Dynamic readout of the Hh gradient in the Drosophila wing disc reveals pattern-specific tradeoffs between robustness and precision
Figures
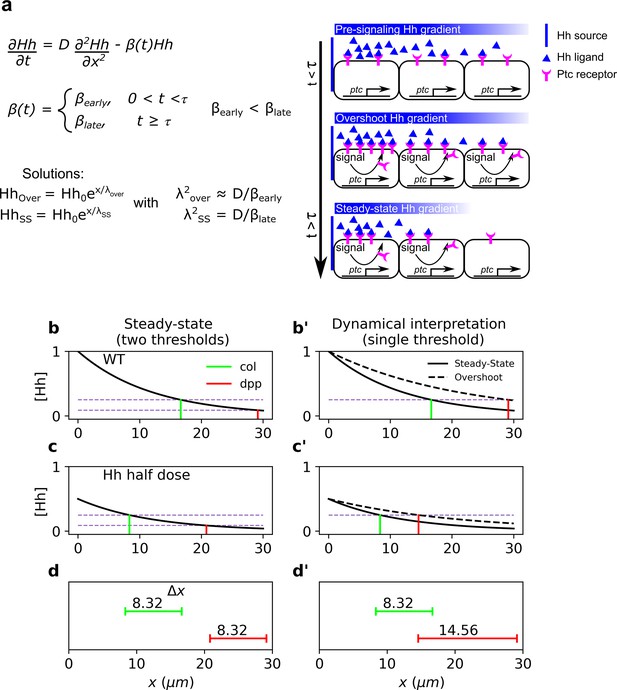
Dynamical interpretation model predicts differential robustness when morphogen dosage is reduced to half.
(a) Simple model of Hh signaling using a time-dependent step-wise degradation function. Diagrams displays a pre steady-state gradient that then retracts upon Hh-dependent ptc upregulation, resulting in a narrower gradient. (b, c) Plots of the analytical solution for the model in a using full (); (b, b’) or half (); (c, c’) Hh dosage. (d, d’) Displacements upon the above perturbation for the steady-state model with two thresholds (dotted horizontal lines corresponding to the locations of col and dpp) d; and for the dynamical interpretation model with a single-threshold readout (single dotted horizontal line) using the overshoot vs. the steady-state gradient predicts different shifts d’. The parameter values used for these plots are: , which approximately correspond to the anterior border positions of col and dpp, respectively. The color coding of dpp in red and col in green, will be used in the rest of the article.
-
Figure 1—source code 1
Code to generate Figure 1.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig1-code1-v2.zip
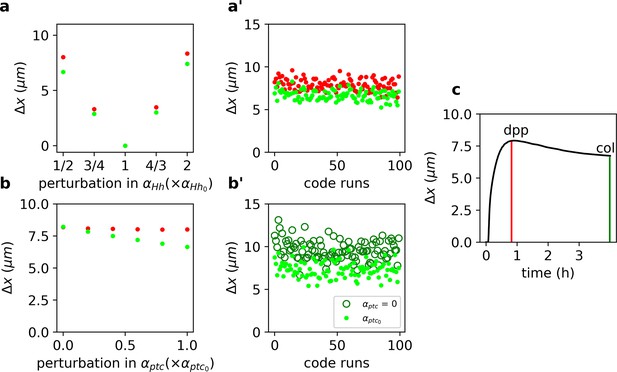
Target-specific robustness still holds in an explicit model of Hh signaling and it is dependent on Hh-dependent Ptc upregulation.
(a) (defined as in Equation 1, but for the function, see Materials and Methods) for overshoot (red) vs. steady-state (green) outputs upon different perturbations in using the values of the parameters reported in Nahmad and Stathopoulos, 2009 (Figure 2—source code 2 and 3 and Figure 2—source data 1, 2, and 6). (a’) defined and color coded as in a, for different combinations of parameter runs, when all parameters (other than ) are varied through a random normal distribution around the mean value with a standard deviation of 10% of the mean value (Figure 2—source data 2). (b) Same as a, but for perturbations in (Figure 2—source data 3). (b’) Comparison of for different parameters runs as in a’ for steady-state outputs (light green dots) and when (dark green empty circles; Figure 2—source data 4). (c) defined as in a, computed for the gradient over time (Figure 2—source data 5). Red and green vertical lines indicate the overshoot and steady-state values corresponding to the anterior borders of dpp and col, respectively (Figure 2—source code 1).
