Calcineurin inhibition enhances Caenorhabditis elegans lifespan by defecation defects-mediated calorie restriction and nuclear hormone signaling
eLife Assessment
This important study reveals insights into how calcineurin influences C. elegans pathogen susceptibility and lifespan through its role in controlling the defecation motor program. The authors provide convincing evidence to support a new mechanism through which calcineurin impacts longevity. This work will be of interest to investigators studying host-pathogen interactions and aging in a number of experimental systems.
https://doi.org/10.7554/eLife.89572.3.sa0Important: Findings that have theoretical or practical implications beyond a single subfield
- Landmark
- Fundamental
- Important
- Valuable
- Useful
Convincing: Appropriate and validated methodology in line with current state-of-the-art
- Exceptional
- Compelling
- Convincing
- Solid
- Incomplete
- Inadequate
During the peer-review process the editor and reviewers write an eLife Assessment that summarises the significance of the findings reported in the article (on a scale ranging from landmark to useful) and the strength of the evidence (on a scale ranging from exceptional to inadequate). Learn more about eLife Assessments
Abstract
Calcineurin is a highly conserved calcium/calmodulin-dependent serine/threonine protein phosphatase with diverse functions. Inhibition of calcineurin is known to enhance the lifespan of Caenorhabditis elegans through multiple signaling pathways. Aiming to study the role of calcineurin in regulating innate immunity, we discover that calcineurin is required for the rhythmic defecation motor program (DMP) in C. elegans. Calcineurin inhibition leads to defects in the DMP, resulting in intestinal bloating, rapid colonization of the gut by bacteria, and increased susceptibility to bacterial infection. We demonstrate that intestinal bloating caused by calcineurin inhibition mimics the effects of calorie restriction, resulting in enhanced lifespan. The TFEB ortholog, HLH-30, is required for lifespan extension mediated by calcineurin inhibition. Finally, we show that the nuclear hormone receptor, NHR-8, is upregulated by calcineurin inhibition and is necessary for the increased lifespan. Our studies uncover a role for calcineurin in the C. elegans DMP and provide a new mechanism for calcineurin inhibition-mediated longevity extension.
eLife digest
Many research efforts currently focus on identifying the dietary, pharmacological or genetic interventions that could help to prolong life. In the process, these investigations often uncover complex or even unexpected relationships between a range of physiological processes. The link between longevity and the immune system, for example, is yet to be fully understood.
To explore these dynamics, Das et al. focused on calcineurin, an enzyme present in organisms across the tree of life. In humans, calcineurin is known to regulate a set of proteins essential for the immune response; these proteins are absent in the microscopic worm Caenorhabditis elegans, in which inhibiting calcineurin extends lifespan. Investigating how calcineurin inhibition impacts the immune system of C. elegans therefore presents a unique opportunity to better understand the complex links between immunity and longevity.
Experiments conducted on worms genetically modified to lack calcineurin showed that these animals lived longer than their ‘normal’ counterparts, but that they were also more susceptible to infection when exposed to a harmful species of bacteria. Further experiments showed that the enzyme was crucial for regulating defecation in C. elegans. Without calcineurin, the worms became bloated and constipated; they could not properly eliminate bacteria, which could then proliferate in the digestive system and cause issues. However, intestinal bloating also activated signalling pathways normally triggered by calorie restriction – an intervention well-known for extending the lifespan of various species.
Taken together, the findings by Das et al. help explain why calcineurin inhibition in C. elegans leads to opposite effects on longevity and resistance to infection. They also align with a recent body of work showing the profound effect of gut bloating on food-seeking behaviors, immunity and lifespan. Further investigations into these mechanisms may one day uncover new ways to improve human health.
Introduction
Interventions that enhance lifespan also impart resistance to multiple stresses (Johnson et al., 2001). Indeed, the positive correlation between improved stress resistance and enhanced lifespan has been exploited to identify long-lived mutants (de Castro et al., 2004; Denzel et al., 2014; Johnson et al., 2001; Muñoz and Riddle, 2003; Wang et al., 2004). Among stress responses, innate immunity appears to be a crucial factor for enhanced lifespan (Campos et al., 2021; Fabian et al., 2021; Soo et al., 2023; Xia et al., 2019). However, the correlation between innate immunity and lifespan is not always positive. Interventions that alter lifespan may not modulate innate immunity, and vice-versa (Labed et al., 2018; Naim et al., 2021; Otarigho and Aballay, 2020; Sun et al., 2011). Some signaling pathways also establish a tradeoff between innate immunity and lifespan. Mutants that have improved immunity but reduced lifespan have been identified (Amrit et al., 2019; Otarigho and Aballay, 2021; Ren and Ambros, 2015). Conversely, mutants with enhanced lifespans but declined immune responses have also been discovered (Kawli et al., 2010). Moreover, the genetic pathways for lifespan and immunity could be uncoupled in mutants exhibiting enhanced lifespan and improved immune responses (Alper et al., 2010; Guerrero et al., 2021). Therefore, the relationship between lifespan and innate immunity appears to be complex and remains to be fully understood.
Calcineurin, a conserved protein from yeast to humans, is a calcium/calmodulin-dependent serine/threonine protein phosphatase that is involved in diverse cellular processes and signal transduction pathways (Chen et al., 2022; Hogan et al., 2003; Schulz and Yutzey, 2004; Ulengin-Talkish and Cyert, 2023). Dephosphorylation of substrate proteins by calcineurin affects several cellular pathways, including transcriptional signaling programs (Ulengin-Talkish and Cyert, 2023). Calcineurin regulates the activity of the transcription factors of the nuclear factor of activated T cells (NFAT) family (Hogan et al., 2003). Dephosphorylation of NFATs by calcineurin triggers their nuclear localization and activates immune responses in vertebrates (Herbst et al., 2015; Hogan et al., 2003; Vandewalle et al., 2014). In the nematode Caenorhabditis elegans, calcineurin regulates thermotaxis, body size, fertility, and lifespan (Bandyopadhyay et al., 2002; Dong et al., 2007; Kuhara et al., 2002; Lee et al., 2013). Knockdown of the catalytic subunit of calcineurin, tax-6, is known to enhance C. elegans lifespan via multiple pathways, including autophagy and CREB-regulated transcriptional coactivators (CRTCs; Dong et al., 2007; Dwivedi et al., 2009; Mair et al., 2011; Tao et al., 2013). However, the role of calcineurin in regulating C. elegans response to pathogen infections has not been studied. Because C. elegans lacks the NFAT transcription factors (Song et al., 2013), it will be intriguing to study how calcineurin inhibition impacts C. elegans innate immunity. These studies could also shed some light on the complex interplay between lifespan and immunity.
In this study, we examined the effect of calcineurin inhibition on C. elegans innate immunity. Surprisingly, we found that the knockdown of tax-6 enhanced the susceptibility of C. elegans to bacterial infection despite enhancing lifespan. We discovered that tax-6 is required for the rhythmic defecation motor program (DMP). The knockdown of tax-6 resulted in intestinal bloating due to defects in the DMP, which enhanced susceptibility to bacterial infection by increasing gut colonization by bacteria. Intestinal bloating resulted in calorie restriction-like phenotypes, including reduced lipid levels, and led to increased lifespan. We discovered that the TFEB ortholog, HLH-30, is required for calcineurin inhibition-mediated lifespan extension. Moreover, we found that the nuclear hormone receptor, NHR-8, is upregulated by calcineurin inhibition and is necessary for increased lifespan. Our studies uncover a new mechanism for calcineurin inhibition-mediated longevity extension.
Results
Calcineurin knockdown enhances C. elegans susceptibility to Pseudomonas aeruginosa infection
To understand the role of calcineurin in the innate immune response of C. elegans, we examined the survival of a hypomorphic allele of tax-6, tax-6(p675), when exposed to the pathogenic bacterium P. aeruginosa PA14. Surprisingly, tax-6(p675) mutants showed significantly reduced survival on P. aeruginosa compared to wild-type N2 animals (Figure 1A). Similarly, animals carrying a null allele of tax-6, tax-6(ok2065), also exhibited a significant decrease in survival on P. aeruginosa compared to N2 animals (Figure 1B). In addition, knockdown of tax-6 by RNA interference (RNAi) increased susceptibility to P. aeruginosa relative to control animals (Figure 1C). The RNAi effects were specific to tax-6, as tax-6(p675) mutants did not show further increased susceptibility to P. aeruginosa when subjected to tax-6 RNAi (Figure 1D). As reported earlier (Dong et al., 2007; Dwivedi et al., 2009; Mair et al., 2011; Tao et al., 2013), we confirmed that both tax-6 loss of function and RNAi knockdown led to an increased lifespan on E. coli (Figure 1E and F). An earlier study demonstrated that calcineurin regulates cAMP response element-binding protein (CREB) and CRTCs to regulate lifespan in C. elegans (Mair et al., 2011). The knockdown of the CREB homolog-1 (crh-1) and crtc-1 enhanced lifespan similar to the tax-6 knockdown (Mair et al., 2011). We investigated whether crh-1 and crtc-1 knockdowns also affected C. elegans survival on P. aeruginosa as severely as tax-6 knockdown. Interestingly, crh-1 and crtc-1 knockdowns did not compromise survival on P. aeruginosa as much as tax-6 knockdown did (Figure 1—figure supplement 1). These findings suggested that, despite the increased lifespan, tax-6 knockdown animals exhibit markedly enhanced susceptibility to P. aeruginosa infection through a mechanism likely independent of crh-1 and crtc-1.
