Survival of mineral-bound peptides into the Miocene
Abstract
Previously, we showed that authentic peptide sequences could be obtained from 3.8-Ma-old ostrich eggshell (OES) from the site of Laetoli, Tanzania (Demarchi et al., 2016). Here, we show that the same sequences survive in a >6.5 Ma OES recovered from a palaeosteppe setting in northwestern China. The eggshell is thicker than those observed in extant species and consistent with the Liushu Struthio sp. ootaxon. These findings push the preservation of ancient proteins back to the Miocene and highlight their potential for paleontology, paleoecology, and evolutionary biology.
Editor's evaluation
This fundamental study substantially pushes the known preservation of protein sequences bound to mineral surfaces. The successful recovery of these sequences from late Miocene fossil eggshell also has important implications for taxonomic classification. This solid work encourages future paleoproteomic research on paleontological remains from deep antiquity and across various taxa. The paper will be of great interest to a wide range of paleoscientists.
https://doi.org/10.7554/eLife.82849.sa0Introduction
The oldest authenticated peptide sequences to date were reported in 2016 from 3.8-Ma-old ostrich eggshell (OES) from the site of Laetoli, Tanzania (Demarchi et al., 2016). This finding had great scientific impact since it integrated computational chemistry (molecular dynamics simulations) as well as experimental data to propose a mechanism of preservation, concluding that mineral binding ensures the survival of protein sequences. Importantly, this study demonstrated that peptide-bound amino acids could survive into deep time even in hot environments. The effect of temperature on the kinetics of protein diagenesis has been described by several authors, both on the basis of actualistic experiments and of the quantification of the extent of degradation in ancient samples of known ages (Crisp et al., 2013; Demarchi et al., 2013; Hendy et al., 2012; Johnson et al., 1997; Kaufman, 2006; Kaufman, 2003; Schroeder and Bada, 1976; Wehmiller, 2013; Wehmiller, 1977; Wehmiller and Belknap, 1978). This discovery has fuelled the analysis of ancient proteins from other mineral matrices, namely tooth enamel (Cappellini et al., 2019; Welker et al., 2020; Welker et al., 2019) in order to reconstruct phylogenies.
The recovery of peptides has so far been limited to biomineral samples of Plio-Pleistocene age: our attempts at retrieving intact peptides from Cretaceous eggshell were unsuccessful, despite the fact that a genuine intracrystalline fraction of amino acids was preserved in the same sample (Saitta et al., 2020). Here, we show that the same peptide sequences that exhibit strong binding to the calcite surface—and that were recovered from the Laetoli OES—also persist into the late Miocene, in a >6.5-Ma-old eggshell sample from the Linxia Basin, northeastern Tibetan Plateau, China, Liushu Formation.
Results and discussion
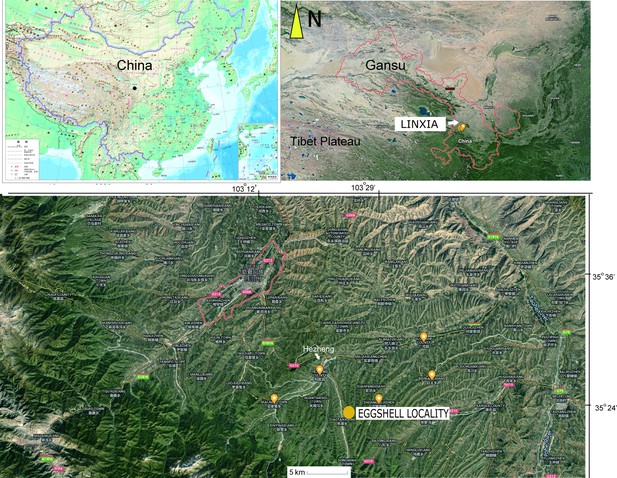
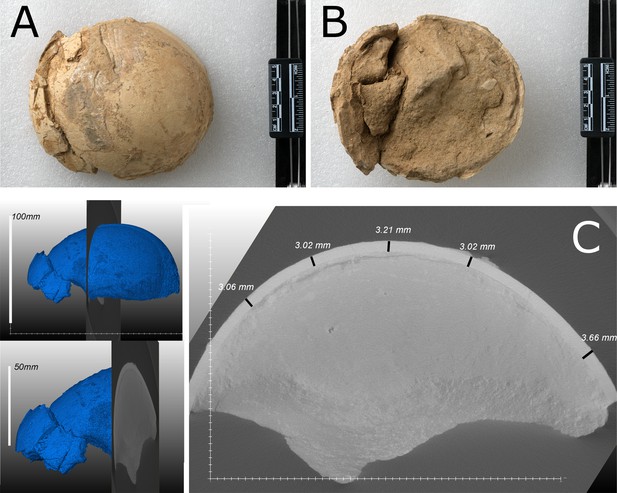
The OES (IVPP V26107) was recovered from an area between the towns of Xinji and Songming, in Hezheng County, close to the border with Guanghe County, from mudstone facies of the Liushu Formation (Figure 1).
