Calcineurin inhibition enhances Caenorhabditis elegans lifespan by defecation defects-mediated calorie restriction and nuclear hormone signaling
Figures
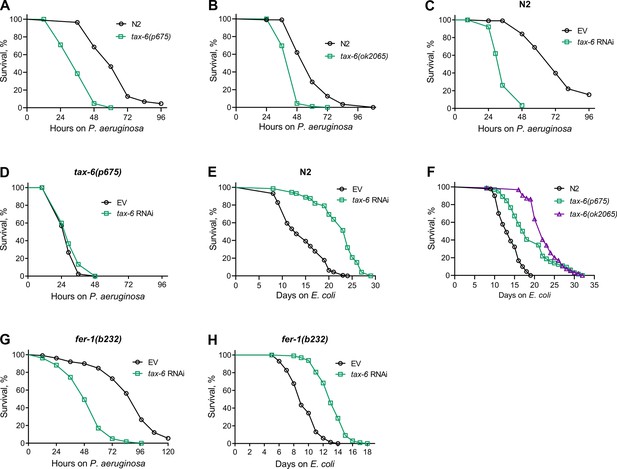
Calcineurin knockdown enhances C. elegans susceptibility to P. aeruginosa.
(A) Representative survival plots of N2 and tax-6(p675) animals on P. aeruginosa PA14 at 25 °C. The animals were grown on E. coli OP50 at 20 °C until 1-day-old adults before transferring to P. aeruginosa PA14 at 25 °C. p<0.001. (B) Representative survival plots of N2 and tax-6(ok2065) animals on P. aeruginosa PA14 at 25 °C. The animals were grown on E. coli OP50 at 20 °C until 1-day-old adults before transferring to P. aeruginosa PA14 at 25 °C. p<0.001. (C) Representative survival plots of N2 animals on P. aeruginosa PA14 at 25 °C after treatment with the empty vector (EV) control and tax-6 RNAi. p<0.001. (D) Representative survival plots of tax-6(p675) animals on P. aeruginosa PA14 at 25 °C after treatment with the EV control and tax-6 RNAi. n.s., nonsignificant. (E) Representative survival plots of N2 animals grown on bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. p<0.001. (F) Representative survival plots of N2, tax-6(p675), and tax-6(ok2065) animals grown on E. coli OP50 at 20 °C. Day 0 represents young adults. p<0.001 for tax-6(p675) and tax-6(ok2065) compared to N2. (G) Representative survival plots of fer-1(b232) animals on P. aeruginosa PA14 at 25 °C after treatment with the EV control and tax-6 RNAi at 25 °C. p<0.001. (H) Representative survival plots of fer-1(b232) animals grown on bacteria for RNAi against tax-6 along with the EV control at 25 °C. The animals were developed at 25 °C. Day 0 represents young adults. p<0.001. For all panels, n=3 biological replicates; animals per condition per replicate >60.
-
Figure 1—source data 1
Calcineurin knockdown enhances C. elegans susceptibility to P. aeruginosa.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig1-data1-v1.xlsx
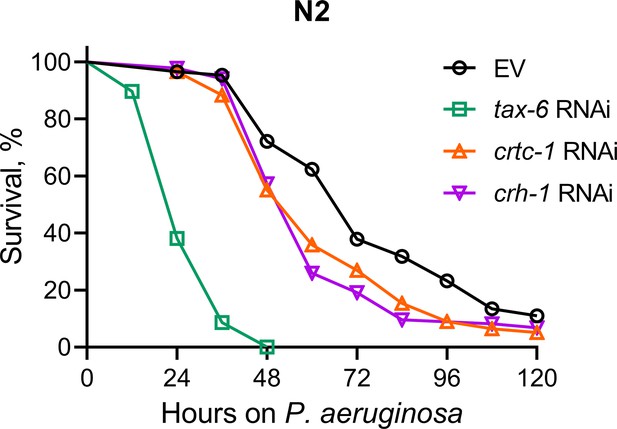
Calcineurin knockdown enhances C. elegans susceptibility to P. aeruginosa.