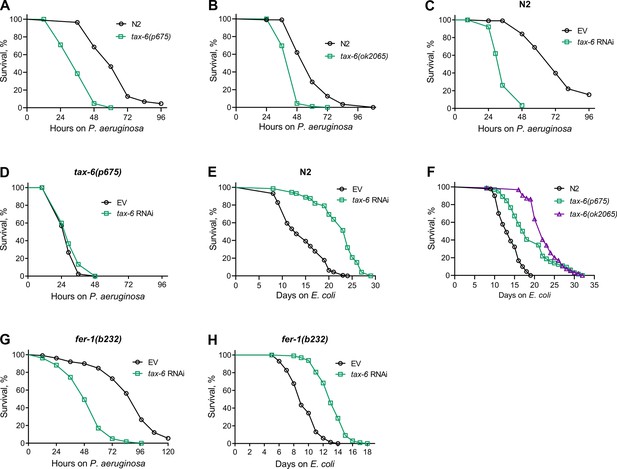
Calcineurin knockdown enhances C. elegans susceptibility to P. aeruginosa.
(A) Representative survival plots of N2 and tax-6(p675) animals on P. aeruginosa PA14 at 25 °C. The animals were grown on E. coli OP50 at 20 °C until 1-day-old adults before transferring to P. aeruginosa PA14 at 25 °C. p<0.001. (B) Representative survival plots of N2 and tax-6(ok2065) animals on P. aeruginosa PA14 at 25 °C. The animals were grown on E. coli OP50 at 20 °C until 1-day-old adults before transferring to P. aeruginosa PA14 at 25 °C. p<0.001. (C) Representative survival plots of N2 animals on P. aeruginosa PA14 at 25 °C after treatment with the empty vector (EV) control and tax-6 RNAi. p<0.001. (D) Representative survival plots of tax-6(p675) animals on P. aeruginosa PA14 at 25 °C after treatment with the EV control and tax-6 RNAi. n.s., nonsignificant. (E) Representative survival plots of N2 animals grown on bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. p<0.001. (F) Representative survival plots of N2, tax-6(p675), and tax-6(ok2065) animals grown on E. coli OP50 at 20 °C. Day 0 represents young adults. p<0.001 for tax-6(p675) and tax-6(ok2065) compared to N2. (G) Representative survival plots of fer-1(b232) animals on P. aeruginosa PA14 at 25 °C after treatment with the EV control and tax-6 RNAi at 25 °C. p<0.001. (H) Representative survival plots of fer-1(b232) animals grown on bacteria for RNAi against tax-6 along with the EV control at 25 °C. The animals were developed at 25 °C. Day 0 represents young adults. p<0.001. For all panels, n=3 biological replicates; animals per condition per replicate >60.
-
Figure 1—source data 1
Calcineurin knockdown enhances C. elegans susceptibility to P. aeruginosa.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig1-data1-v1.xlsx
We observed enhanced matricidal hatching in tax-6 knockdown animals on P. aeruginosa. Therefore, we asked whether the enhanced susceptibility of tax-6 knockdown animals to P. aeruginosa was because of enhanced matricidal hatching. To this end, we studied the effects of tax-6 knockdown in fer-1(b232) temperature-sensitive mutants. When grown at 25 °C, fer-1(b232) animals have unfertilized oocytes (Argon and Ward, 1980), thus eliminating the possibility of matricidal hatching. As shown in Figure 1G, even in the absence of matricidal hatching, fer-1(b232) animals exhibited reduced survival on P. aeruginosa following tax-6 knockdown, indicating that the increased susceptibility is not due to enhanced matricidal hatching. Moreover, fer-1(b232) animals showed an extended lifespan on E. coli when tax-6 was knocked down (Figure 1H). These results suggested that tax-6 knockdown leads to both increased lifespan and enhanced susceptibility to pathogen infection.
We then explored whether the increased susceptibility to bacterial infection following tax-6 knockdown was mediated by any known C. elegans innate immunity pathways, including the MAP kinase pathway mediated by NSY-1/SEK-1/PMK-1 (Kim et al., 2002), the MLK-1/MEK-1/KGB-1 c-Jun kinase pathway (Kim et al., 2004), the TGF-β/DBL-1 pathway (Mallo et al., 2002), and the bZIP transcription factor ZIP-2 pathway (Estes et al., 2010). In mutants deficient in each of these pathways, tax-6 knockdown further increased susceptibility to P. aeruginosa (Figure 1—figure supplement 2A–D). The knockdown of tax-6 appeared to have a more pronounced effect in pmk-1(km25) mutants than in other mutants, suggesting that inhibition of tax-6 might exacerbate the adverse effects observed in pmk-1(km25) mutants. Additionally, the increased susceptibility to P. aeruginosa seen in tax-6 knockdown animals was independent of the FOXO transcription factor DAF-16 (Figure 1—figure supplement 2E). Thus, the enhanced susceptibility to P. aeruginosa resulting from tax-6 knockdown appears to operate independently of established C. elegans innate immunity pathways.
Calcineurin is required for the C. elegans defecation motor program (DMP)
To understand why tax-6 knockdown animals had enhanced susceptibility to P. aeruginosa infection, we studied the colonization of the intestine of tax-6 RNAi animals by P. aeruginosa. Knockdown of tax-6 led to enhanced intestinal colonization by P. aeruginosa compared to the control animals (Figure 2A and B). Similarly, tax-6(ok2065) and tax-6(p675) animals also showed increased intestinal colonization by P. aeruginosa compared to N2 animals (Figure 2C and D and Figure 2—figure supplement 1A, B). Animals with bloated intestinal lumens exhibit enhanced bacterial colonization of the gut, an improved lifespan, and an increased susceptibility to pathogens (Kumar et al., 2019; Singh and Aballay, 2019a). Because tax-6 knockdown animals displayed all of these phenotypes, we asked whether tax-6 knockdown led to the bloating of the intestinal lumen. Indeed, we found that tax-6 knockdown resulted in bloated intestinal lumens (Figure 2E and F).
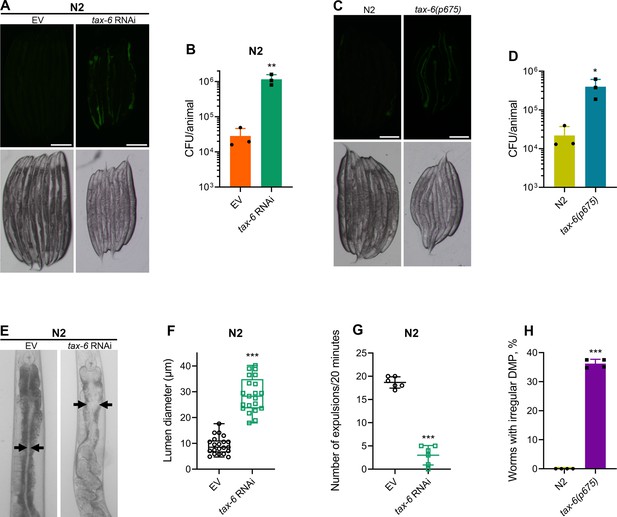
Calcineurin is required for the C. elegans defecation motor program (DMP).
