-supp1-v2.xlsx