-
Figure 2—source code 1
Code to generate Figure 2.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-code1-v2.zip
-
Figure 2—source code 2
Code to solve steady-state solution of Equation 18.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-code2-v2.zip
-
Figure 2—source code 3
Code to solve transient solution of Equations 12–17.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-code3-v2.zip
-
Figure 2—source data 1
Raw data to generate Figure 2a.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-data1-v2.csv
-
Figure 2—source data 2
Raw data to generate Figure 2a’.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-data2-v2.csv
-
Figure 2—source data 3
Raw data to generate Figure 2b.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-data3-v2.csv
-
Figure 2—source data 4
Raw data to generate Figure 2b’.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-data4-v2.csv
-
Figure 2—source data 5
Raw data to generate Figure 2c.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-data5-v2.csv
-
Figure 2—source data 6
Parameters used to solve Equations 12–17 (same values as in Nahmad and Stathopoulos, 2009).
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig2-data6-v2.csv
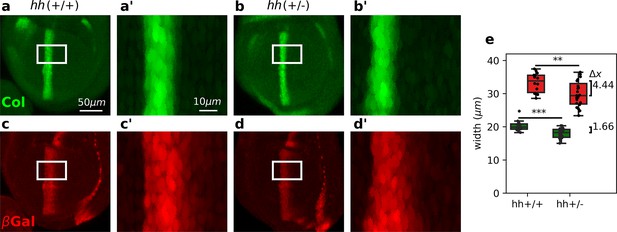
Differential robustness of Hh targets to hh dosage.
(a-d) Representative third-instar wild-type, hh(+/+) (a, c), and hh heterozygous hh(+/−) (b, d) wing discs immunostained with Col (a, b) and β-galactosidase (c, d) antibodies. Both hh(+/+) and hh(+/−) flies carry a transgene with a dppLacZ enhancer trap, so β-galactosidase marks the pattern of dpp expression. The scale bars in a, a’ apply to b, b’; c, c’; and d, d’ panels, respectively. (a’-d’) Enlarged areas of the white boxes shown in (a-d). (e) Widths of the col and dppLacZ patterns (color coded as in a–d) measured in the region marked by the white rectangle (see Figure 3—source data 1 and Figure 3—source code 1). The brackets on the right represent the difference between the medians of both groups. A non-parametric Mann–Whitney U test was applied in both cases (Figure 3—source data 1). Statistical p-values are for Col (**) and for dppLacZ (***). hh(+/−) discs (n = 14). hh(+/+) discs (n = 23). See Figure 3—source code 1.
-
Figure 3—source code 1
Code to generate Figure 3.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig3-code1-v2.zip
-
Figure 3—source data 1
Raw data represented in Figure 3e .
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig3-data1-v2.csv
-
Figure 3—source data 2
Raw data represented in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig3-data2-v2.csv
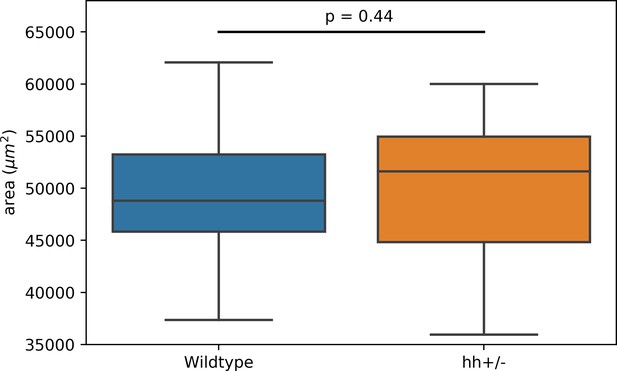
The wing disc pouch area does not change in mutant discs.
The pouch area was calculated as the area enclosed by the hinge-pouch folds. Statistical p-value after a Student t-test.
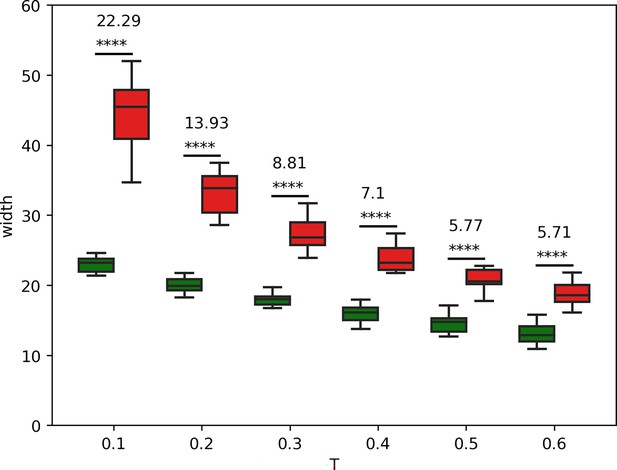
Differences in the width of col and dppLacZ patterns in hh(+/+) discs at different threshold values.