(A) Representative fluorescence (top) and the corresponding brightfield (bottom) images of N2 animals incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on the empty vector (EV) control and tax-6 RNAi bacteria. Scale bar=200 µm. (B) Colony-forming units (CFU) per animal of N2 worms incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on the EV control and tax-6 RNAi bacteria. **p<0.01 via the t-test (n=3 biological replicates). (C) Representative fluorescence (top) and the corresponding brightfield (bottom) images of N2 and tax-6(p675) animals incubated on P. aeruginosa-GFP for 6 hours at 25 °C after growth on E. coli OP50 at 20 °C. Scale bar=200 µm. (D) CFU per animal of N2 and tax-6(p675) worms incubated on P. aeruginosa-GFP for 6 hours at 25 °C after growth on E. coli OP50 at 20 °C. *p<0.05 via the t-test (n=3 biological replicates). (E) Representative photomicrographs of N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. Arrows point to the border of the intestinal lumen. (F) Quantification of the diameter of the intestinal lumen of N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. ***p<0.001 via the t-test (n=21 worms each). (G) The number of expulsion events observed in 20 min in 1-day-old adult N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C. ***p<0.001 via the t-test (n=6 worms each). (H) Percent of 1-day-old adult worms having irregular DMP. ***p<0.001 via the t-test (n=4 biological replicates).
-
Figure 2—source data 1
Calcineurin is required for the C. elegans defecation motor program.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig2-data1-v1.xlsx
Intestinal bloating could result from either defects in pharyngeal pumping (Kumar et al., 2019) or defects in the defecation motor program (DMP) (Singh and Aballay, 2019b). We found that tax-6 knockdown did not affect the pharyngeal pumping rate (Figure 2—figure supplement 2A). This prompted us to explore whether calcineurin is necessary for the DMP. The C. elegans DMP is a highly coordinated rhythmic behavior required for the regular expulsion of intestinal contents, occurring approximately once per minute. The DMP involves a series of muscle contractions, including posterior body muscle contraction (pBoc), anterior body muscle contraction (aBoc), and expulsion muscle contraction (EMC), to release intestinal contents (Thomas, 1990). When we counted expulsion events over a 20-min period, we found that tax-6 knockdown animals had a drastically reduced number of expulsion events (Figure 2G). The DMP-defect phenotype had low penetrance in tax-6(p675) mutants, which exhibited both regular and irregular DMPs (Figure 2—figure supplement 2B). At the 1-day-old adult stage, about 36% of tax-6(p675) animals showed irregular and slowed DMP, while the remainder had regular DMP (Figure 2H), suggesting that tax-6(p675) is a weak allele. The fraction of the animals with irregular DMP appeared to increase with age, indicating that this phenotype might be age-dependent. This may also explain why tax-6(p675) animals were reported to have a normal defecation cycle in an earlier study (Lee et al., 2005).
Next, we investigated whether the reduced number of expulsion events was due to regular intervals with longer cycle lengths or if rhythmicity was entirely disrupted upon tax-6 knockdown. To assess this, we obtained ethograms of the DMP for N2 animals grown on control and tax-6 RNAi. While animals on control RNAi displayed regular cycles of pBoc, aBoc, and EMC, the tax-6 RNAi animals exhibited disrupted rhythmicity (Figure 3A and Figure 3—figure supplement 1). Most tax-6 knockdown animals lacked the pBoc and aBoc steps and had sporadic expulsion events. Isolated pBoc events were occasionally observed, indicating a complete loss of rhythmicity in tax-6 knockdown animals. Ethograms for tax-6(ok2065) animals also showed disrupted rhythmicity (Figure 3B and Figure 3—figure supplement 2). Although the number of expulsion events appeared higher in tax-6(ok2065) animals compared to tax-6 RNAi animals (Figure 3—figure supplements 1 and 2), these expulsion events seemed superficial, releasing little to no gut content. This suggested slow movement of gut content in tax-6(ok2065) animals, leading to constipation and intestinal bloating. We examined gut content movement by measuring the clearance of blue dye (erioglaucine disodium salt) from the gut. The clearance was significantly slower in tax-6(ok2065) animals compared to N2 animals (Figure 3C), indicating impaired gut content movement due to the loss of tax-6. Similarly, tax-6 knockdown animals also showed significantly slowed gut content movement (Figure 3D).
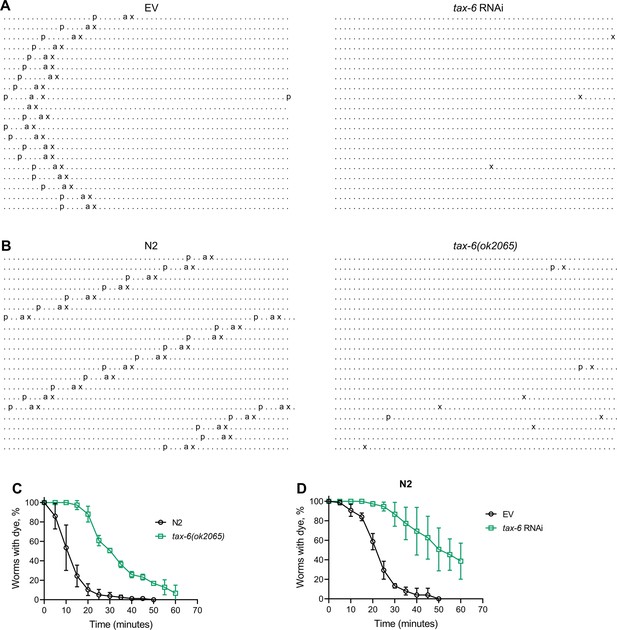
Calcineurin inhibition disrupts the C. elegans defecation motor program (DMP).
(A) Representative DMP ethograms of N2 animals after growth on the empty vector (EV) control and tax-6 RNAi bacteria till 1-day-old stage. Each dot represents a second, and each row represents a minute. ‘p’, ‘a’, and ‘x’ represent posterior body-wall muscle contraction, anterior body-wall muscle contraction, and expulsion muscle contraction, respectively. (B) Representative DMP ethograms of N2 and tax-6(ok2065) animals after their growth on E. coli OP50 at 20 °C till the 1-day-old adult stage. (C) Plots of the change in the percent of animals with blue dye in their gut with time for 1-day-old adult N2 and tax-6(ok2065) animals (n=3 biological replicates). (D) Plots of the change in the percent of animals with blue dye in their gut with time for 1-day-old adult N2 animals grown on EV control and tax-6 RNAi bacteria (n=3 biological replicates).
-
Figure 3—source data 1
Calcineurin inhibition disrupts the C. elegans defecation motor program.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig3-data1-v1.xlsx
We then explored whether the disruption of DMP rhythmicity due to tax-6 knockdown affected P. aeruginosa responses similarly to longer but regular DMP cycles. To do this, we studied P. aeruginosa colonization in clk-1(qm30) and isp-1(qm150) mutants, which have regular but extended DMP cycles (Feng et al., 2001; Wong et al., 1995). Interestingly, both clk-1(qm30) and isp-1(qm150) mutants showed significantly reduced intestinal colonization by P. aeruginosa compared to N2 animals (Figure 3—figure supplement 3A–D). This reduced colonization could be attributed to their significantly decreased pharyngeal pumping rates (Wong et al., 1995; Yee et al., 2014), suggesting a lower intake of bacterial food in these mutants. While the survival of clk-1(qm30) animals on P. aeruginosa was comparable to N2 animals (Figure 3—figure supplement 3E), isp-1(qm150) animals exhibited significantly improved survival (Figure 3—figure supplement 3F). Conversely, knockdown of flr-1, nhx-2, and pbo-1 in N2 animals resulted in significantly reduced survival on P. aeruginosa compared to control RNAi (Figure 3—figure supplement 3G). Knockdown of these genes causes complete disruption of DMP rhythmicity, increasing gut colonization by P. aeruginosa (Singh and Aballay, 2019b). Overall, these findings demonstrated that calcineurin is crucial for maintaining the DMP ultradian clock, and its inhibition increases susceptibility to P. aeruginosa by disrupting the DMP.
Calcineurin inhibition enhances lifespan via DMP defects-induced calorie restriction
Defects in the C. elegans DMP lead to reduced nutrient absorption, resulting in decreased intestinal fat levels (Sheng et al., 2015). We investigated whether tax-6 knockdown similarly reduces fat levels. To test this, we performed oil-red-O (ORO) staining on tax-6 RNAi animals. The knockdown of tax-6 significantly reduced fat levels (Figure 4A and B). Next, we examined whether the reduced fat levels in tax-6 knockdown animals mimic calorie restriction-like phenotypes. Calorie restriction is known to upregulate the expression of the FoxA transcription factor PHA-4 (Panowski et al., 2007). Consistent with this, tax-6 knockdown also resulted in the upregulation of pha-4 expression levels (Figure 4C). Because calorie restriction enhances lifespan (Kaeberlein et al., 2006; Lakowski and Hekimi, 1998; Panowski et al., 2007), we explored whether calcineurin inhibition could similarly increase lifespan through calorie restriction mechanisms. The knockdown of tax-6 in the genetic model of calorie restriction, eat-2(ad465) (Lakowski and Hekimi, 1998), did not increase the lifespan (Figure 4D).