Both eggshell and skeletal remains are previously known from the Late Miocene Liushu Formation, Linxia Basin, of Gansu Province, where the mean annual temperature is ∼11◦C (Hou et al., 2005; Liu et al., 2016; Li et al., 2021; Wang, 2008). Age control on the highly fossiliferous units of the Lishu Formation is good (Deng et al., 2019; Deng et al., 2013; Zhang et al., 2012), with detailed correlations across China as well as with Neogene deposits globally. These units have been assessed via new magnetostratigraphic data; most of the Hipparion fauna is estimated to be earlier than 6.4–6.5 Ma, with a lower Liushu transition to a Platybelodon fauna yielding age estimates of 11.1–12.5 Ma (Zhang et al., 2012). These dates agree with the prior estimates from biostratigraphic and magnetostratigraphic data (Deng et al., 2019; Deng et al., 2013). The recovery site is closer to the southern part of the basin near the Heilinding section of Zhang et al., 2012, which had an estimated minimum age of 6.4–6.5 Ma (Chron 3An). In contrast, the candidate stratotype section, Guoniguo, that exposes lower parts of the Liushu, is north of the recovery area (Deng et al., 2019; Zhang et al., 2012).
Ostrich (Struthio) remains to have a long history of recovery from the late Miocene of northwest China (Buffetaut and Angst, 2021; Hou et al., 2005; Li et al., 2021; Lowe, 1931; Mikhailov and Zelenkov, 2020). The first reported ostrich fossils from China were from units of the ‘Hipparion Clay’ (Red Clay) dated between 6.54 and 7.18 Ma (Lowe, 1931; Zhu et al., 2008). Mikhailov and Zelenkov, 2020 concluded that the Wang, 2008 Liushu taxon is referable to Struthio and not presently supported as a distinct but related genus; it is proposed to be from a taxon larger than extant Struthio species with a thickness of ~2.4 mm consistent with the eggshell sampled here (Figure 2C).
Given the extreme antiquity of the samples, protein extraction was performed in an ultra-clean facility at the University of Copenhagen in order to minimize any chance of contamination, following Hendy et al., 2018. Furthermore, OES powders were bleached extensively to isolate the intracrystalline fraction only (as described by Demarchi et al., 2016), samples were analysed by LC-MS/MS using a new LC column, and six blanks (analytical and procedural) were included in the run, flanking the sample. Data analysis was equally cautious: raw tandem mass spectrometry data were used to reconstruct potential peptide sequences starting from raw product ion spectra; only de novo peptides with an ALC (Average Local Confidence) score ≥80% were considered. This is the most stringent threshold that can be applied, and it signifies that all potential peptide sequences with lower confidence are discarded. The de novo peptides were searched against the Uniprot/Swissprot database (containing 565,254 manually annotated and reviewed protein sequences). A database of common laboratory contaminants (common Repository of Adventitious Protein [cRAP]) was included in the search. Eleven unique peptide sequences were found to match the sequence of struthiocalcin-1 (Table 1), all containing the typical Asp-rich motif ‘DDDD’ (Figures 3 and 4), which had been shown in our previous paper to be the mineral-binding peptide belonging to the sequence of struthiocalcin-1 (SCA-1). Annotated tandem mass spectra are shown in Figure 3 (ALDDDDYPKG) and Figure 3—figure supplement 1 (ALDDDYPK), Figure 3—figure supplement 2 (SALDDDDYPKG), Figure 3—figure supplement 3 (DDDDYPKGKH), Figure 3—figure supplement 4 (LDDDDYPKGK), Figure 3—figure supplement 5 (SALDDDDYPK), Figure 3—figure supplement 6 (DDDDYPKGK), Figure 3—figure supplement 7 (LDDDDYPKG), Figure 3—figure supplement 8 (DDDYPKGK), Figure 3—figure supplement 9 (DDDDYPK), and Figure 3—figure supplement 10 (DDYPKGK).