Representative survival plots of N2 animals on P. aeruginosa PA14 at 25 °C after treatment with the empty vector (EV) control, tax-6, crtc-1, and crh-1 RNAi. p<0.001 for tax-6 and p<0.01 for crtc-1 and crh-1 compared to EV (n=3 biological replicates; animals per condition per replicate >85).
-
Figure 1—figure supplement 1—source data 1
Calcineurin knockdown enhances C. elegans susceptibility to P. aeruginosa.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig1-figsupp1-data1-v1.xlsx
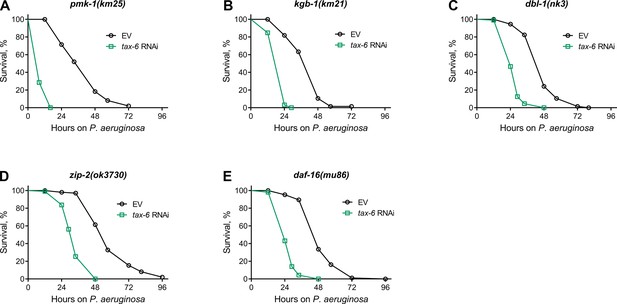
Calcineurin inhibition enhances susceptibility to P. aeruginosa independent of known immunity pathways.
(A–E) Representative survival plots of pmk-1(km25) (A), kgb-1(km21) (B), dbl-1(nk3) (C), zip-2(ok3730) (D), and daf-16(mu86) (E) animals on P. aeruginosa PA14 at 25 °C after treatment with the empty vector (EV) control and tax-6 RNAi. p<0.001 for all the plots (n=3 biological replicates; animals per condition per replicate >90).
-
Figure 1—figure supplement 2—source data 1
Calcineurin inhibition enhances susceptibility to P. aeruginosa independent of known immunity pathways.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig1-figsupp2-data1-v1.xlsx
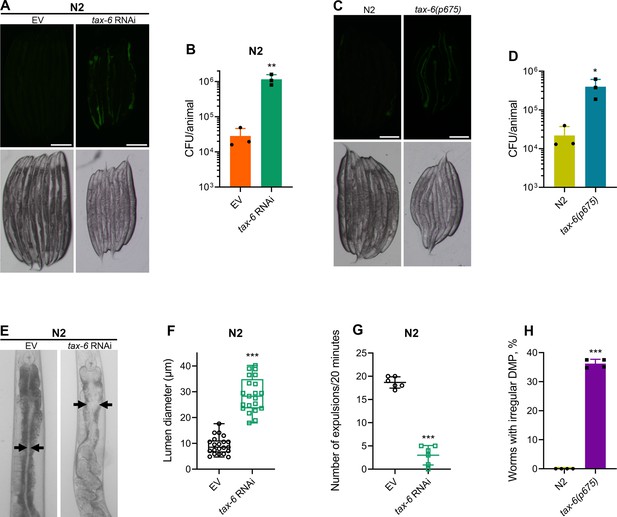
Calcineurin is required for the C. elegans defecation motor program (DMP).
(A) Representative fluorescence (top) and the corresponding brightfield (bottom) images of N2 animals incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on the empty vector (EV) control and tax-6 RNAi bacteria. Scale bar=200 µm. (B) Colony-forming units (CFU) per animal of N2 worms incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on the EV control and tax-6 RNAi bacteria. **p<0.01 via the t-test (n=3 biological replicates). (C) Representative fluorescence (top) and the corresponding brightfield (bottom) images of N2 and tax-6(p675) animals incubated on P. aeruginosa-GFP for 6 hours at 25 °C after growth on E. coli OP50 at 20 °C. Scale bar=200 µm. (D) CFU per animal of N2 and tax-6(p675) worms incubated on P. aeruginosa-GFP for 6 hours at 25 °C after growth on E. coli OP50 at 20 °C. *p<0.05 via the t-test (n=3 biological replicates). (E) Representative photomicrographs of N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. Arrows point to the border of the intestinal lumen. (F) Quantification of the diameter of the intestinal lumen of N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. ***p<0.001 via the t-test (n=21 worms each). (G) The number of expulsion events observed in 20 min in 1-day-old adult N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C. ***p<0.001 via the t-test (n=6 worms each). (H) Percent of 1-day-old adult worms having irregular DMP. ***p<0.001 via the t-test (n=4 biological replicates).