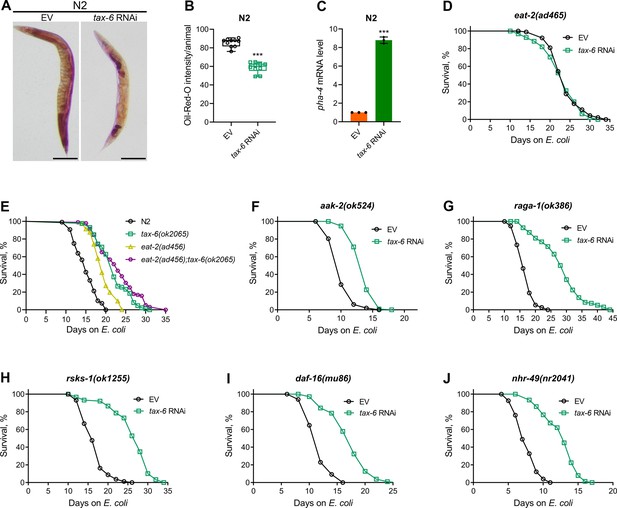
Calcineurin knockdown enhances lifespan via DMP defects-mediated calorie restriction.
(A) Representative photomicrographs of oil-red-O (ORO) stained 1-day-old adult N2 animals grown on the empty vector (EV) control and tax-6 RNAi bacteria at 20 °C. Scale bar=200 µm. (B) Quantification of ORO intensity per animal of 1-day-old adult N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C. The values are area normalized for each animal. ***p<0.001 via the t-test (n=10 worms each). (C) Quantitative reverse transcription-PCR (qRT-PCR) for pha-4 mRNA levels in N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. ***p<0.001 (n=3 biological replicates). (D) Representative survival plots of eat-2(ad465) animals grown on the bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. n.s., nonsignificant (n=3 biological replicates; animals per condition per replicate >80). (E) Representative survival plots of N2, tax-6(ok2065), eat-2(ad465), and eat-2(ad465);tax-6(ok2065) animals grown on E. coli OP50 at 20 °C. Day 0 represents young adults. p<0.001 for tax-6(ok2065), eat-2(ad465), and eat-2(ad465);tax-6(ok2065) compared to N2. p<0.05 for eat-2(ad465);tax-6(ok2065) compared to tax-6(ok2065) (n=3 biological replicates; animals per condition per replicate >65). (F–J) Representative survival plots of aak-2(ok524) (E), raga-1(ok386) (F), rsks-1(ok1255) (G), daf-16(mu86) (H), and nhr-49(nr2041) (I) animals grown on the bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. p<0.001 for all the plots (n=3 biological replicates; animals per condition per replicate >60).
-
Figure 4—source data 1
Calcineurin knockdown enhances lifespan via DMP defects-mediated calorie restriction.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig4-data1-v1.xlsx
Since eat-2(ad465) animals consume fewer bacteria, it is possible that their feeding-based RNAi efficiency is reduced, leading to a lack of responsiveness to tax-6 RNAi. To assess RNAi efficiency in eat-2(ad465) animals, we exposed them to act-5 and bli-3 RNAi, which are known to cause developmental arrest and a blistered cuticle, respectively (Gahlot and Singh, 2024). Similar to N2 animals, eat-2(ad465) mutants were sensitive to the toxic effects of act-5 and bli-3 knockdown (Figure 4—figure supplement 1), indicating that eat-2(ad465) animals are not defective in RNAi. To further confirm that calcineurin inhibition enhances lifespan via calorie restriction, we examined the lifespan of eat-2(ad465);tax-6(ok2065) double mutants. While eat-2(ad465) animals displayed increased lifespan compared to N2 controls, eat-2(ad465);tax-6(ok2065) double mutants had a lifespan similar to tax-6(ok2065) animals (Figure 4E). These results suggested that calcineurin inhibition promotes lifespan extension through calorie restriction mechanisms.
The mechanisms of lifespan enhancement by dietary or calorie restriction are complex and involve several different downstream pathways (Chamoli et al., 2014; Greer and Brunet, 2009; Hansen et al., 2008). Further, different pathways are operational under different dietary regimens (Greer and Brunet, 2009). Therefore, we investigated the involvement of pathways associated with calorie restriction-mediated lifespan extension in response to calcineurin inhibition. Specifically, we analyzed the interactions of the AMP-activated kinase, aak-2 (Greer and Brunet, 2009), the mTOR pathway, raga-1 (Robida-Stubbs et al., 2012; Zhang et al., 2019), ribosomal S6 kinase, rsks-1 (Chen et al., 2009; Selman et al., 2009; Zhang et al., 2019), the FOXO transcription factor, daf-16 (Greer and Brunet, 2009), and the nuclear hormone receptor, nhr-49 (Chamoli et al., 2014) with calcineurin inhibition in regulating lifespan. Mutants defective in aak-2 appeared to have only a partial increase in lifespan upon calcineurin inhibition (Figure 4F). In contrast, loss-of-function mutants of raga-1, rsks-1, daf-16, and nhr-49 exhibited enhanced lifespan upon calcineurin inhibition (Figure 4G–J).
Calcineurin knockdown enhances lifespan via HLH-30 and NHR-8
To further investigate longevity pathways regulated by calorie restriction that may enhance lifespan downstream of calcineurin inhibition, we examined the role of the TFEB ortholog HLH-30. HLH-30 is essential for the starvation response and triggers lipid depletion under nutrient-deprived conditions (O’Rourke and Ruvkun, 2013). Knockdown of tax-6 did not extend the lifespan of hlh-30(tm1978) animals (Figure 5A), indicating that HLH-30 is required for the increased lifespan observed with calcineurin inhibition. To determine whether HLH-30 is necessary for DMP defect-mediated lifespan extension, we assessed the lifespan of hlh-30(tm1978) animals upon knockdown of flr-1, nhx-2, and pbo-1. Knockdown of these genes did not alter the lifespan of hlh-30(tm1978) animals compared to control RNAi (Figure 5A), except for a slight increase in lifespan with nhx-2 knockdown. The hlh-30(tm1978) animals were found to be sensitive to RNAi (Figure 5—figure supplement 1), suggesting that the lack of lifespan change with tax-6, flr-1, and pbo-1 RNAi was not due to reduced RNAi efficiency in hlh-30(tm1978) animals. Additionally, we confirmed that tax-6 knockdown caused DMP defects and intestinal bloating in hlh-30(tm1978) animals (Figure 5—figure supplement 2A–C). We also explored whether HLH-30 affected fat depletion due to DMP defects. The ORO levels declined significantly in hlh-30(tm1978) animals upon the knockdown of tax-6 and other DMP-regulating genes (Figure 5B and C). The reduction in ORO levels was less pronounced in tax-6 knockdown animals compared to those with other DMP-regulating gene knockdowns.
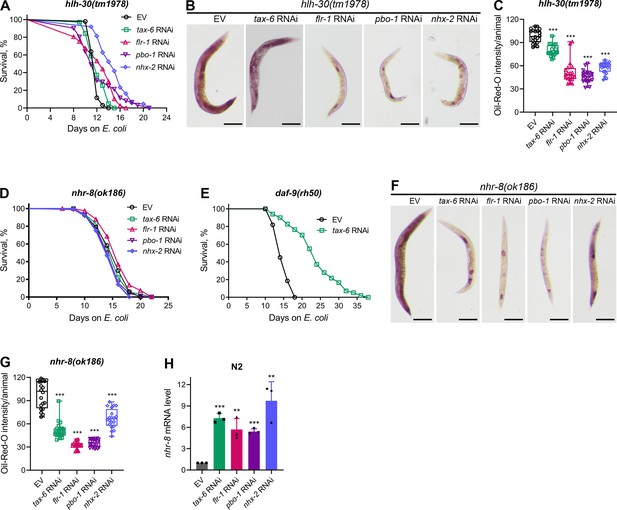
Calcineurin inhibition enhances lifespan via HLH-30 and NHR-8.