Peptide sequences identified in sample IVPP V26107.
| Scan | Precursor mass | z | m/z | Peptide sequence | Peptide mass | Error (ppm) | Peptide –10lgP |
|---|---|---|---|---|---|---|---|
| 5401 | 1107.472 | 2 | 555.246 | ALDDDDYPKG | 1107.472 | 5.7 | 27.24 |
| 5268 | 1050.457 | 2 | 526.236 | ALDDDDYPK | 1050.451 | 6.1 | 24.83 |
| 5051 | 1194.517 | 2 | 598.266 | SALDDDDYPKG | 1194.504 | 10.9 | 21.11 |
| 4984 | 1137.494 | 2 | 569.754 | SALDDDDYPK | 1137.483 | 10.1 | 29.94 |
| 4727 | 1137.494 | 2 | 569.755 | SALDDDDYPK | 1137.483 | 9.7 | 16.76 |
| 4565 | 1107.483 | 2 | 554.749 | ALDDDDYPKG | 1107.472 | 9.8 | 32.57 |
| 4565 | 1107.484 | 2 | 554.749 | ALDDDDYPKG | 1107.472 | 10.6 | 28.7 |
| 4443 | 1050.459 | 2 | 526.237 | ALDDDDYPK | 1050.451 | 7.6 | 17.62 |
| 4313 | 1050.461 | 2 | 526.235 | ALDDDDYPK | 1050.451 | 9.9 | 15.13 |
| 4199 | 1050.458 | 2 | 526.236 | ALDDDDYPK | 1050.451 | 7.2 | 16.35 |
| 4037 | 1036.442 | 2 | 519.230 | LDDDDYPKG | 1036.435 | 7.2 | 16.57 |
| 2724 | 1164.539 | 2 | 583.277 | LDDDDYPKGK | 1164.53 | 8.2 | 30.91 |
| 2502 | 866.335 | 2 | 434.176 | DDDDYPK | 866.335 | 6.9 | 18.46 |
| 2176 | 866.335 | 2 | 434.175 | DDDDYPK | 866.335 | 7.1 | 21.18 |
| 2026 | 866.338 | 2 | 434.176 | DDDDYPK | 866.329 | 10.1 | 19.07 |
| 1396 | 1051.454 | 2 | 526.734 | DDDDYPKGK | 1051.446 | 7.8 | 28.62 |
| 1138 | 936.427 | 2 | 469.221 | DDDYPKGK | 936.419 | 8.4 | 27.8 |
| 1046 | 1188.513 | 3 | 397.178 | DDDDYPKGKH | 1188.505 | 7.2 | 30.65 |
| 1042 | 821.398 | 2 | 411.706 | DDYPKGK | 821.392 | 7.6 | 17.39 |
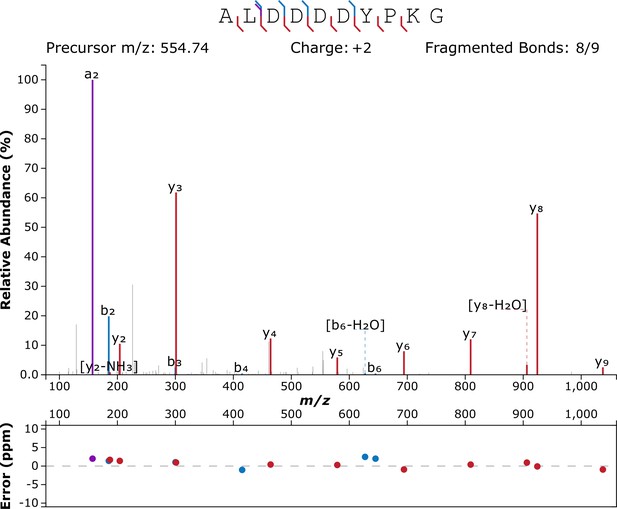
Annotated product ion spectrum of peptide ALDDDDYPKG, m/z 554.749, −10lgP=32.57.