-
Figure 2—source data 1
Calcineurin is required for the C. elegans defecation motor program.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig2-data1-v1.xlsx
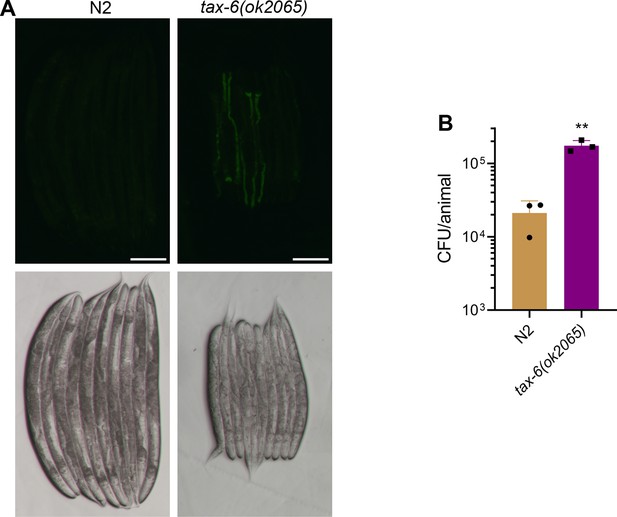
Calcineurin is required for the C. elegans defecation motor program (DMP).
(A) Representative fluorescence (top) and the corresponding brightfield (bottom) images of N2 and tax-6(ok2065) animals incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on E. coli OP50 at 20 °C. Scale bar=200 µm. (B) CFU per animal of N2 and tax-6(ok2065) worms incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on E. coli OP50 at 20 °C. **p<0.01 via the t-test (n=3 biological replicates).
-
Figure 2—figure supplement 1—source data 1
Calcineurin is required for the C. elegans defecation motor program.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig2-figsupp1-data1-v1.xlsx
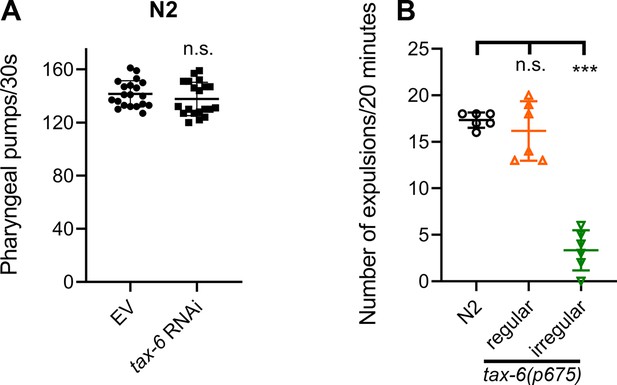
Calcineurin inhibition leads to defects in the defecation motor program (DMP) without affecting the pharyngeal pumping rate.
(A) Pharyngeal pumps per 30 s of 1-day-old adult N2 animals grown on the empty vector (EV) control and tax-6 RNAi bacteria at 20 °C. n.s., nonsignificant via the t-test (n=20 worms each). (B) The number of expulsion events observed in 20 min in 1-day-old adult N2 and regular and irregular tax-6(p675) animals grown on E. coli OP50 at 20 °C. ***p<0.001, n.s., nonsignificant via the t-test (n=6 worms each).