(A) Representative survival plots of hlh-30(tm1978) animals grown on bacteria for RNAi against tax-6, flr-1, pbo-1, and nhx-2 along with the empty vector (EV) control at 20 °C. Day 0 represents young adults. p values for tax-6, flr-1, pbo-1, and nhx-2 compared to EV are <0.01,<0.001, nonsignificant, and <0.001, respectively (n=3 biological replicates; animals per condition per replicate >60). (B) Representative photomicrographs of oil-red-O (ORO) stained 1-day-old adult hlh-30(tm1978) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. Scale bar=200 µm. (C) Quantification of ORO intensity per animal of 1-day-old adult hlh-30(tm1978) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. The values are area normalized for each animal. ***p<0.001 via the t-test (n=20 worms each). (D) Representative survival plots of nhr-8(ok186) animals grown on bacteria for RNAi against tax-6, flr-1, pbo-1, and nhx-2 along with the EV control at 20 °C. Day 0 represents young adults. p values for tax-6, flr-1, pbo-1, and nhx-2 compared to EV are nonsignificant,<0.05, nonsignificant, and <0.05, respectively (n=3 biological replicates; animals per condition per replicate >70). (E) Representative survival plots of daf-9(rh50) animals grown on the bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. p<0.001 (n=3 biological replicates; animals per condition per replicate >80). (F) Representative photomicrographs of ORO stained 1-day-old adult nhr-8(ok186) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. Scale bar=200 µm. (G) Quantification of ORO intensity per animal of 1-day-old adult nhr-8(ok186) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. The values are area normalized for each animal. ***p<0.001 via the t-test (n=20 worms each). (H) Quantitative reverse transcription-PCR (qRT-PCR) for nhr-8 mRNA levels in N2 animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C until 1-day-old adults. ***p<0.001 via the t-test (n=3 biological replicates).
-
Figure 5—source data 1
Calcineurin inhibition enhances lifespan via HLH-30 and NHR-8.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig5-data1-v1.xlsx
Steroid hormone signaling mediated by NHR-8 is known to regulate lifespan extension under calorie restriction (Thondamal et al., 2014). We tested whether NHR-8 was necessary for lifespan extension induced by calcineurin inhibition. Indeed, tax-6 knockdown did not extend the lifespan of nhr-8(ok186) animals (Figure 5D). While the specific ligands of NHR-8 are not well-defined (Magner et al., 2013), steroid hormones produced by the cytochrome P450 enzyme DAF-9 have been shown to enhance lifespan via NHR-8 under calorie restriction (Thondamal et al., 2014). Therefore, we investigated whether DAF-9 mediated the enhanced lifespan downstream of calcineurin inhibition. We found that tax-6 knockdown enhanced the lifespan of daf-9(rh50) animals (Figure 5E), indicating that DAF-9-produced steroid hormones were not required for lifespan increment by calcineurin inhibition. We also examined the lifespan of nhr-8(ok186) animals upon knockdown of flr-1, nhx-2, and pbo-1. Knockdown of these genes did not affect the lifespan of nhr-8(ok186) animals compared to control RNAi (Figure 5D). As with hlh-30(tm1978) animals, the lack of lifespan change in nhr-8(ok186) animals was not due to RNAi insensitivity, as they were responsive to RNAi (Figure 5—figure supplement 1). Furthermore, tax-6 knockdown resulted in DMP defects in nhr-8(ok186) animals (Figure 5—figure supplement 2D). We investigated whether NHR-8 influenced fat depletion upon DMP defects. A significant reduction in fat levels was observed in nhr-8(ok186) animals upon knockdown of tax-6, flr-1, nhx-2, and pbo-1 (Figure 5F and G), suggesting that NHR-8 functions downstream of or in parallel with fat depletion.
Since calorie restriction can enhance the expression of genes that promote lifespan, such as pha-4 (Panowski et al., 2007), we examined whether tax-6 knockdown modulates nhr-8 mRNA levels. We observed a significant increase in nhr-8 mRNA levels following tax-6 knockdown (Figure 5H). To determine whether this upregulation was due to intestinal bloating, we assessed nhr-8 mRNA levels in flr-1, nhx-2, and pbo-1 knockdown animals. Knockdown of these genes also increased nhr-8 mRNA levels (Figure 5H), indicating that intestinal bloating induces nhr-8 upregulation. Thus, calcineurin inhibition likely enhances lifespan via NHR-8 by increasing its expression.
Given that HLH-30 and NHR-8 are essential for lifespan extension upon calcineurin inhibition, we investigated whether these pathways also influence survival in response to P. aeruginosa infection following calcineurin knockdown. Both hlh-30(tm1978) and nhr-8(ok186) animals showed significantly reduced survival upon tax-6 RNAi (Figure 6A and B). These findings suggested that the reduced survival on P. aeruginosa following calcineurin inhibition is independent of HLH-30 and NHR-8 and is more likely due to increased gut colonization by P. aeruginosa resulting from DMP defects (Figure 6C).
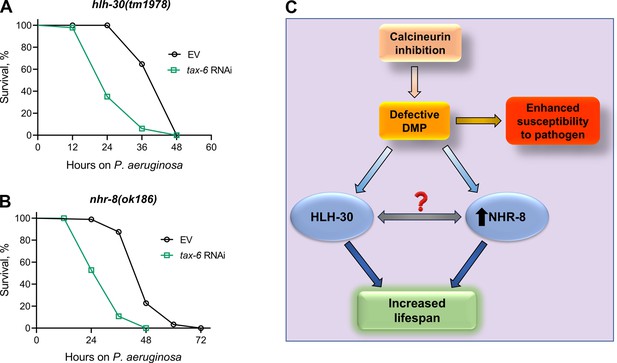
Calcineurin inhibition-mediated effects on lifespan and survival on P. aeruginosa are mediated by distinct mechanisms.
(A)-(B) Representative survival plots of hlh-30(tm1978) (A) and nhr-8(ok186) (B) animals on P. aeruginosa PA14 at 25 °C after treatment with the empty vector (EV) control and tax-6 RNAi. p<0.001 for both the plots (n=3 biological replicates; animals per condition per replicate >85). (C) Model depicting the mechanism of increased lifespan and enhanced susceptibility to pathogen upon inhibition of calcineurin via the defects in the defecation motor program (DMP).
-
Figure 6—source data 1
Calcineurin inhibition-mediated effects on lifespan and survival on P. aeruginosa are mediated by distinct mechanisms.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig6-data1-v1.xlsx
Discussion
We discovered that calcineurin is required for the C. elegans DMP. Calcineurin activity is regulated by intracellular calcium levels. Increased amounts of calcium ions activate the phosphatase activity of calcineurin via the binding of the calcium-sensing protein calmodulin to calcineurin (Klee et al., 1998). The rhythmic DMP cycle in C. elegans is known to be regulated by rhythmic calcium waves (Dal Santo et al., 1999; Teramoto and Iwasaki, 2006). The calcium waves are regulated by the endoplasmic reticulum calcium channel, ITR-1, and mutations in the itr-1 gene affect defecation by preventing cytoplasmic calcium release (Dal Santo et al., 1999). It is likely that calcium waves regulate DMP via calcineurin activities. Indeed, calcineurin is expressed in the enteric muscles that are required for contractions for DMP (Lee et al., 2005). It is interesting to note that tax-6 gain-of-function mutants are also known to have DMP defects (Lee et al., 2005). Therefore, optimum calcineurin activity appears to be crucial for maintaining a rhythmic DMP.
Calcineurin inhibition has been shown to extend C. elegans lifespan via multiple mechanisms (Dong et al., 2007; Dwivedi et al., 2009; Mair et al., 2011; Tao et al., 2013). Activation of autophagy has been shown to be one of the mechanisms of extended lifespan upon calcineurin inhibition (Dwivedi et al., 2009). We showed that defects in the DMP caused by calcineurin inhibition reduce lipid levels and mimic calorie restriction. Calorie restriction is known to induce autophagy (Morselli et al., 2010), and autophagy is required for calorie restriction-mediated lifespan extension (Hansen et al., 2008; Jia and Levine, 2007). Therefore, it is likely that reduced lipid levels because of intestinal bloating result in the activation of autophagy upon calcineurin inhibition. Importantly, the TFEB ortholog, HLH-30, is required for lipolysis and autophagy under starvation conditions (Lapierre et al., 2013; O’Rourke and Ruvkun, 2013). HLH-30 is also required for lifespan enhancement via autophagy and multiple longevity pathways (Lapierre et al., 2013; O’Rourke and Ruvkun, 2013). We observed that HLH-30 is also required for the increase in lifespan upon calcineurin inhibition. Because autophagy may regulate lifespan via multiple longevity pathways (Hansen et al., 2008; Lapierre et al., 2013), this could explain why calcineurin inhibition enhances lifespan via multiple mechanisms.