Figure created using http://www.interactivepeptidespectralannotator.com/PeptideAnnotator.html (Brademan et al., 2019).
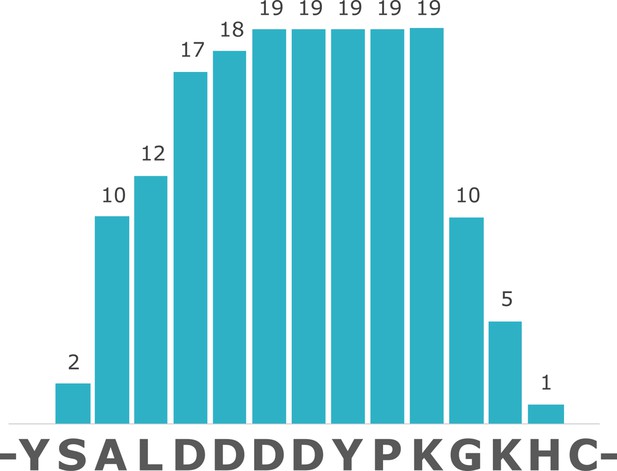
Coverage of struthiocalcin-1 (SCA-1) in Miocene OES specimen IVPP V26107.
Numbers indicate spectrum matches for each position in the sequence.
Conclusions
We present the first evidence for peptide survival into the Miocene, confirming our previous (Pliocene) data and providing further support to the mechanism of preservation based on the binding of Asx-rich peptides to calcite surfaces (Demarchi et al., 2016). The sequence recovered from the Chinese specimen is identical to the peptides found in the 3.8 Ma Laetoli OES, thus also supporting the attribution of the Liushu ootaxon to genus Struthio. While the four Asp residues are conserved across several avian taxa, in some species, including other ratites, Asp can be substituted by Glu and some of the flanking residues are also variable (Demarchi et al., 2022, fig. S1); therefore, variability within this sequence could be informative for evolutionary relationships between extinct and extant taxa. The absolute age of the Chinese sample is greater than that of the African material (Demarchi et al., 2016), but, given the latitudinal differences between the two sites, it is likely to have experienced lower temperatures throughout its burial history. This suggests that sequences of even greater antiquity may be recovered from biominerals harboring a closed system of proteins, particularly from sites in cold environments (e.g., high altitude and/or latitude).
Materials and methods
Sample preparation and analysis
Request a detailed protocolA subsample of OES specimen IVPP V26107 was prepared in the ultra-clean facility at the University of Copenhagen, following the protocol of Demarchi et al., 2016; Demarchi et al., 2022 and omitting the digestion step. In brief, the fragment was powdered, bleached for 72 hr (NaOCl, 15% w/v) and demineralized in cold weak hydrochloric acid (0.6 M HCl). The acid was added in 200 µl increments until it stopped reacting, for a total volume of 1400 µl 0.6 M HCl. The extracts were exchanged in ammonium bicarbonate buffer (pH=7.8) using 3 kDa MWCO ultrafilters and the peptides were purified and concentrated using C18 Stage Tips (Cappellini et al., 2019; Rappsilber et al., 2007).