-
Figure 2—figure supplement 2—source data 1
Calcineurin inhibition leads to defects in the defecation motor program without affecting the pharyngeal pumping rate.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig2-figsupp2-data1-v1.xlsx
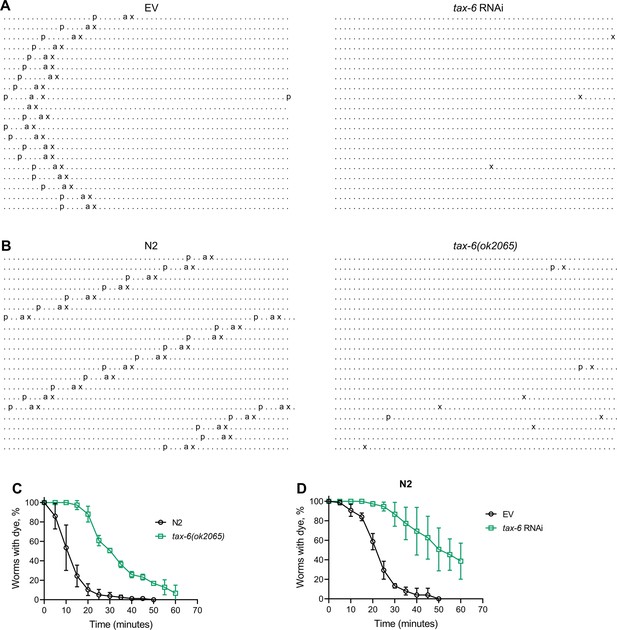
Calcineurin inhibition disrupts the C. elegans defecation motor program (DMP).
(A) Representative DMP ethograms of N2 animals after growth on the empty vector (EV) control and tax-6 RNAi bacteria till 1-day-old stage. Each dot represents a second, and each row represents a minute. ‘p’, ‘a’, and ‘x’ represent posterior body-wall muscle contraction, anterior body-wall muscle contraction, and expulsion muscle contraction, respectively. (B) Representative DMP ethograms of N2 and tax-6(ok2065) animals after their growth on E. coli OP50 at 20 °C till the 1-day-old adult stage. (C) Plots of the change in the percent of animals with blue dye in their gut with time for 1-day-old adult N2 and tax-6(ok2065) animals (n=3 biological replicates). (D) Plots of the change in the percent of animals with blue dye in their gut with time for 1-day-old adult N2 animals grown on EV control and tax-6 RNAi bacteria (n=3 biological replicates).
-
Figure 3—source data 1
Calcineurin inhibition disrupts the C. elegans defecation motor program.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig3-data1-v1.xlsx

Calcineurin inhibition disrupts the C. elegans defecation motor program (DMP).
Three different DMP ethograms of N2 animals after growth on each empty vector (EV) control and tax-6 RNAi bacteria till 1-day-old stage. Each dot represents a second, and each row represents a minute. ‘p’, ‘a’, and ‘x’ represent posterior body-wall muscle contraction, anterior body-wall muscle contraction, and expulsion muscle contraction, respectively.

Calcineurin inhibition disrupts the C. elegans defecation motor program (DMP).
Three different DMP ethograms of N2 and tax-6(ok2065) animals each after their growth on E. coli OP50 at 20 °C till the 1-day-old adult stage. Each dot represents a second, and each row represents a minute. ‘p’, ‘a’, and ‘x’ represent posterior body-wall muscle contraction, anterior body-wall muscle contraction, and expulsion muscle contraction, respectively.
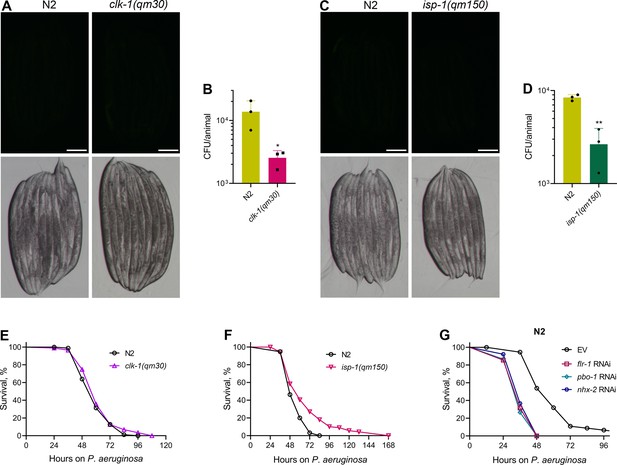
Slow DMP versus disrupted DMP lead to distinct phenotypes on P. aeruginosa exposure.