Dietary restriction-mediated lifespan extensions are complex and context-dependent (Greer and Brunet, 2009). Multiple longevity pathways have been identified under different paradigms of dietary restriction (Chamoli et al., 2020; Chamoli et al., 2014; Chen et al., 2009; Greer and Brunet, 2009; Lapierre et al., 2013; Matai et al., 2019; Selman et al., 2009; Thondamal et al., 2014). We found that tax-6 knockdown enhanced lifespan independent of multiple dietary restriction pathways, including raga-1, rsks-1, daf-16, and nhr-49. An earlier study identified that tax-6 knockdown enhanced lifespan via NHR-49 (Burkewitz et al., 2015). We currently do not understand the reasons for this discrepancy. We identified that tax-6 knockdown resulted in the upregulation of nhr-8 mRNA levels, and nhr-8 was required for the increased lifespan in tax-6 knockdown animals. NHR-8 has been shown to mediate calorie restriction-dependent lifespan via the regulation of xenobiotic responses (Chamoli et al., 2014; Verma et al., 2018). It remains to be studied whether NHR-8 and HLH-30 work in the same or different pathways downstream of calcineurin inhibition (Figure 6C).
Recent studies in different organisms have shown that gut bloating has profound effects on food-seeking behaviors, immunity, and lifespan (Duvall et al., 2019; Filipowicz et al., 2021; Kumar et al., 2019; Min et al., 2021; Singh and Aballay, 2019b; Singh and Aballay, 2019c). Several C. elegans mutants with defects in the DMP and bloated intestinal lumens are known to have dampened nutrient absorption, leading to reduced lipid deposition in the gut, mimicking calorie restriction (Sheng et al., 2015). It is possible that some of the effects of intestinal bloating on the host physiology are mediated by calorie restriction. Indeed, the neuropeptide Y receptors, which control a diverse set of behaviors, including appetite, are activated by gut bloating (Singh and Aballay, 2019c). Calorie restriction is known to induce neuropeptide Y, which might trigger feeding behaviors (Aveleira et al., 2015; de Rijke et al., 2005; Ferreira-Marques et al., 2016). The complete characterization of the physiological changes downstream of gut bloating may provide broad insights into the effects of gut physiology on behaviors, immunity, and lifespan.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | OP50 | Caenorhabditis Genetics Center (CGC) | OP50 | |
| Strain, strain background (E. coli) | HT115(DE3) | Source BioScience | HT115(DE3) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 | Frederick M Ausubel laboratory | PA14 | |
| Strain, strain background (P. aeruginosa) | PA14-GFP | Frederick M Ausubel laboratory | PA14-GFP | |
| Strain, strain background (Caenorhabditis elegans) | N2 Bristol | CGC | N2 | |
| Strain, strain background (C. elegans) | tax-6(p675) | CGC | PR675 | |
| Strain, strain background (C. elegans) | tax-6(ok2065) | CGC | RB1667 | |
| Strain, strain background (C. elegans) | fer-1(b232) | CGC | HH142 | |
| Strain, strain background (C. elegans) | eat-2(ad465) | CGC | DA465 | |
| Strain, strain background (C. elegans) | aak-2(ok524) | CGC | RB754 | |
| Strain, strain background (C. elegans) | raga-1(ok386) | CGC | VC222 | |
| Strain, strain background (C. elegans) | rsks-1(ok1255) | CGC | RB1206 | |
| Strain, strain background (C. elegans) | daf-16(mu86) | CGC | CF1038 | |
| Strain, strain background (C. elegans) | nhr-49(nr2041) | CGC | STE68 | |
| Strain, strain background (C. elegans) | hlh-30(tm1978) | CGC | JIN1375 | |
| Strain, strain background (C. elegans) | nhr-8(ok186) | CGC | AE501 | |
| Strain, strain background (C. elegans) | daf-9(rh50) | CGC | RG1228 | |
| Strain, strain background (C. elegans) | pmk-1(km25) | CGC | KU25 | |
| Strain, strain background (C. elegans) | kgb-1(km21) | CGC | KU21 | |
| Strain, strain background (C. elegans) | dbl-1(nk3) | CGC | NU3 | |
| Strain, strain background (C. elegans) | zip-2(ok3730) | CGC | VC3056 | |
| Strain, strain background (C. elegans) | clk-1(qm30) | CGC | MQ130 | |
| Strain, strain background (C. elegans) | isp-1(qm150) | CGC | MQ887 | |
| Strain, strain background (C. elegans) | sid-1(qt9) | CGC | HC196 | |
| Strain, strain background (C. elegans) | eat-2(ad465);tax-6(ok2065) | This study | Materials and methods section | |
| Sequence-based reagent | Pan-act_qPCR_F | This study | qPCR primers | TCGGTATGGGACAGAAGGAC |
| Sequence-based reagent | Pan-act_qPCR_R | This study | qPCR primers | CATCCCAGTTGGTGACGATA |
| Sequence-based reagent | pha-4_qPCR_F | This study | qPCR primers | CAAAGAGGAGCCAGAGTCGG |
| Sequence-based reagent | pha-4_qPCR_R | This study | qPCR primers | TGTTTCTGCTCGCGTTTTCG |
| Sequence-based reagent | nhr-8_qPCR_F | This study | qPCR primers | CTACACAGTTTCTCCGGCGT |
| Sequence-based reagent | nhr-8_qPCR_R | This study | qPCR primers | GCCATTTGGGCCATAACACC |
| Sequence-based reagent | tax-6(ok2065)_genotyping_F1 | This study | Genotyping primers | CTCCTTTGAGGGAGCCAGTG |
| Sequence-based reagent | tax-6(ok2065)_genotyping_F2 | This study | Genotyping primers | CTGGGGACAATCCACCATGAA |
| Sequence-based reagent | tax-6(ok2065)_genotyping_R1 | This study | Genotyping primers | TGTGTCCTGTATCTGTGGGC |
| Sequence-based reagent | eat-2(ad465) _genotyping_F | This study | Genotyping primers | CGGTGCAAAGAGCACATCTC |
| Sequence-based reagent | eat-2(ad465) _genotyping_R | This study | Genotyping primers | TTAAGGCGTACGAGCCTTCC |
| Software, algorithm | GraphPad Prism 8 | GraphPad Software | RRID:SCR_002798 | https://www.graphpad.com/scientificsoftware/prism/ |
| Software, algorithm | Photoshop CS5 | Adobe | RRID:SCR_014199 | https://www.adobe.com/products/photoshop.html |
| Software, algorithm | ImageJ | NIH | RRID:SCR_003070 | https://imagej.nih.gov/ij/ |
Bacterial strains
Request a detailed protocolThe following bacterial strains were used: Escherichia coli OP50, E. coli HT115(DE3), Pseudomonas aeruginosa PA14, and P. aeruginosa PA14 expressing green fluorescent protein (P. aeruginosa PA14-GFP). The cultures for these bacteria were grown in Luria-Bertani (LB) broth at 37 °C.
C. elegans strains and growth conditions
Request a detailed protocolC. elegans hermaphrodites were maintained on nematode growth medium (NGM) plates seeded with E. coli OP50 at 20 °C unless otherwise indicated. Bristol N2 hermaphrodites were used as the wild-type control unless otherwise indicated. The strains used in the study are provided in the Key Resources Table. Some of the strains were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). The eat-2(ad465);tax-6(ok2065) strain was generated using a standard genetic cross. The fer-1(b232) hermaphrodites were maintained on E. coli OP50 at 15 °C and were grown at 25 °C prior to P. aeruginosa killing assays and longevity assays to obtain animals with unfertilized oocytes.
RNA interference (RNAi)
Request a detailed protocolRNAi was used to generate loss-of-function phenotypes by feeding nematodes E. coli strain HT115(DE3) expressing double-stranded RNA homologous to a target gene. RNAi was carried out as described previously (Gokul and Singh, 2022; Ravi et al., 2023). Briefly, E. coli with the appropriate vectors were grown in LB broth containing ampicillin (100 μg/mL) at 37 °C overnight and plated onto NGM plates containing 100 μg/mL ampicillin and 3 mM isopropyl β-D-thiogalactoside (IPTG) (RNAi plates). RNAi-expressing bacteria were allowed to grow overnight at 37 °C. Gravid adults were transferred to RNAi-expressing bacterial lawns and allowed to lay eggs for 2 hr. The gravid adults were removed, and the eggs were allowed to develop at 20 °C to 1-day-old adults for subsequent assays. The RNAi clones were from the Ahringer RNAi library and were verified by sequencing.
RNAi efficiency test
Request a detailed protocolThe RNAi efficiency test was carried out as described earlier (Gahlot and Singh, 2024; Ghosh and Singh, 2024). Briefly, the wild-type N2, eat-2(ad456), nhr-8(ok186), and hlh-30(tm1978) worms were synchronized by egg laying and grown on empty vector control, act-5, and bli-3 RNAi plates at 20 °C. The sid-1(qt9) worms were used as RNAi-defective controls. After 72 hr, the animals were monitored for development defects and blisters on the cuticle for act-5 and bli-3 RNAi, respectively.