Prepared StageTips of the procedural blank and sample were eluted with 30 µl of 40% acetonitrile (ACN) 0.1% formic acid into a 96-well plate prior to LC-MS/MS analysis. To remove the ACN, the plate was vacuum centrifuged until approximately 5 µl remained. Samples were then resuspended with 6 µl of 0.1% trifluoroacetic acid (TFA) 5% ACN. Based on protein concentration results at 205 nm (NanoDrop, Thermo Fisher Scientific), 5 µl of each sample and procedural blank was then separated over a 77 min gradient by an EASY-nLC 1200 (Proxeon, Odense, Denmark) attached to a Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Germany) using a 15 cm column. The column (75 μm inner diameter) was made in-house, laser pulled, and packed with 1.9 μm C18 beads (Dr. Maisch, Germany). Parameters were the same as those already published for historical samples (Mackie et al., 2018). In short, MS1: 120k resolution, maximum injection time (IT) 25 ms, scan target 3E6. MS2: 60k resolution, top 10 mode, maximum IT 118 ms, minimum scan target 3E3, normalized collision energy of 28, dynamic exclusion 20 s, and isolation window of 1.2 m/z. Wash-blanks consisting of 0.1% TFA 5% ACN were also run in order to hinder cross-contamination. The LC-MS/MS run included, in this order: two wash blanks, one procedural blank, one wash blank, the OES sample, and two wash blanks. The data sets have been deposited to the ProteomeXchange Consortium via the Proteomics Identifications Database (PRIDE) partner repository with the identifier PXD035872.
Data analysis
Request a detailed protocolBioinformatic analysis was carried out using PEAKS Studio 8.5 (Bioinformatics Solutions Inc; Zhang et al., 2012). The Uniprot_swissprot database (downloaded 11/08/2021) was used for carrying out the searches and common contaminants were included (cRAP: http://www.thegpm.org/crap/). No enzyme was specified for the digestion and the tolerance was set to 10 ppm on the precursor and 0.05 Da on the fragments. The thresholds for peptide and protein identification were set as follows: peptide score −10lgP≥15, protein score −10lgP≥20, and de novo sequences scores (ALC%)≥80.
Data availability
Tandem mass spectra supporting peptide sequence identification are reported in Figure 3 and Figure 3 - Supplement 1 to 10. Raw mass spectrometry data and results of bioinformatics analysis are available via ProteomeXchange with identifier PXD035872.
-
PRIDEID PXD035872. Ostrich eggshell peptides survival into the Miocene (6-9 Ma).
References
-
Interactive peptide spectral annotator: a versatile web-based tool for proteomic applicationsMolecular & Cellular Proteomics 18:S193–S201.https://doi.org/10.1074/mcp.TIR118.001209
-
BookLate Cenozoic Biostratigraphy of the Linxia Basin, Northwestern China. Fossil Mammals of AsiaColumbia University Press.
-
Neogene integrative stratigraphy and timescale of ChinaScience China Earth Sciences 62:310–323.https://doi.org/10.1007/s11430-017-9155-4
-
Assessing amino acid racemization variability in coral intra-crystalline protein for geochronological applicationsGeochimica Cosmochimica Acta 86:338–353.https://doi.org/10.1016/j.gca.2012.02.020
-
A guide to ancient protein studiesNature Ecology & Evolution 2:791–799.https://doi.org/10.1038/s41559-018-0510-x
-
A Miocene ostrich fossil from Gansu province, northwest ChinaChinese Science Bulletin 50:1808.https://doi.org/10.1360/982005-575
-
The determination of late quaternary paleoenvironments at Equus cave, South Africa, using stable isotopes and amino acid racemization in ostrich eggshellPalaeogeography, Palaeoclimatology, Palaeoecology 136:121–137.https://doi.org/10.1016/S0031-0182(97)00043-6
-
Amino acid paleothermometry of Quaternary ostracodes from the Bonneville basin, UtahQuaternary Science Reviews 22:899–914.https://doi.org/10.1016/S0277-3791(03)00006-4
-
Exceptional preservation of an extinct ostrich from the Late Miocene Linxia basin of ChinaVertebrata PalAsiatica 59:229–244.
-
BookStruthious remains from northern China and Mongolia: with descriptions of Struthio wimani, Struthio anderssoni and Struthio mongolicus spp. novGeological Survey of China.
-
A review of the geochemical applications of the amino acid racemization reactionEarth-Science Reviews 12:347–391.https://doi.org/10.1016/0012-8252(76)90011-8
-
Reexamination of taxonomic assignment of Struthio linxiaensis Hou et al., 2005Acta Palaeontol Sin 47:362–368.