(A) Representative fluorescence (top) and the corresponding brightfield (bottom) images of N2 and clk-1(qm30) animals incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on E. coli OP50 at 20 °C. Scale bar=200 µm. (B) CFU per animal of N2 and clk-1(qm30) worms incubated on P. aeruginosa-GFP for 6 hr at 25 °C after growth on E. coli OP50 at 20 °C. *p<0.05 via the t-test (n=3 biological replicates). (C) Representative fluorescence (top) and the corresponding brightfield (bottom) images of N2 and isp-1(qm150) animals incubated on P. aeruginosa-GFP for 6 hours at 25 °C after growth on E. coli OP50 at 20 °C. Scale bar=200 µm. (D) CFU per animal of N2 and isp-1(qm150) worms incubated on P. aeruginosa-GFP for 6 hours at 25 °C after growth on E. coli OP50 at 20 °C. **p<0.01 via the t-test (n=3 biological replicates). (E) Representative survival plots of N2 and clk-1(qm30) animals on P. aeruginosa PA14 at 25 °C. The animals were grown on E. coli OP50 at 20 °C until 1-day-old adults before transferring to P. aeruginosa PA14 at 25 °C. n.s., nonsignificant (n=3 biological replicates; animals per condition per replicate >90). (F) Representative survival plots of N2 and isp-1(qm150) animals on P. aeruginosa PA14 at 25 °C. The animals were grown on E. coli OP50 at 20 °C until 1-day-old adults before transferring to P. aeruginosa PA14 at 25 °C. p<0.001 (n=3 biological replicates; animals per condition per replicate >90). (G) Representative survival plots of N2 animals on P. aeruginosa PA14 at 25 °C after treatment with the empty vector (EV) control, flr-1, pbo-1, and nhx-2 RNAi. p<0.001 for flr-1, pbo-1, and nhx-2 RNAi compared to EV (n=3 biological replicates; animals per condition per replicate >90).
-
Figure 3—figure supplement 3—source data 1
Slow DMP versus disrupted DMP lead to distinct phenotypes on P. aeruginosa exposure.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig3-figsupp3-data1-v1.xlsx
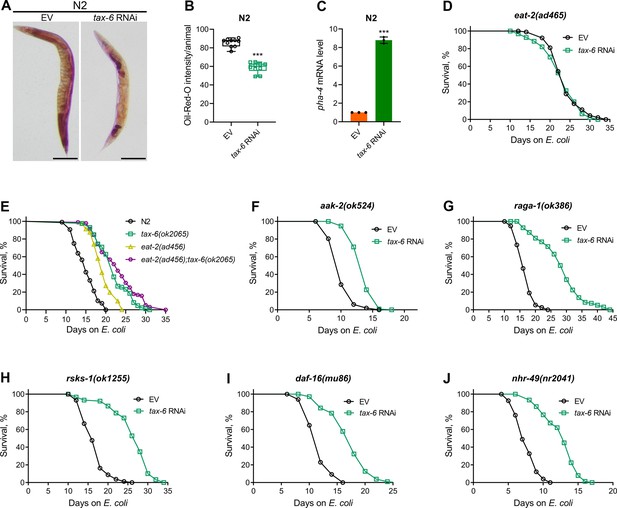
Calcineurin knockdown enhances lifespan via DMP defects-mediated calorie restriction.
(A) Representative photomicrographs of oil-red-O (ORO) stained 1-day-old adult N2 animals grown on the empty vector (EV) control and tax-6 RNAi bacteria at 20 °C. Scale bar=200 µm. (B) Quantification of ORO intensity per animal of 1-day-old adult N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C. The values are area normalized for each animal. ***p<0.001 via the t-test (n=10 worms each). (C) Quantitative reverse transcription-PCR (qRT-PCR) for pha-4 mRNA levels in N2 animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. ***p<0.001 (n=3 biological replicates). (D) Representative survival plots of eat-2(ad465) animals grown on the bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. n.s., nonsignificant (n=3 biological replicates; animals per condition per replicate >80). (E) Representative survival plots of N2, tax-6(ok2065), eat-2(ad465), and eat-2(ad465);tax-6(ok2065) animals grown on E. coli OP50 at 20 °C. Day 0 represents young adults. p<0.001 for tax-6(ok2065), eat-2(ad465), and eat-2(ad465);tax-6(ok2065) compared to N2. p<0.05 for eat-2(ad465);tax-6(ok2065) compared to tax-6(ok2065) (n=3 biological replicates; animals per condition per replicate >65). (F–J) Representative survival plots of aak-2(ok524) (E), raga-1(ok386) (F), rsks-1(ok1255) (G), daf-16(mu86) (H), and nhr-49(nr2041) (I) animals grown on the bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. p<0.001 for all the plots (n=3 biological replicates; animals per condition per replicate >60).