C. elegans longevity assays
Request a detailed protocolLifespan assays were performed on RNAi plates containing E. coli HT115(DE3) with the empty vector control and tax-6 RNAi clone in the presence of 50 µg/mL of 5-fluorodeoxyuridine (FUdR). Animals were synchronized on RNAi plates without FUdR and incubated at 20 °C. At the late L4 larval stage, the animals were transferred onto the corresponding RNAi plates containing 50 µg/mL of FUdR and incubated at 20 °C. For fer-1(b232) lifespan assays, the animals were synchronized on RNAi plates without FUdR and incubated at 25 °C. The fer-1(b232) lifespan assays were carried out at 25 °C without FUdR. Animals were scored every other day as live, dead, or gone. Animals that failed to display touch-provoked movement were scored as dead. Animals that crawled off the plates were censored. Young adult animals were considered as day 0 for the lifespan analysis. Three independent experiments were performed. The exact number of animals per condition per replicate is provided in the source data files.
C. elegans killing assays on P. aeruginosa PA14
Request a detailed protocolBacterial cultures were prepared by inoculating individual bacterial colonies of P. aeruginosa into 2 mL of LB and growing them for 10–12 hr on a shaker at 37 °C. Bacterial lawns were prepared by spreading 20 µL of the culture on the entire surface of 3.5-cm-diameter modified NGM agar plates (0.35% instead of 0.25% peptone). The plates were incubated at 37 °C for 12–16 hr and then cooled to room temperature for at least 30 min before seeding with synchronized 1-day-old adult animals. The killing assays were performed at 25 °C, and live animals were transferred daily to fresh plates. Animals were scored at times indicated and were considered dead when they failed to respond to touch. The exact number of animals per condition per replicate is provided in the source data files.
P. aeruginosa-GFP colonization assay
Request a detailed protocolBacterial cultures were prepared by inoculating individual bacterial colonies of P. aeruginosa-GFP into 2 mL of LB and growing them for 10–12 hr on a shaker at 37 °C. Bacterial lawns were prepared by spreading 20 µL of the culture on the entire surface of 3.5-cm-diameter modified NGM agar plates (0.35% instead of 0.25% peptone) containing 50 μg/mL of kanamycin. The plates were incubated at 37 °C for 12 hr and then cooled to room temperature for at least 30 min before seeding with 1-day-old adult gravid adults. The assays were performed at 25 °C. After 6 hr of incubation, the animals were transferred from P. aeruginosa-GFP plates to fresh E. coli OP50 plates and visualized within 5 min under a fluorescence microscope.
Quantification of intestinal bacterial loads
Request a detailed protocolThe intestinal bacterial loads were quantified by measuring colony-forming units (CFU) as described earlier (Singh and Aballay, 2019c; Singh and Aballay, 2019b) with appropriate modifications. Briefly, P. aeruginosa-GFP lawns were prepared as described above. After 6 hr of exposure, the animals were transferred from P. aeruginosa-GFP plates to the center of fresh E. coli OP50 plates thrice for 10 min each to eliminate bacteria stuck to their body. Afterward, 10 animals/condition were transferred into 50 μL of PBS containing 0.01% triton X-100 and ground using glass beads. Serial dilutions of the lysates (101, 102, 103, 104) were seeded onto LB plates containing 50 μg/mL of kanamycin to select for P. aeruginosa-GFP cells and grown overnight at 37 °C. Single colonies were counted the next day and represented as the number of bacterial cells or CFU per animal. At least three independent experiments were performed for each condition.
Fluorescence imaging
Request a detailed protocolFluorescence imaging was carried out as described previously (Gokul and Singh, 2022; Ravi et al., 2023). Briefly, the animals were anesthetized using an M9 salt solution containing 50 mM sodium azide and mounted onto 2% agarose pads. The animals were then visualized using a Nikon SMZ-1000 fluorescence stereomicroscope.
Quantification of intestinal lumen bloating
Request a detailed protocolThe 1-day-old adult animals grown on the empty vector control and tax-6 RNAi were anesthetized using an M9 salt solution containing 50 mM sodium azide, mounted onto 2% agarose pads, and imaged. The diameter of the intestinal lumen was measured using the Zeiss Zen Pro 2011 software. At least ten animals were used for each condition.
Measurement of defecation motor program (DMP) rate
Request a detailed protocolWild-type N2 and tax-6(p675) animals were synchronized and grown at 20 °C on E. coli OP50 for 4 and 5 days, respectively, before measuring the DMP rate. For RNAi experiments, N2 worms were synchronized on EV and tax-6 RNAi plates and grown for 4 days at 20 °C before measuring the DMP rate. The DMP cycle length was scored by assessing the time between expulsions (which are preceded by posterior and anterior body wall muscle contraction and the contraction of enteric muscles in a normal regular pattern; Thomas, 1990). The number of expulsion events in 20 min was measured for each worm. The DMP rate was recorded for 5–6 worms/condition.
Ethogram for DMP
Request a detailed protocolWild type N2 and tax-6(ok2065) worms were synchronized by egg laying and grown at 20 °C on E. coli OP50 for 4 and 5 days, respectively. For RNAi experiments, N2 worms were synchronized and grown on empty vector control and tax-6 RNAi at 20 °C for 4 days. The assay was performed at 25 °C. The defecation motor program consists of three steps- posterior body-wall muscle contraction (p), anterior body-wall muscle contraction (a), and expulsion muscle contraction (x). Animals were observed for the exact time of these three contractions on a dissecting scope for a continuous period of 20 min. A computer program was developed to plot the ethogram where we manually input the timings of ‘p’, ‘a’, and ‘x’ contractions.
Blue dye assay to assess functional defecation rate
Request a detailed protocolWild type N2 and tax-6(ok2065) worms were synchronized by egg laying and grown at 20 °C on E. coli OP50 for 4 and 5 days, respectively, before measuring the functional defecation rate. For RNAi experiments, N2 worms were synchronized and grown on empty vector control and tax-6 RNAi at 20 °C for 4 days. The functional defecation rate was assessed using a blue dye, erioglaucine disodium salt (5% wt/v), as an indicator of gut content. The time required by worms to clear the blue dye from the gut was used to calculate a functional defecation rate. Briefly, 50 worms from each condition were incubated in a solution containing blue dye and E. coli OP50 culture in a 1:1 ratio for 2 hr. Then, the worms were transferred to an NGM plate seeded with E. coli OP50. About 25–27 worms from each condition were transferred to a fresh NGM plate seeded with E. coli OP50. The functional defecation rate was assessed by observing the clearance of blue dye from the intestine of worms every 5 min over a period of 60 min. Three independent experiments were performed for each condition.
Pharyngeal pumping assay
Request a detailed protocolFor the pharyngeal pumping assay, 1-day-old adult animals grown on the empty vector control and tax-6 RNAi were used. The number of contractions of the terminal bulb of the pharynx was counted for 30 s per worm. The pumping rates for 20 worms were recorded for each condition.
Lipid staining
Request a detailed protocolThe oil-red-O (ORO) staining was carried out as described earlier (Lynn et al., 2015) with appropriate modification. Briefly, wild-type N2, hlh-30(tm1978), and nhr-8(ok186) animals were synchronized on empty vector control and tax-6 RNAi bacteria and grown at 20 °C for 4 days. About 300–400 synchronized gravid adult worms from experimental plates were washed three times with 1 X PBS plus 0.01% triton X-100 (PBST). To permeabilize the cuticle, 600 µL of 40% isopropanol was added to 100 µL of worm pellet and was rocked for 3 min. The animals were spun down at 500 rpm for 30 s, and then 600 µL of supernatant was removed. Subsequently, 600 µL of ORO working stock was added to the worm pellet to stain the worms and incubated at room temperature for 1 hr on a shaker. ORO working stock was freshly prepared by diluting the stock 0.5% ORO in isopropanol to 60% in water and rocked at room temperature for 2 hr, followed by the removal of debris with a 0.22 µm filter. Afterward, worm samples were pelleted, 600 µL of supernatant was removed, 600 µL of PBST was added, and the samples were rocked for 30 min at room temperature. After that, the worms were mounted on a 2% agarose pad and imaged. At least 10 worms/condition were imaged. Three independent biological replicates were performed. The ORO intensity was quantified using Image J software.