-
Amino acid studies of the Del Mar, California, midden site: apparent rate constants, ground temperature models, and chronological implicationsEarth and Planetary Science Letters 37:184–196.https://doi.org/10.1016/0012-821X(77)90163-7
-
PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identificationMolecular & Cellular Proteomics 11:M111.https://doi.org/10.1074/mcp.M111.010587
-
A new magnetostratigraphic framework for Late Neogene Hipparion Red Clay in the eastern loess plateau of ChinaPalaeogeography, Palaeoclimatology, Palaeoecology 268:47–57.https://doi.org/10.1016/j.palaeo.2008.08.001
Article and author information
Author details
Funding
Ministry of University and Research - Italy (Young Researchers - Rita Levi Montalcini)
- Beatrice Demarchi
Danish National Research Foundation (PROTEIOS (DNRF128))
- Meaghan Mackie
- Matthew J Collins
Alexander von Humboldt Foundation
- Julia Clarke
University of Texas at Austin
- Julia Clarke
National Natural Science Foundation of China
- Zhiheng Li
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
BD’s work was supported by a “Young Researchers - Rita Levi Montalcini” grant from the Italian Ministry of University and Research. MM and MJC were supported by the Danish National Research Foundation Award PROTEIOS (DNRF128). JC acknowledges the Alexander von Humboldt Foundation and the Jackson School of Geosciences and ZL and DT the Chinese National Science Foundation. The authors thank Prof. Jesper Velgaard Olsen at the Novo Nordisk Center for Protein Research for providing access and resources, which were also funded in part by a donation from the Novo Nordisk Foundation (Grant no. NNF14CC0001). For the purpose of open access, MJC is part of the Rights Retention Pilot at the University of Cambridge and has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising from this submission. The authors wish to thank the reviewers and editors for their feedback on this manuscript.
Copyright
© 2022, Demarchi et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,352
- views
-
- 158
- downloads
-
- 14
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Biochemistry and Chemical Biology
In healthy cells, cyclin D1 is expressed during the G1 phase of the cell cycle, where it activates CDK4 and CDK6. Its dysregulation is a well-established oncogenic driver in numerous human cancers. The cancer-related function of cyclin D1 has been primarily studied by focusing on the phosphorylation of the retinoblastoma (RB) gene product. Here, using an integrative approach combining bioinformatic analyses and biochemical experiments, we show that GTSE1 (G-Two and S phases expressed protein 1), a protein positively regulating cell cycle progression, is a previously unrecognized substrate of cyclin D1–CDK4/6 in tumor cells overexpressing cyclin D1 during G1 and subsequent phases. The phosphorylation of GTSE1 mediated by cyclin D1–CDK4/6 inhibits GTSE1 degradation, leading to high levels of GTSE1 across all cell cycle phases. Functionally, the phosphorylation of GTSE1 promotes cellular proliferation and is associated with poor prognosis within a pan-cancer cohort. Our findings provide insights into cyclin D1’s role in cell cycle control and oncogenesis beyond RB phosphorylation.
-
- Biochemistry and Chemical Biology
- Microbiology and Infectious Disease
Teichoic acids (TA) are linear phospho-saccharidic polymers and important constituents of the cell envelope of Gram-positive bacteria, either bound to the peptidoglycan as wall teichoic acids (WTA) or to the membrane as lipoteichoic acids (LTA). The composition of TA varies greatly but the presence of both WTA and LTA is highly conserved, hinting at an underlying fundamental function that is distinct from their specific roles in diverse organisms. We report the observation of a periplasmic space in Streptococcus pneumoniae by cryo-electron microscopy of vitreous sections. The thickness and appearance of this region change upon deletion of genes involved in the attachment of TA, supporting their role in the maintenance of a periplasmic space in Gram-positive bacteria as a possible universal function. Consequences of these mutations were further examined by super-resolved microscopy, following metabolic labeling and fluorophore coupling by click chemistry. This novel labeling method also enabled in-gel analysis of cell fractions. With this approach, we were able to titrate the actual amount of TA per cell and to determine the ratio of WTA to LTA. In addition, we followed the change of TA length during growth phases, and discovered that a mutant devoid of LTA accumulates the membrane-bound polymerized TA precursor.