-
Figure 4—source data 1
Calcineurin knockdown enhances lifespan via DMP defects-mediated calorie restriction.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig4-data1-v1.xlsx
The eat-2(ad465) animals are not defective in RNAi.
Representative images of wild-type N2, eat-2(ad465), and sid-1(qt9) worms on the empty vector (EV) control, act-5, and bli-3 RNAi. Scale bar = 1 mm.
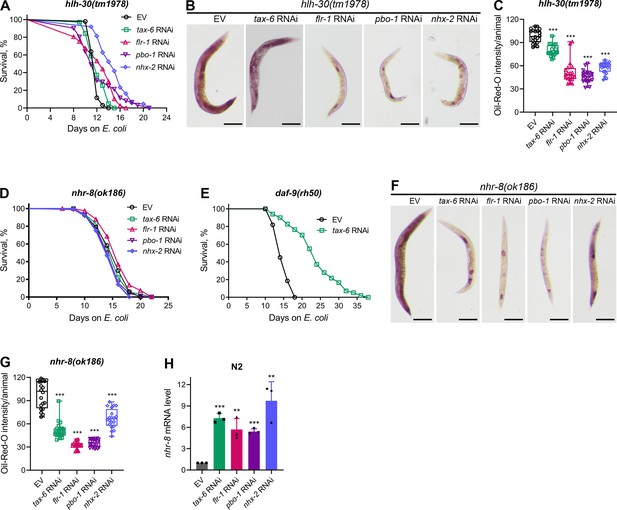
Calcineurin inhibition enhances lifespan via HLH-30 and NHR-8.
(A) Representative survival plots of hlh-30(tm1978) animals grown on bacteria for RNAi against tax-6, flr-1, pbo-1, and nhx-2 along with the empty vector (EV) control at 20 °C. Day 0 represents young adults. p values for tax-6, flr-1, pbo-1, and nhx-2 compared to EV are <0.01,<0.001, nonsignificant, and <0.001, respectively (n=3 biological replicates; animals per condition per replicate >60). (B) Representative photomicrographs of oil-red-O (ORO) stained 1-day-old adult hlh-30(tm1978) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. Scale bar=200 µm. (C) Quantification of ORO intensity per animal of 1-day-old adult hlh-30(tm1978) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. The values are area normalized for each animal. ***p<0.001 via the t-test (n=20 worms each). (D) Representative survival plots of nhr-8(ok186) animals grown on bacteria for RNAi against tax-6, flr-1, pbo-1, and nhx-2 along with the EV control at 20 °C. Day 0 represents young adults. p values for tax-6, flr-1, pbo-1, and nhx-2 compared to EV are nonsignificant,<0.05, nonsignificant, and <0.05, respectively (n=3 biological replicates; animals per condition per replicate >70). (E) Representative survival plots of daf-9(rh50) animals grown on the bacteria for RNAi against tax-6 along with the EV control at 20 °C. Day 0 represents young adults. p<0.001 (n=3 biological replicates; animals per condition per replicate >80). (F) Representative photomicrographs of ORO stained 1-day-old adult nhr-8(ok186) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. Scale bar=200 µm. (G) Quantification of ORO intensity per animal of 1-day-old adult nhr-8(ok186) animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C. The values are area normalized for each animal. ***p<0.001 via the t-test (n=20 worms each). (H) Quantitative reverse transcription-PCR (qRT-PCR) for nhr-8 mRNA levels in N2 animals grown on the EV control, tax-6, flr-1, pbo-1, and nhx-2 RNAi bacteria at 20 °C until 1-day-old adults. ***p<0.001 via the t-test (n=3 biological replicates).