RNA isolation and quantitative reverse transcription-PCR (qRT-PCR)
Request a detailed protocolAnimals were synchronized by egg laying. Approximately 40 N2 gravid adult animals were transferred to 9 cm RNAi plates seeded with E. coli HT115 expressing the appropriate vectors and allowed to lay eggs for 4–5 hr. The gravid adults were then removed, and the eggs were allowed to develop at 20 °C for 96 hr. Subsequently, the animals were collected, washed with M9 buffer, and frozen in TRIzol reagent (Life Technologies, Carlsbad, CA). Total RNA was extracted using the RNeasy Plus Universal Kit (QIAGEN, Netherlands). Residual genomic DNA was removed using TURBO DNase (Life Technologies, Carlsbad, CA). A total of 6 μg of total RNA was reverse-transcribed with random primers using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA).
qRT-PCR was conducted using the Applied Biosystems One-Step Real-time PCR protocol using SYBR Green fluorescence (Applied Biosystems) on an Applied Biosystems 7900HT real-time PCR machine in 96-well-plate format. Twenty-five-microliter reactions were analyzed as outlined by the manufacturer (Applied Biosystems). The relative fold-changes of the transcripts were calculated using the comparative CT(2-ΔΔCT) method and normalized to pan-actin (act-1,–3, –4) as described earlier (Singh and Aballay, 2019b; Singh and Aballay, 2017). The cycle thresholds of the amplification were determined using StepOnePlus software (Applied Biosystems). All samples were run in triplicate. The primer sequences are provided in the Key Resources Table.
Quantification and statistical analysis
Request a detailed protocolThe statistical analysis was performed with Prism 8 (GraphPad). All error bars represent mean ± standard deviation (SD). The unpaired, two-tailed, two-sample t-test was used when needed, and the data were judged to be statistically significant when p<0.05. In the figures, asterisks (*) denote statistical significance as follows: *, p<0.05, **, p<0.01, ***, p<0.001, as compared with the appropriate controls. The Kaplan-Meier method was used to calculate the survival fractions, and statistical significance between survival curves was determined using the log-rank test. All experiments were performed in triplicate. The exact number of animals per condition per replicate is provided in the source data files.
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files; source data files have been provided for Figures 1, 2, 3, 4, 5, 6, Figure 1—figure supplement 1 and 2, Figure 2—figure supplement 1 and 2, Figure 3—figure supplement 3, and Figure 5—figure supplement 2.
References
-
The Caenorhabditis elegans germ line regulates distinct signaling pathways to control lifespan and innate immunityThe Journal of Biological Chemistry 285:1822–1828.https://doi.org/10.1074/jbc.M109.057323
-
Calcineurin in development and diseaseGenes & Diseases 9:915–927.https://doi.org/10.1016/j.gendis.2021.03.002
-
Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to jugloneFree Radical Biology & Medicine 37:139–145.https://doi.org/10.1016/j.freeradbiomed.2004.04.021
-
Hypothalamic neuropeptide expression following chronic food restriction in sedentary and wheel-running ratsJournal of Molecular Endocrinology 35:381–390.https://doi.org/10.1677/jme.1.01808
-
Transcriptional regulation by calcium, calcineurin, and NFATGenes & Development 17:2205–2232.https://doi.org/10.1101/gad.1102703
-
Regulation of the calmodulin-stimulated protein phosphatase, calcineurinThe Journal of Biological Chemistry 273:13367–13370.https://doi.org/10.1074/jbc.273.22.13367
-
Calcineurin regulates enteric muscle contraction through EXP-1, excitatory GABA-gated channel, in C. elegansJournal of Molecular Biology 352:313–318.https://doi.org/10.1016/j.jmb.2005.07.032
-
Inducible antibacterial defense system in C. elegansCurrent Biology 12:1209–1214.https://doi.org/10.1016/S0960-9822(02)00928-4
-
MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availabilityNature Cell Biology 15:668–676.https://doi.org/10.1038/ncb2741
-
Thiol reductive stress activates the hypoxia response pathwayThe EMBO Journal 42:e114093.https://doi.org/10.15252/embj.2023114093
-
A cellular atlas of calcineurin signalingBiochimica et Biophysica Acta. Molecular Cell Research 1870:119366.https://doi.org/10.1016/j.bbamcr.2022.119366
-
Calcineurin/NFAT signaling and innate host defence: a role for NOD1-mediated phagocytic functionsCell Communication and Signaling 12:8.https://doi.org/10.1186/1478-811X-12-8
-
A novel gene-diet pair modulates C. elegans agingPLOS Genetics 14:e1007608.https://doi.org/10.1371/journal.pgen.1007608
Article and author information
Author details
Funding
Department of Biotechnology, Ministry of Science and Technology, India (HRD-17011/2/2023-HRD-DBT)
- Jogender Singh
Department of Biotechnology, Ministry of Science and Technology, India (BT/RLF/Re-entry/50/2020)
- Jogender Singh
Ministry of Education, India (MoE-STARS/STARS-2/2023-0116)
- Jogender Singh
Council of Scientific and Industrial Research, India (37/1741/23/EMR-II)
- Jogender Singh
Science and Engineering Research Board (CRG/2023/001136)
- Jogender Singh
Science and Engineering Research Board (SRG/2020/000022)
- Jogender Singh
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Sagar Santosh Gawande for help in plotting the DMP ethogram. Some strains used in this study were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Funding: This work was supported by the following grants: Har-Gobind Khorana-Innovative Young Biotechnologist Fellowship (File No. HRD-17011/2/2023-HRD-DBT) and Ramalingaswami Re-entry Fellowship (Ref. No. BT/RLF/Re-entry/50/2020) awarded by the Department of Biotechnology, India; STARS grant (File No. MoE-STARS/STARS-2/2023–0116) awarded by the Ministry of Education, India; Research Grant (Ref. No. 37/1741/23/EMR-II) awarded by the Council of Scientific & Industrial Research (CSIR), India; Science and Engineering Research Board (SERB) Startup Research Grant (Ref. No. SRG/2020/000022) and Core Research Grant (Ref. No. CRG/2023/001136) awarded by DST, India; and IISER Mohali intramural funds. PD was supported by a senior research fellowship from the Council of Scientific & Industrial Research (CSIR), India.
Version history
- Sent for peer review:
- Preprint posted:
- Reviewed Preprint version 1:
- Reviewed Preprint version 2:
- Version of Record published:
Cite all versions
You can cite all versions using the DOI https://doi.org/10.7554/eLife.89572. This DOI represents all versions, and will always resolve to the latest one.
Copyright
© 2023, Das et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,229
- views
-
- 81
- downloads
-
- 10
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
G protein-coupled receptors (GPCRs) are integral membrane proteins which closely interact with their plasma membrane lipid microenvironment. Cholesterol is a lipid enriched at the plasma membrane with pivotal roles in the control of membrane fluidity and maintenance of membrane microarchitecture, directly impacting on GPCR stability, dynamics, and function. Cholesterol extraction from pancreatic beta cells has previously been shown to disrupt the internalisation, clustering, and cAMP responses of the glucagon-like peptide-1 receptor (GLP-1R), a class B1 GPCR with key roles in the control of blood glucose levels via the potentiation of insulin secretion in beta cells and weight reduction via the modulation of brain appetite control centres. Here, we unveil the detrimental effect of a high cholesterol diet on GLP-1R-dependent glucoregulation in vivo, and the improvement in GLP-1R function that a reduction in cholesterol synthesis using simvastatin exerts in pancreatic islets. We next identify and map sites of cholesterol high occupancy and residence time on active vs inactive GLP-1Rs using coarse-grained molecular dynamics (cgMD) simulations, followed by a screen of key residues selected from these sites and detailed analyses of the effects of mutating one of these, Val229, to alanine on GLP-1R-cholesterol interactions, plasma membrane behaviours, clustering, trafficking and signalling in INS-1 832/3 rat pancreatic beta cells and primary mouse islets, unveiling an improved insulin secretion profile for the V229A mutant receptor. This study (1) highlights the role of cholesterol in regulating GLP-1R responses in vivo; (2) provides a detailed map of GLP-1R - cholesterol binding sites in model membranes; (3) validates their functional relevance in beta cells; and (4) highlights their potential as locations for the rational design of novel allosteric modulators with the capacity to fine-tune GLP-1R responses.
-
- Cell Biology
- Immunology and Inflammation
Macrophages are crucial in the body’s inflammatory response, with tightly regulated functions for optimal immune system performance. Our study reveals that the RAS–p110α signalling pathway, known for its involvement in various biological processes and tumourigenesis, regulates two vital aspects of the inflammatory response in macrophages: the initial monocyte movement and later-stage lysosomal function. Disrupting this pathway, either in a mouse model or through drug intervention, hampers the inflammatory response, leading to delayed resolution and the development of more severe acute inflammatory reactions in live models. This discovery uncovers a previously unknown role of the p110α isoform in immune regulation within macrophages, offering insight into the complex mechanisms governing their function during inflammation and opening new avenues for modulating inflammatory responses.