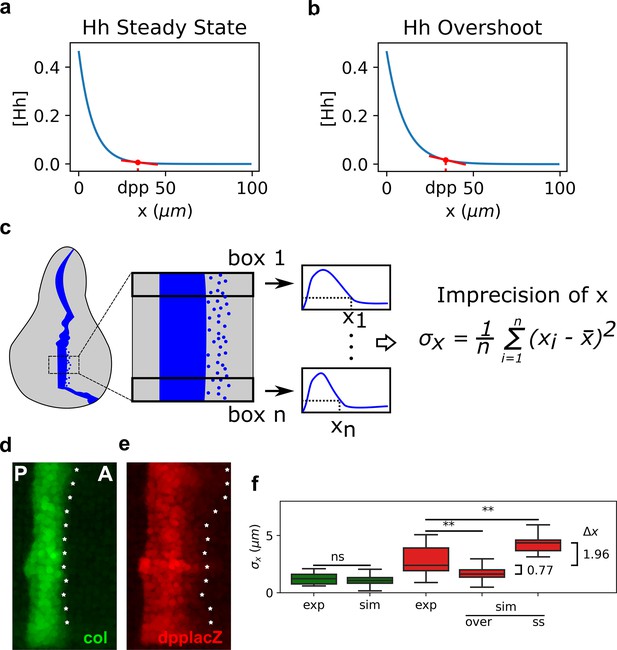
The overshoot model predicts more precision of the anterior border of dpp than the steady-state model.
(a, b). Representation of the steady-state (a) and overshoot (b) Hh gradients. At the location of the dpp anterior border, the slope of the gradient is steeper for the overshoot gradient than for the steady-state gradient. (c) Schematic representation of how we define our measure of precision for a patterning border (both in experimental and in simulated patterns). First, a box defines the region of interest (ROI) in the pattern. Then, this ROI is subdivided in boxes, each of which define a position . The measure of precision is the standard deviation of all the values. (d, e) Representative Col (d) and dppLacZ (e) patterns in which the for each ROI as defined in c is measured and marked with an asterisk along the anterior border. (f) Quantification of in several experimental (exp) and simulated (sim) patterns of col (green) and dpp (red). In the simulated patterns, noise levels are adjusted so that the distributions of col are not statistically significant and these noise levels are used to computed the simulated of dpp as determined by the steady state (ss) or overshot (over) models. exp sample sizes as in Figure 3. sim sample sizes is n = 50 in all cases. For the statistical comparison, a Mann–Whitney U tests were applied in all cases. Statistical p-value for col was . For experimental vs. overshoot dpp: (**), and for experimental vs. simulated steady-state dpp: (**).
-
Figure 4—source code 1
Code to generate Figure 4.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig4-code1-v2.zip
-
Figure 4—source data 1
Raw data represented in Figure 4f.
- https://cdn.elifesciences.org/articles/85755/elife-85755-fig4-data1-v2.csv
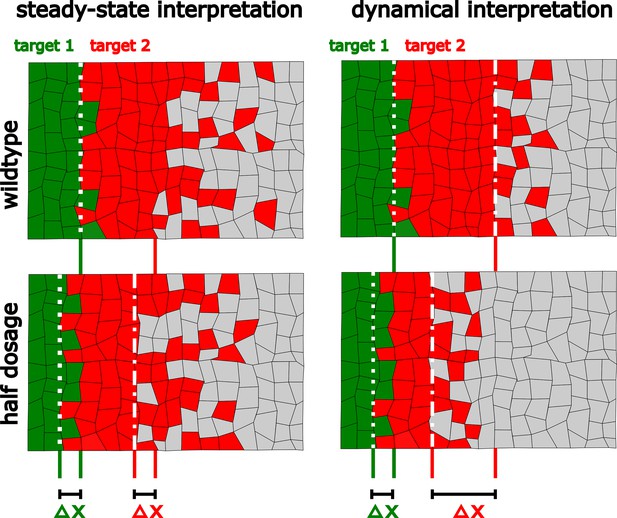
The dynamical interpretation of the Hh gradient trades off robustness for higher precision in a target-specific manner.
In the steady-state interpretation, all the target genes are established with the same robustness () upon perturbations in the amount of ligand. In the overshoot model interpretation one of the target genes (red) is established with less robustness than the other (green). However, it allows the less robust gene to be defined with greater precision than the steady state would define it (compare the sharpness of the boundaries of these patterns).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (Drosophila melanogaster) | hh(+/−) allele | Bloomington Drosophila Stock Center | 1749 | ry[506] hh[AC]/TM3, Sb. hh[AC] is an amorphic allele |
| strain, strain background (Drosophila melanogaster) | dppLacZ | Bloomington Drosophila Stock Center | 12379 | cn[1] dpp[10638]/CyO; ry[506]. dpp[10638] is a lacZ is a dpp enhancer trap. |
| antibody | anti-Col (mouse monoclonal) | Gift from M. Crozatier Vervoort et al., 1999 | 1:250; overnight incubation | |
| antibody | anti-β-gal (rabbit polyclonal) | MP Biomedicals | Cat. # 55976 | 1:250; overnight incubation |
| software, algorithm | Python | this paper | pandas; numpy; OpenCV; matplotlib; seaborn; odeint; solve_bvp | Customized source codes (available from this paper) |