-
Figure 5—source data 1
Calcineurin inhibition enhances lifespan via HLH-30 and NHR-8.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig5-data1-v1.xlsx
The hlh-30(tm1978) and nhr-8(ok186) animals are not defective in RNAi.
Representative images of wild-type N2, hlh-30(tm1978), nhr-8(ok186), and sid-1(qt9) worms on the empty vector (EV) control, act-5, and bli-3 RNAi. Scale bar=1 mm.
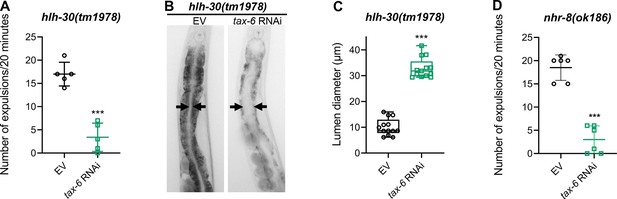
Calcineurin inhibition leads to defects in the defecation motor program (DMP) in hlh-30(tm1978) and nhr-8(ok186) animals.
(A) The number of expulsion events observed in 20 min in 1-day-old adult hlh-30(tm1978) animals grown on the empty vector (EV) control and tax-6 RNAi bacteria at 20 °C. ***p<0.001 via the t-test (n=5 worms each). (B) Representative photomicrographs of hlh-30(tm1978) animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. Arrows point to the border of the intestinal lumen. (C) Quantification of the diameter of the intestinal lumen of hlh-30(tm1978) animals grown on the EV control and tax-6 RNAi bacteria at 20 °C until 1-day-old adults. ***p<0.001 via the t-test (n=13 worms each). (D) The number of expulsion events observed in 20 min in 1-day-old adult nhr-8(ok186) animals grown on the EV control and tax-6 RNAi bacteria at 20 °C. ***p<0.001 via the t-test (n=6 worms each).
-
Figure 5—figure supplement 2—source data 1
Calcineurin inhibition leads to defects in the defecation motor program in hlh-30(tm1978) and nhr-8(ok186) animals.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig5-figsupp2-data1-v1.xlsx
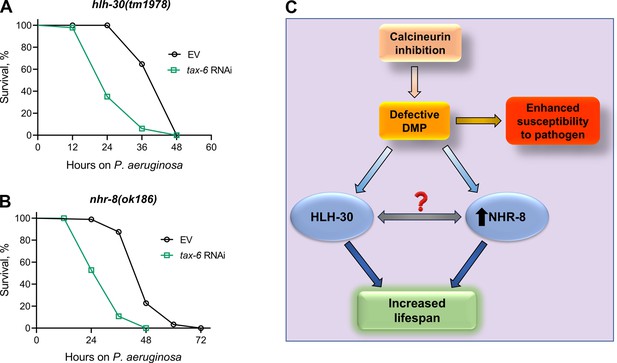
Calcineurin inhibition-mediated effects on lifespan and survival on P. aeruginosa are mediated by distinct mechanisms.
(A)-(B) Representative survival plots of hlh-30(tm1978) (A) and nhr-8(ok186) (B) animals on P. aeruginosa PA14 at 25 °C after treatment with the empty vector (EV) control and tax-6 RNAi. p<0.001 for both the plots (n=3 biological replicates; animals per condition per replicate >85). (C) Model depicting the mechanism of increased lifespan and enhanced susceptibility to pathogen upon inhibition of calcineurin via the defects in the defecation motor program (DMP).
-
Figure 6—source data 1
Calcineurin inhibition-mediated effects on lifespan and survival on P. aeruginosa are mediated by distinct mechanisms.
- https://cdn.elifesciences.org/articles/89572/elife-89572-fig6-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | OP50 | Caenorhabditis Genetics Center (CGC) | OP50 | |
| Strain, strain background (E. coli) | HT115(DE3) | Source BioScience | HT115(DE3) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 | Frederick M Ausubel laboratory | PA14 | |
| Strain, strain background (P. aeruginosa) | PA14-GFP | Frederick M Ausubel laboratory | PA14-GFP | |
| Strain, strain background (Caenorhabditis elegans) | N2 Bristol | CGC | N2 | |
| Strain, strain background (C. elegans) | tax-6(p675) | CGC | PR675 | |
| Strain, strain background (C. elegans) | tax-6(ok2065) | CGC | RB1667 | |
| Strain, strain background (C. elegans) | fer-1(b232) | CGC | HH142 | |
| Strain, strain background (C. elegans) | eat-2(ad465) | CGC | DA465 | |
| Strain, strain background (C. elegans) | aak-2(ok524) | CGC | RB754 | |
| Strain, strain background (C. elegans) | raga-1(ok386) | CGC | VC222 | |
| Strain, strain background (C. elegans) | rsks-1(ok1255) | CGC | RB1206 | |
| Strain, strain background (C. elegans) | daf-16(mu86) | CGC | CF1038 | |
| Strain, strain background (C. elegans) | nhr-49(nr2041) | CGC | STE68 | |
| Strain, strain background (C. elegans) | hlh-30(tm1978) | CGC | JIN1375 | |
| Strain, strain background (C. elegans) | nhr-8(ok186) | CGC | AE501 | |
| Strain, strain background (C. elegans) | daf-9(rh50) | CGC | RG1228 | |
| Strain, strain background (C. elegans) | pmk-1(km25) | CGC | KU25 | |
| Strain, strain background (C. elegans) | kgb-1(km21) | CGC | KU21 | |
| Strain, strain background (C. elegans) | dbl-1(nk3) | CGC | NU3 | |
| Strain, strain background (C. elegans) | zip-2(ok3730) | CGC | VC3056 | |
| Strain, strain background (C. elegans) | clk-1(qm30) | CGC | MQ130 | |
| Strain, strain background (C. elegans) | isp-1(qm150) | CGC | MQ887 | |
| Strain, strain background (C. elegans) | sid-1(qt9) | CGC | HC196 | |
| Strain, strain background (C. elegans) | eat-2(ad465);tax-6(ok2065) | This study | Materials and methods section | |
| Sequence-based reagent | Pan-act_qPCR_F | This study | qPCR primers | TCGGTATGGGACAGAAGGAC |
| Sequence-based reagent | Pan-act_qPCR_R | This study | qPCR primers | CATCCCAGTTGGTGACGATA |
| Sequence-based reagent | pha-4_qPCR_F | This study | qPCR primers | CAAAGAGGAGCCAGAGTCGG |
| Sequence-based reagent | pha-4_qPCR_R | This study | qPCR primers | TGTTTCTGCTCGCGTTTTCG |
| Sequence-based reagent | nhr-8_qPCR_F | This study | qPCR primers | CTACACAGTTTCTCCGGCGT |
| Sequence-based reagent | nhr-8_qPCR_R | This study | qPCR primers | GCCATTTGGGCCATAACACC |
| Sequence-based reagent | tax-6(ok2065)_genotyping_F1 | This study | Genotyping primers | CTCCTTTGAGGGAGCCAGTG |
| Sequence-based reagent | tax-6(ok2065)_genotyping_F2 | This study | Genotyping primers | CTGGGGACAATCCACCATGAA |
| Sequence-based reagent | tax-6(ok2065)_genotyping_R1 | This study | Genotyping primers | TGTGTCCTGTATCTGTGGGC |
| Sequence-based reagent | eat-2(ad465) _genotyping_F | This study | Genotyping primers | CGGTGCAAAGAGCACATCTC |
| Sequence-based reagent | eat-2(ad465) _genotyping_R | This study | Genotyping primers | TTAAGGCGTACGAGCCTTCC |
| Software, algorithm | GraphPad Prism 8 | GraphPad Software | RRID:SCR_002798 | https://www.graphpad.com/scientificsoftware/prism/ |
| Software, algorithm | Photoshop CS5 | Adobe | RRID:SCR_014199 | https://www.adobe.com/products/photoshop.html |
| Software, algorithm | ImageJ | NIH | RRID:SCR_003070 | https://imagej.nih.gov/ij/ |





